Abstract
A potential role of Insulin-like growth factor 1 (IGF1) receptor/phosphatidylinositol-3 kinase (PI3k)/Akt signaling in the initiation and progression of Lumbar disc degeneration (LDD) has been recently reported. However, the regulation of IGF1 receptor (IGF1R) at post-transcriptional levels in the development of LDD remains unknown. Here, we studied the effects of microRNA-4458 on the expression of IGF1R. We examined the IGF1R levels and microRNA-4458 (miR-4458) levels in the resected LDD discs, compared with the traumatized, non-LDD discs. We analyzed the binding of miR-4458 to the 3’-UTR of IGF1R mRNA and its effects on IGF1R translation by bioinformatics analysis and by luciferase-reporter assay, respectively. We modified miR-4458 levels in a human nucleus pulposus SV40 cell line (HNPSV), and examined the effects of miR-4458 on the expression of IGF1R and Akt, as well as their phosphorylation. We found that the levels of miR-4458 were significantly higher and the levels of IGF1R were significantly lower in LDD discs, compared with the control non-LDD discs. The levels of IGF1R inversely correlated with the levels of miR-4458 in LDD discs. Moreover, miR-4458 was found to bind to the 3’-UTR of IGF1R mRNA to prevent its translation. In miR-4458-modified HNPSV cells, we found that miR-4458 decreased both total IGF1R and phosphorylated IGF1R, resulting in deceases in phosphorylated Akt. Thus, these data suggest that miR-4458 may suppress PI3k/Akt signaling pathway through 3’-UTR-inhibtion of IGF1R mRNA to promote development of LDD.
Keywords: Lumbar disc degeneration (LDD), miR-4458, insulin-like growth factor 1 (IGF1), insulin-like growth factor 1 receptor (IGF1R)
Introduction
Lumbar disc degeneration (LDD) is a major cause of low back pain and neck pain, which frequently results from a degenerated disc in the spine, and is a product of lifelong degradation with synchronized remodeling of discs and neighboring vertebrae, including concomitant adaptation of the disc structures to the alterations in physical loading and responses to the occasional injury [1-3]. The clinical diagnosis of LDD is based on a summary of symptoms, radiographic magnetic resonance imaging, microscopic, histologic and biochemical analysis, macroscopic measurement of degeneration in surgical and autopsy samples [1-3]. However, radiography has been widely used for diagnosis of LDD epidemiologically, particularly before the advent of magnetic resonance imaging qualitative methods of evaluating LDD from magnetic resonance imaging for screening of the disease in a large population [1-3].
The pathogenesis of LDD involves disc cell dysfunction and degradation. Disc cells actively regulate matrix homeostasis through activities modulated by a variety of cytokines and growth factors acting in a paracrine and/or autocrine fashion [4]. While normal disc cells maintain the matrix at a steady state, an imbalance between the anabolic and catabolic processes in the pathological processes result in LDD [4]. Disc damage caused by mechanical injury, inflammation, or aging may modulate the structure of the discs through regulating disc homeostasis [5].
The insulin-like growth factor 1 (IGF1) receptor signaling pathway initiates with binding of the ligand, IGF1 toits cell-surface receptor IGF1R [6-8]. Although IGF1 binds to both IGF1R (high affinity) and the insulin receptor (low affinity), the IGF1R has been shown to be the major functional receptor for IGFI, which is a receptor tyrosine kinase to activate phosphatidylinositol-3 kinase (PI3k)/Akt signaling pathway upon stimulation [6-8]. Phosphorylated Akt is a natural stimulator of cell growth and proliferation, and a potent inhibitor of programmed cell death [6-8], and Akt functions through its downstream targets, the mammalian target of rapamycin (mTOR) and forkhead box protein O1 (FoxO1) [9] as we recently reported [10]. However, the regulation of IGF1R at post-transcriptional level in the development of LDD remains unknown.
