Abstract
The pathogenesis of allergen-related inflammation in the intestine is to be further understood. Micro RNA (miR) can regulate immune responses. This study aims to investigate the role of miR-17-92 cluster in the induction of food allergen-related inflammation in the intestine. In this study, a mouse model of food allergen-related intestinal inflammation was developed. Expression of miR-17-92 cluster in B cells of the intestinal mucosa was analyzed by real time quantitative RT-PCR. The results showed that the levels of miR-19a, one of the members of the miR-17-92 cluster, were detected in the B cells of the intestine of mice sensitized to ovalbumin, which was significantly higher than that in naïve control mice. The expression of IL-10 by B cells was significantly lower in the sensitized mice as compared with naive control mice. Exposure to IL-4 in the culture increased the expression of miR-19a as well as suppression the expression of IL-10 in B cells via remolding DNA structure at the IL-10 promoter locus. We conclude that B cells from sensitized mice show higher levels of miR-19a, which plays an important role in the suppression of IL-10 in the B cells.
Keywords: Allergy, intestine, B cell, interleukin-10, micro RNA
Introduction
Interleukin-10 (IL-10) is an anti-inflammatory cytokine. In humans, IL-10 is encoded by the IL10 gene [1]. In humans, IL-10 is encoded by the IL10 gene, which is located on chromosome 1 and is primarily produced by monocytes and other immune cells such as lymphocytes, mast cells, regulatory T cells, and activated B cells [1]. IL-10 has multiple effects in immunoregulation and inflammation. It down regulates the expression of T helper (Th)1 cytokines, co-stimulatory molecules on dendritic cells (DC) and macrophages. It enhances B cell survival, proliferation, and antibody production [2]. Yet, in terms of the cellular and molecular aspect, the regulation of IL-10 expression has not been fully understood.
It is reported that IL-10-production B cells have immune regulatory function based on that IL-10 inhibits synthesis of pro-inflammatory cytokines such as IL-4, IFN-γ, IL-2, IL-3, TNFα and GM-CSF made by cells such as macrophages and CD4 T cells [3]. Thus, IL-10-production B cells are proposed as a fraction of immune regulatory B cells [4]. The compromise of frequency or function of IL-10-production B cells in immune disorder has been reported [5,6]. Cumulative evidence indicates that micro RNA (miR) can regulate lymphocyte function [7].
It is suggested that miR-17-92 regulates B cell function [8]. miR-17-92 cluster encodes six hairpin transcripts carrying six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a), located on human chromosome 13, within the third intron of the primary transcript C13 or f25 [9]. Elevated expression of miR-17-92 has been observed in a variety of immune disorders, such as cancer [10] and allergic asthma [11]. Whether miR-17-92 cluster regulates the IL-10 expression in B cells has not been investigated yet.
In this study, we observed that B cells isolated from the intestine of miR-17-92 mediated the effect of IL-4 on suppression of IL-10 expression in B cells.
Materials and methods
Mice
Male C57BL/6 mice (6-8 week old) were purchased from the Guangdong Experimental Animal Center. The miR-17-92fl/fl mice (B6 background) were purchased from the Jackson Laboratory. The mice were maintained in a pathogen free environment with accessing food and water freely. The experimental procedures were approved by the Animal Ethic Committee at Shenzhen University. All the experiments were carried out in accordance with the approved guidelines.
Development of FA mouse model
Following our established procedures, mice were gavage-fed with ovalbumin (OVA; 1 mg/mouse; Sigma Aldrich) and cholera toxin (CT; 20 μg/mouse; Sigma Aldrich) in 0.3 ml saline weekly for 4 weeks. The mice were sacrificed in week 5.
Assessment of serum IL-4 and specific IgE
Blood samples were collected from each mouse at sacrifice. The sera were isolated by centrifugation for 10 min at 4°C and analyzed by enzyme-linked immunosorbent assay (ELISA) with reagent kits of IL-4 and OVA-specific IgE (Biomart, Shanghai, China) following the manufacturer’s instructions.