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that regulate various biological processes [11-13]. Interestingly, bioinformatics approaches have predicted one-third of all mammalian genes to be targeted and regulated by miRNAs [11-13]. Among miRNAs, miR-4458 has been rarely studied. Only very recently, miR-4458 was shown to be a tumor suppressor by targeting I-kappa-B kinase epsilon in human hepatocellular carcinoma cells, and might be potential predictors of prognosis of hepatocellular carcinoma [14]. In another study, miR-4458 was shown to inhibit the progression of colon cancer cells by suppressing glycolysis and lactate production via hexokinase 2 mRNA [15]. However, a role of miR-4458 in the development of LDD has not been reported.
Here, we studied the effects of microRNA-4458 on the expression of IGF1R. We examined the IGF1R levels and miR-4458 levels in the resected LDD discs, compared withthe traumatized, non-LDD discs. We analyzed the binding of miR-4458 to the 3’-UTR of IGF1R mRNA and its effects on IGF1R translation by bioinformatics analysis and by luciferase-reporter assay, respectively. We modified miR-4458 levels in a human nucleus pulposus SV40 cell line (HNPSV), and examined the effects of miR-4458 on the expression of IGF1R and Akt, as well as their phosphorylation. Our data showed that the levels of miR-4458 were significantly higher and the levels of IGF1R were significantly lower in LDD discs, compared with the control non-LDD discs. The levels of IGF1R inversely correlated with the levels of miR-4458 in LDD discs. Moreover, miR-4458 was found to bind to the 3’-UTR of IGF1R mRNA to prevent its translation. In miR-4458-modified HNPSV cells, we found that miR-4458 decreased both total IGF1R and phosphorylated IGF1R, resulting in deceases in phosphorylated Akt. Thus, miR-4458 may suppress PI3k/Akt signaling pathway through 3’-UTR-inhibtion of IGF1R mRNA to promote development of LDD.
Materials and methods
Specimens from patients
A total of 46 subjects (24 with resected LDD discs and 22 traumatized, non-LDD subjects with fractured discs as controls) were included in this study. All the subjects had no accompanying diseases (e.g. diabetes) that may affect systemic IGF-1 levels. All specimens had been histologically and clinically diagnosed at Qingpu Branch of Zhongshan Hospital affiliated to Fudan Universityfrom 2008 to 2015. For the use of these clinical materials for research purposes, prior patient’s consents and approval from the Institutional Research Ethics Committee were obtained (NO: GDR452). IGF-1R (pIGF-1R) levels were measured by Western blot and normalized to α-tubulin.
Human disc cell line, reagents and transfection
A human disc cell line, nucleus pulposus SV40 (HNPSV)has been described [16], and used in our previous study [10]. The HNPSV cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco; Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Sigma-Aldrich, St Louis, MO, USA), penicillin (100 μg/ml) and streptomycin (250 ng/ml) at 37°C, in a 5% CO2 atmosphere. Cells were analyzed after stimulated with 100 ng/ml recombinant human IGF-1 (Sigma-Aldrich). HNPSV cells were transfected with a miR-4458 construct, or an antisense (as) of miR-4458, or a null sequence (null) as a control. Transfection was performed with Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions.
Western blot
Protein was extracted from the cultured cells by RIPA buffer (Sigma-Aldrich) for Western Blot. The supernatants were collected after centrifugation at 12000×g at 4°C for 20 min. Protein concentration was determined using BCA protein assay, and whole lysates were mixed with 4×SDS loading buffer (125 mmol/l Tris-HCl, 4% SDS, 20% glycerol, 100 mmol/l DTT, and 0.2% bromophenol blue) at a ratio of 1:3. Samples were heated at 100°C for 5 min and were separated on SDS-polyacrylamide gels. The separated proteins were then transferred to a PVDF membrane. The membrane blots were first probed with a primary antibody. After incubation with horseradish peroxidase-conjugated second antibody, autoradiograms were prepared using the enhanced chemiluminescent system to visualize the protein antigen. The signals were recorded using X-ray film. Primary antibodies for Western Blot are anti-IGF1R, anti-phosphorylated-IGF1R (pIGF1R), anti-Akt, anti-phosphorylated-Akt (pAkt) and anti-α-tubulin (all purchased from Cell Signaling, San Jose, CA, USA). Secondary antibody is HRP-conjugated anti-rabbit (Jackson ImmunoResearch Labs, West Grove, PA, USA). Images shown in the figure were representative from 5 repeats. Densitometry of Western blots was quantified with NIH ImageJ software. The protein was first normalized to α-tubulin, and then normalized to experimental controls.
Quantitative PCR (RT-qPCR)
Total RNAs were extracted from the cultured cells with RNeasy kit (Qiagen, Hilden, Germany), and used for cDNA synthesis. Quantitative PCR was performed in duplicates with QuantiTect SYBR Green PCR Kit (Qiagen). All primers were purchased from Qiagen. Data were analyzed with 2-ΔΔCt method for quantification of the relative mRNA expression levels. Values of genes were first normalized against α-tubulin, and then compared with controls.
MicroRNA target prediction and 3’-UTR luciferase-reporter assay
MiRNAs targets were predicted with the algorithms TargetSan (https://www.targetscan.org) [17]. Luciferase-reporters were successfully constructed using molecular cloning technology. The IGF1R 3’-UTR reporter plasmid (IGF1R 3’-UTR) and IGF1R 3’-UTR reporter plasmid with a mutant at the miR-4458 binding site (IGF1R 3’-UTR mut) were purchased from Creative Biogene (Shirley, NY, USA). Cells were co-transfected with IGF1R 3’-UTR/IGF1R 3’-UTR mut and miR-4458/as-miR-4458/null by Lipofectamine 2000 (5×104 cells per well). Cells were collected 24 hours after transfection for dual-luciferase reporter assay (Promega, Beijing, China), according to the manufacturer’s instructions. The normalized control was null-transfected cells with 3’-UTR of IGF1R mRNA (wild type).
Statistical analysis
All statistical analyses were carried out using GraphPad prism 6.0 (GraphPad Software Inc. La Jolla, CA, USA). All values are depicted as mean ± SD and are considered significant if p<0.05. All data were statistically analyzed using one-way ANOVA with a Bonferroni correction, followed by Fisher’s Exact Test for comparison of two groups. Bivariate correlations were calculated by Spearman’s Rank Correlation Coefficients.
Results
Lower IGF1R and high miR-4458 are inversely correlated in LDD specimens
We examined the IGF1R and miR-4458 levels in the 24 LDD discs and 22 traumatized, non-LDD discs (CTL) from the patients. We detected significantly higher levels of miR-4458 (Figure 1A) and significantly lower levels of IGF1R (Figure 1B) in LDD by RT-qPCR and Western blot, respectively. Then we studied the relationship between IGF1R and miR-4458. We found a strong inverse correlation between IGF1R and miR-4458 in LDD specimens (Figure 1C, R=-0.90; p<0.0001), suggesting a causal relationship.
Figure 1.
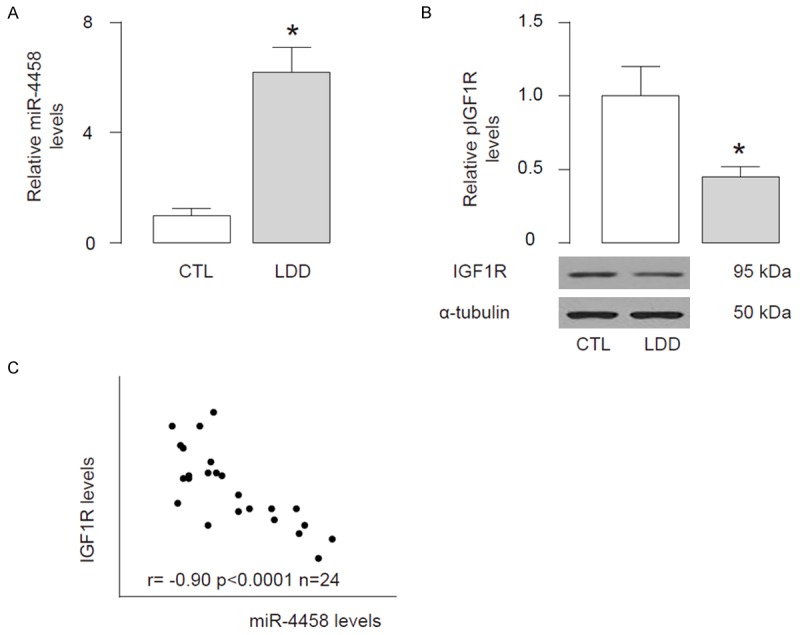
Lower IGF1R and high miR-4458 are inversely correlated in LDD specimens. We examined the IGF1R and miR-4458 levels in the 24 LDD discs and 22 traumatized, non-LDD discs (CTL) from the patients. (A, B) We detected significantly higher levels of miR-4458 by RT-qPCR (A) and significantly lower levels of IGF1R by Western blot (B) in LDD specimens, compared with CTL. (C) We found a strong inverse correlation between IGF1R and miR-4458 in LDD specimens (r=-0.90; p<0.0001; n=24). *p<0.05.