Observation of mast cell and eosinophil infiltration in the intestinal mucosa
A jejunal segment was excised from each mouse at sacrifice. The tissue was fixed with 4% formalin overnight and processed for paraffin sections. The sections were stained with 0.5% toluidine blue (for mast cells) and hematoxylin/eosin (for eosinophils) respectively. Mast cells and eosinophils in the sections were counted on 20 randomly selected fields/mouse. The sections were coded. The observers were not aware of the code to avoid the observer bias.
Assessment of allergen-specific CD4+ T cells in the intestine
The small intestine was collected from each mouse at sacrifice. The lamina propria mononuclear cells (LPMC) were isolated with our established procedures [12]. CD4+ T cells and dendritic cells (DC) were isolated from the LPMCs with the magnetic cell sorting (MACS) with a reagent kit (Miltenyi Biotech) following the manufacturer’s instructions. The cell purity was checked by flow cytometry (95%). The CD4+ T cells were labeled with CFSE (carboxyfluorescein diacetatesuccinimidyl ester) and cultured with DCs (DC: T cell = 2 × 104 cells: 105 cells/well) for 3 days. The cells were collected and analyzed by flow cytometry.
Isolation of B cells
CD19+ B cells were isolated from LPMCs by MACS with a reagent kit (Miltenyi Biotech) following the manufacturer’s instructions. The cell purity was 95% as assessed by flow cytometry.
Real time quantitative RT-PCR (RT-qPCR)
The isolated B cells were processed with TRIzol Reagent (Invitrogen) following the manufacturer’s instructions to isolated total RNA, including miRNA. For miR-17-92 cluster detection, extracted RNA was reverse transcribed to cDNA using the PrimeScript™ RT reagent Kit (Invitrogen); the resulting cDNA was subjected to real-time PCR using SYBR Green ER qPCR Mix (Invitrogen). Reference gene RNA U6B (Invitrogen) was analyzed as an internal control. The Universal qPCR Primer was provided in the VILO kit and the forward primer for miR-17, 18a, 19a, 19b and 20a were purchasing from Qiagen. For mRNA detection, cDNA was synthesized using a reverse transcription reagent kit (Invitrogen). PCR was performed using SYBR Green Masture Mix (Invitrogen). The results were normalized to the β-actin. The sequences of IL-10 primers for PCR are ggtgagaagctgaagaccct and tgtctaggtcctggagtcca.
RNA interference (RNAi) of miR-19a
The RNAi reagent kit of miR-19a was purchased from Beijing Yijie Biotech (Beijing, China). The miR-19a RNAi was performed with B cells with the reagent kit following the manufacturer’s instructions. The RNAi effect on the B cells was assessed with RT-qPCR.
Preparation of cytosolic and nuclear extracts from B cells
B cells were isolated and incubated with lysis buffer at 4°C for 15 min, and centrifuged at 500 ×g for 10 min at 4°C. The supernatant was collected as the cytosolic extract. The pellet was added with nuclear extract buffer and incubated for 15 min at 4°C, followed by centrifugation at 13,000 ×g for 10 min at 4°C. The supernatant was collected as the nuclear extract. The protein concentrations were determined by the Bradford method.
Chromatin immunoprecipitation assay (ChIP)
ChIP assay was performed with a reagent kit purchased from Sigma Aldrich following the manufacturer’s instructions. B cells were fixed with 1% formaldehyde for 15 min. The cells were then lysed and sonicated to shear the chromatin DNA. Cell lysate was precleared with protein G-agarose beads for 2 h at 4°C. The supernatant was incubated overnight at 4°C with 2 μg of specific antibodies (Santa Cruz Biotech) or isotype IgG (a negative control). The precipitated antibody-chromatin complex was collected by incubation with protein G-agarose beads for 1 h at 4°C. The beads were collected by centrifugation and then washed and eluted in elution buffer. DNA was recovered from the precipitated samples by reverse crosslinking at 65°C for 4 h and digested with proteinase K for 1 h at 45°C to remove proteins, then the immunoprecipitated DNA was recovered by phenol/chloroform extraction and ethanol precipitation. The DNA or input (10%, collected before antibody precipitation) was analyzed by qPCR with the following miR-19a primers (purchased from Beijing Yijie Biotech; Beijing, China). The results were presented as folds of input.