MiR-4458 targets 3’-UTR of IGF1R to inhibit its expression
Based on our findings in the patients’ samples, we performed bioinformatics analysis of IGF1R targeting sequence, which showed that miR-4458 binds to 3’-UTR of IGF1R mRNA (Figure 2A). To prove that this binding has a biological effect, we modified miR-4458 levels in a human disc cell line, HNPSV. We transfected HNPSV cells with either miR-4458 or antisense for miR-4458 (as-miR-4458). HNPSV cells were also transfected with a null sequence as a control (null). Modulation of miR-4458 levels in HNPSV cells was confirmed by RT-qPCR (Figure 2B). Then, we used these miR-4458-modified HNPSV cells in the experiments that examine the functional binding of miR-4458 to IGF1R mRNA. The intact 3’-UTR of IGF1R mRNA (IGF1R 3’-UTR), together with a 3’-UTR with mutant at miR-4458-binding site of IGF1R mRNA (IGF1R 3’-UTR mut), was then cloned into luciferase reporter plasmids, and used for co-transfection with miR-4458-modified plasmids into HNPSV cells. First, HNPSV cells were co-transfected with 1 μg as-miR-4458/null plasmids and 1 μg IGF1R 3’-UTR plasmids, showing that miR-4458 depletion significantly increased luciferase activities. Next, HNPSV cells were co-transfected with 1 μg miR-4458/null plasmids and 1 μg IGF1R 3’-UTR or IGF1R 3’-UTR mut plasmids, showing that that IGF1R 3’-UTR plus miR-4458 had the most repression for IGF1R, and the IGF1R 3’-UTR mut failed to decrease luciferase activities by miR-4458 (Figure 2C). These data demonstrate that miR-4458 specifically targets 3’-UTR of IGF1R mRNA to inhibit its translation in HNPSV cells.
Figure 2.
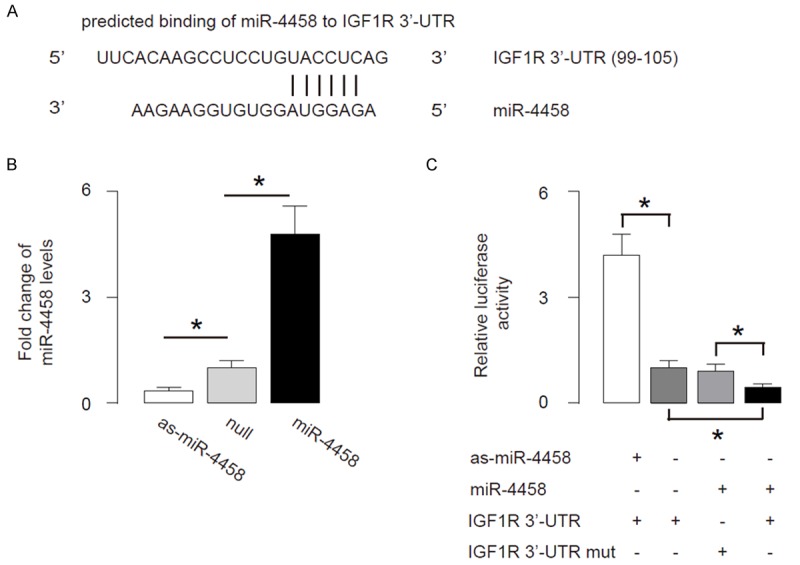
MiR-4458 targets 3’-UTR of IGF1R to inhibit its expression. A. Bioinformatics analysis showed that miR-4458 binds to 3’-UTR of IGF1R mRNA. B. We modified miR-4458 levels in a human disc cell line, HNPSV. We transfected HNPSV cells with either miR-4458 or antisense for miR-4458 (as-miR-4458). HNPSV cells were also transfected with a null sequence as a control (null). Modulation of miR-4458 levels in HNPSV cells was confirmed by RT-qPCR. C. The intact 3’-UTR of IGF1R mRNA (IGF1R 3’-UTR), together with a 3’-UTR with mutant at miR-4458-binding site of IGF1R mRNA (IGF1R 3’-UTR mut), was then cloned into luciferase reporter plasmids, and used for co-transfection with miR-4458-modified plasmids into HNPSV cells. First, HNPSV cells were co-transfected with 1 μg as-miR-4458/null plasmids and 1 μg IGF1R 3’-UTR plasmids, showing that miR-4458 depletion significantly increased luciferase activities. Next, HNPSV cells were co-transfected with 1 μg miR-4458/null plasmids and 1 μg IGF1R 3’-UTR or IGF1R 3’-UTR mut plasmids, showing that that IGF1R 3’-UTR plus miR-4458 had the most repression for IGF1R, and the IGF1R 3’-UTR mut failed to decrease luciferase activities by miR-4458. *p<0.05. N=5.
MiR-4458 decreases IGF1R protein but does not affect its phosphorylation ratio
We found that although the IGF1R transcripts did not change by miR-4458 levels (Figure 3A), the protein levels of total IGF1R in miR-4458-overexpressing HNPSV cells was significantly decreased, while the protein levels of total IGF1R in miR-4458-depleted HNPSV cells was significantly increased (Figure 3B, 3C). Similarly, the protein levels of phosphorylated IGF1R (pIGF1R) in miR-4458-overexpressing HNPSV cells was significantly decreased, while the protein levels of pIGF1R in miR-4458-depleted HNPSV cells was significantly increased (Figure 3B, 3D). Interestingly, the ratio of pIGF1R vs IGF1R in miR-4458-modified HNPSV cells was not different (Figure 3B, 3E). These data suggest that miR-4458 decreases IGF1R protein but does not affect its phosphorylation ratio.
Figure 3.
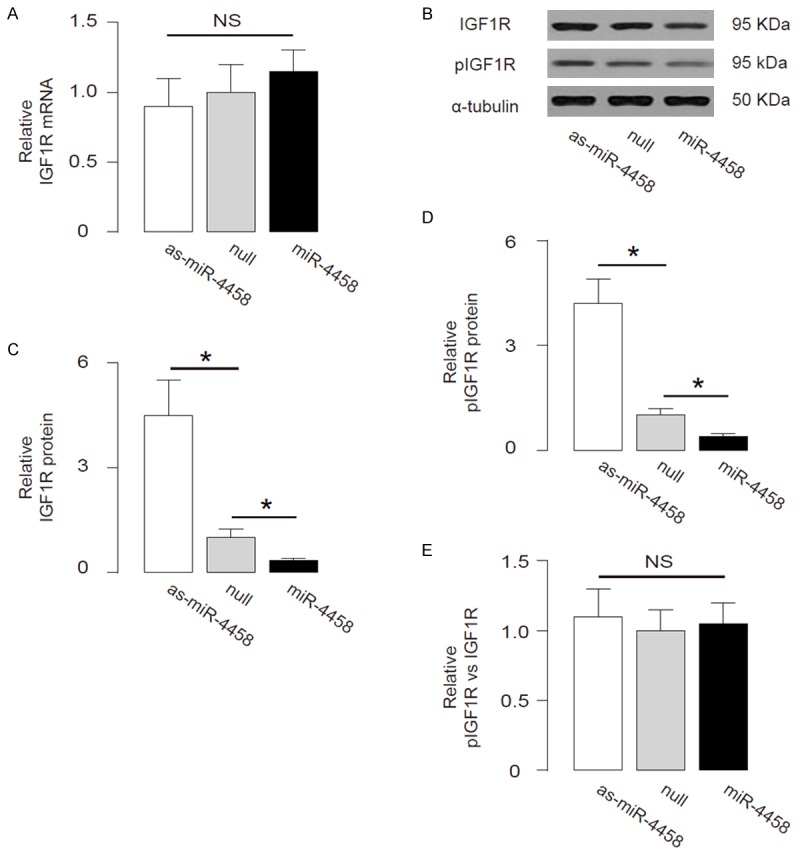
MiR-4458 decreases IGF1R protein but does not affect its phosphorylation ratio. (A) IGF1R levels in miR-4458-modified HNPSV cells by RT-qPCR. (B) The representative images of IGF1R, pIGF1R protein by Western blot. (C-E) The protein levels of total IGF1R (C), pIGF1R (D), and the protein ratio of pIGF1R vs IGF1R (E) in miR-4458-overexpressing HNPSV cells. *p<0.05. N=5.
MiR-4458 regulates Akt signaling to affect LDD
We further analyzed the activation of Akt (phosphorylation of Akt (pAkt) compared with total Akt) in miR-4458-modified HNPSV cells. We found that protein levels of total Akt were not changed in miR-4458-modified HNPSV cells (Figure 4A, 4B), but the protein levels of pAkt in miR-4458-overexpressing HNPSV cells was significantly decreased, while the protein levels of pAkt in miR-4458-depleted HNPSV cells was significantly increased (Figure 4B, 4C). These data suggest that the absolute pIGF1R may regulate the levels of pAkt, which subsequently affects the development of LDD. The findings in the current study were thus summarized in a schematic, showing that miR-4458 may suppress PI3k/Akt signaling pathway through 3’-UTR-inhibtion of IGF1R mRNA to promote development of LDD (Figure 5).
Figure 4.
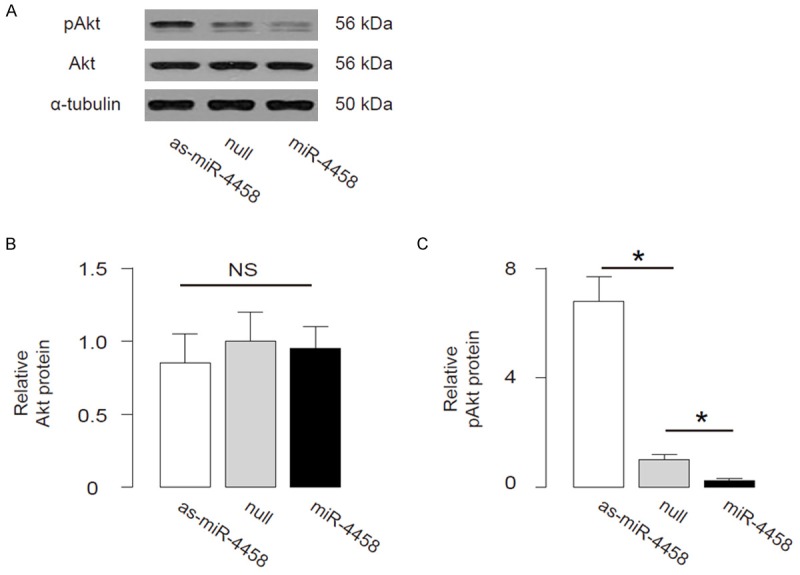
MiR-4458 regulates Akt signaling to affect LDD. (A) The representative images of Akt, pAkt protein by Western blot. (B, C) The protein levels of total Akt (B), pAkt (C) in miR-4458-overexpressing HNPSV cells. *p<0.05. N=5.
Figure 5.
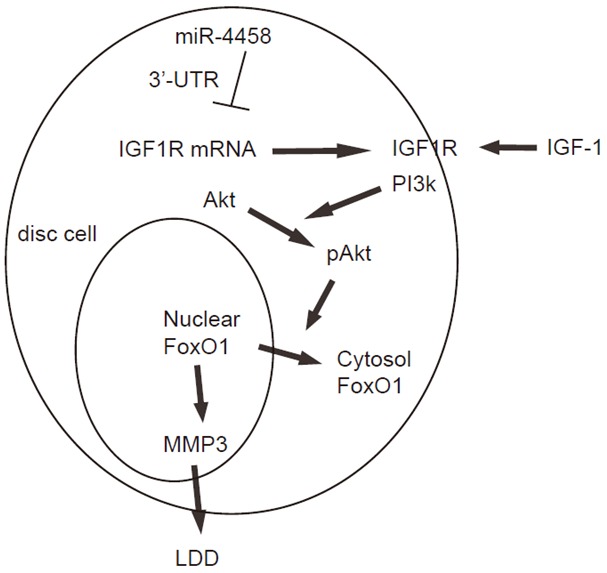
Schematic of the model. IGF-1 suppresses MMP3-mediated development of LDD through Akt-induced nuclear exclusion of FoxO1. MiR-4458 may suppress PI3k/Akt signaling pathway through 3’-UTR-inhibtion of IGF1R mRNA to promote development of LDD.
Discussion
The molecular basis underlying the development of LDD has been extensively studied [4,18], and we recently reported a role of IGF1R/PI3k/Akt signaling in the disease initiation and progression of LDD [10]. We have shown that LDD patients had significantly lower levels of serum IGF1, and LDD discs had significantly lower levels of activated IGF1R. Since the IGF1R signaling has been well-studied in other cell types and tissue, showing a regulatory axis through receptor, PI3k, Akt and its downstream targets including mTor and FoxO1, we examined the effects pfIGF1 induced phosphorylation of IGF1R and subsequently its downstream factor Akt on the suppression of MMP3 in the human disc cells. We also showed that FoxO1 nuclear retention in human disc cells completely abolished the effects of IGF1 on MMP3, suggesting that Akt-induced phosphorylation and nuclear exclusion FoxO1 is responsible for the IGF-1-induced MMP3 suppression. These data highlight a specific molecular regulation of MMP3 during LDD development. Nevertheless, the exact molecular mechanisms by which MMP3 regulates LDD have not been clarified. However, the regulation of IGF1 receptor (IGF1R) at post-transcriptional levels in the development of LDD remains unknown.
Here, we studied the effects of miR-4458 on the expression of IGF1R. We specifically studied miR-4458, since we have screened all the microRNAs that target IGF1R with a high binding affinity, and examined 3 candidates, miR-4458, miR-98-5p and miR-4500, among which only the levels of miR-4458 significantly altered in LDD specimens and found to functionally control the protein levels of IGF1R. In the current study, we found that the levels of miR-4458 were significantly higher and the levels of IGF1R were significantly lower in LDD discs, compared with the control non-LDD discs. The levels of IGF1R inversely correlated with the levels of miR-4458 in LDD discs. Moreover, miR-4458 was found to bind to the 3’-UTR of IGF1R mRNA to prevent its translation. In miR-4458-modified HNPSV cells, we found that miR-4458 decreased both total IGF1R and pIGF1R, resulting in deceases in phosphorylated Akt. However, the ratio of pIGF1R vs IGF1R remained unaltered in miR-4458-modiifed disc cells, suggesting that miR-4458 changed the total IGF1R levels through post-transcriptional control of IGF1R, but did not alter the phosphorylation of IGF1R. Thus, the absolute increases in pIGF1R may result from increases in total IGF1R protein, which may scale-up phosphorylated protein.
Combined with results from our two studies on IGF1R/PI3k/Akt signaling in development of LDD, we proposed a model in which IGF-1R/Akt/FoxO1/MMP3 axis as a novel therapeutic target for inhibiting the development of LDD.This pathway at least has two critical control points, one is the initial stimulator, IGF1, as a trigger of activation of the system, and it functions mainly through induction of receptor phosphorylation. One the other hand, the receptor can be regulated at the protein translation step, specifically by miR-4458. This regulation network is a signature of typical biological processes, and elucidation of such a system that regulates development of LDD may provide useful information for clinical treatment and for generating effective therapy.
Acknowledgements
Thestudy was supported by Shanghai Municipal Science and Technology Commission, No: 1441197300.
Disclosure of conflict of interest
None.
References
- 1.Wang SZ, Rui YF, Lu J, Wang C. Cell and molecular biology of intervertebral disc degeneration: current understanding and implications for potential therapeutic strategies. Cell Prolif. 2014;47:381–390. doi: 10.1111/cpr.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HQ, Samartzis D. Clarifying the nomenclature of intervertebral disc degeneration and displacement: from bench to bedside. Int J Clin Exp Pathol. 2014;7:1293–1298. [PMC free article] [PubMed] [Google Scholar]
- 3.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng C, Liu H, Yang Y, Huang B, Zhou Y. Growth and differentiation factor-5 contributes to the structural and functional maintenance of the intervertebral disc. Cell Physiol Biochem. 2015;35:1–16. doi: 10.1159/000369670. [DOI] [PubMed] [Google Scholar]
- 5.Chen JW, Ni BB, Li B, Yang YH, Jiang SD, Jiang LS. The responses of autophagy and apoptosis to oxidative stress in nucleus pulposus cells: implications for disc degeneration. Cell Physiol Biochem. 2014;34:1175–1189. doi: 10.1159/000366330. [DOI] [PubMed] [Google Scholar]
- 6.Tao Y, Pinzi V, Bourhis J, Deutsch E. Mechanisms of disease: signaling of the insulin-like growth factor 1 receptor pathway--therapeutic perspectives in cancer. Nat Clin Pract Oncol. 2007;4:591–602. doi: 10.1038/ncponc0934. [DOI] [PubMed] [Google Scholar]
- 7.Ewing GP, Goff LW. The insulin-like growth factor signaling pathway as a target for treatment of colorectal carcinoma. Clin Colorectal Cancer. 2010;9:219–223. doi: 10.3816/CCC.2010.n.032. [DOI] [PubMed] [Google Scholar]
- 8.Tognon CE, Sorensen PH. Targeting the insulin-like growth factor 1 receptor (IGF1R) signaling pathway for cancer therapy. Expert Opin Ther Targets. 2012;16:33–48. doi: 10.1517/14728222.2011.638626. [DOI] [PubMed] [Google Scholar]
- 9.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Zhou K, Fu W, Zhang H. Insulin-Like Growth Factor 1 Activates PI3k/Akt Signaling to Antagonize Lumbar Disc Degeneration. Cell Physiol Biochem. 2015;37:225–232. doi: 10.1159/000430347. [DOI] [PubMed] [Google Scholar]
- 11.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavano F, di Mola FF, Piepoli A, Panza A, Copetti M, Burbaci FP, Latiano T, Pellegrini F, Maiello E, Andriulli A, di Sebastiano P. Changes in miR-143 and miR-21 expression and clinicopathological correlations in pancreatic cancers. Pancreas. 2012;41:1280–1284. doi: 10.1097/MPA.0b013e31824c11f4. [DOI] [PubMed] [Google Scholar]
- 13.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Tang D, Sun B, Yu H, Yang Z, Zhu L. Tumor-suppressing effect of miR-4458 on human hepatocellular carcinoma. Cell Physiol Biochem. 2015;35:1797–1807. doi: 10.1159/000373991. [DOI] [PubMed] [Google Scholar]
- 15.Qin Y, Cheng C, Lu H, Wang Y. miR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cells. Biochem Biophys Res Commun. 2016;469:37–43. doi: 10.1016/j.bbrc.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 16.Sakai D, Mochida J, Yamamoto Y, Toh E, Iwashina T, Miyazaki T, Inokuchi S, Ando K, Hotta T. Immortalization of human nucleus pulposus cells by a recombinant SV40 adenovirus vector: establishment of a novel cell line for the study of human nucleus pulposus cells. Spine (Phila Pa 1976) 2004;29:1515–1523. doi: 10.1097/01.brs.0000131419.25265.23. [DOI] [PubMed] [Google Scholar]
- 17.Coronnello C, Benos PV. ComiR: Combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41:W159–164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Cui G, Shao C, Zhou X, Xiao Y, Zhou J. [Effects of recombinant adenovirus vector carrying human insulin-like growth factor 1 gene on the apoptosis of nucleus pulposus cells in vitro] . Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2013;27:1375–1379. [PubMed] [Google Scholar]