Western blotting
The B cell cytosolic extracts were fractioned by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and transferred onto a PVDF membrane. After blocking with 5% skim milk for 30 min, the membrane was incubated with the anti-IL-10 antibody overnight at 4°C and followed by incubating with peroxidase-labeled secondary antibody for 1 h at room temperature. The membrane was washed between incubation with TBST (Tris-buffered saline Tween 20). The membrane was developed with ECL (enhanced chemiluminescence; Invitrogen). The results were photographed with an image station (UVI, Cambridge, UK).
Statistics
Data are presented as mean ± SD. Difference between two groups was determined by Student t test. P<0.05 was set as the significant criterion.
Results
B cells from FA mouse intestine show higher levels of miR-19a
The FA mice showed high serum levels of IL-4 and OVA-specific IgE, mast cell and eosinophil infiltration in the intestinal mucosa, and high frequency of OVA-specific CD4+ T cell proliferation in the culture after stimulated with OVA (Figure 1). The data demonstrate that the allergen-related inflammation was induced in the intestine.
Figure 1.
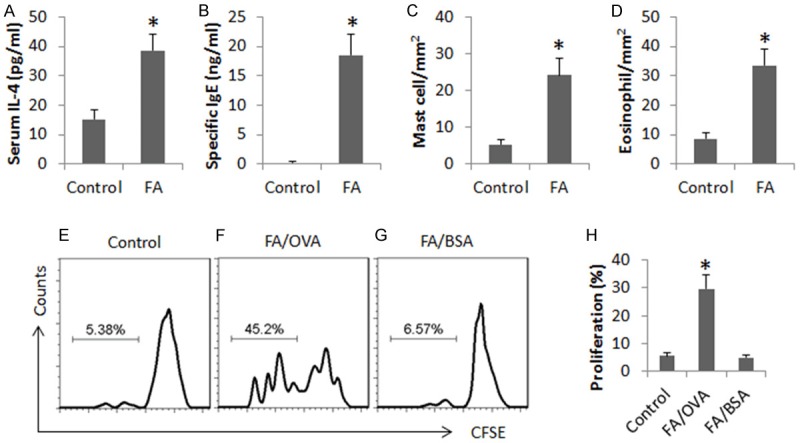
Assessment of allergen-related inflammation in the intestine. (A-D) The bars indicate the levels of serum IL-4 (A) and OVA-specific IgE (B, by ELISA), the frequency of mast cell (C) and eosinophil (D) in the intestinal mucosa (by histology) in samples collected from control mice (control) and FA mice (FA). (E-G) The gated histograms indicate the frequency of proliferating CD4+ T cells isolated from the mouse intestine (by CFSE-dilution assay). (H) The bars show the summarized data of (E-G). OVA: OVA (5 µg/ml) in the culture. BSA: BSA (5 µg/ml) in the culture. Data are presented as mean ± SD. *p<0.01, compared with the control group. Each group consists of 10 mice. Samples from individual mice were analyzed separately.
We also isolated B cells from the intestine. The cytosolic extracts of B cells were analyzed by RT-qPCR to determine the expression of miR-17-92 cluster and IL-10. As shown by Figure 2A-E, the levels of miR-19a, but not the miR-17, miR-18a, miR-19b and miR-92a, were significantly higher in FA mice than that in naïve mice. The levels of IL-10 mRNA were detected in the B cells, which were higher in control mice than in the FA mice (Figure 2F). The data of miR-19a and IL-10 mRNA were analyzed by correlation assay. A significant negative correlation (r = -0.7156, p<0.01) was identified between miR-19a and IL-10 in B cells (Figure 2G). The data implicate that IL-4 and miR-19a may be associated with the suppression of IL-10 in B cells of the FA mouse intestine.
Figure 2.
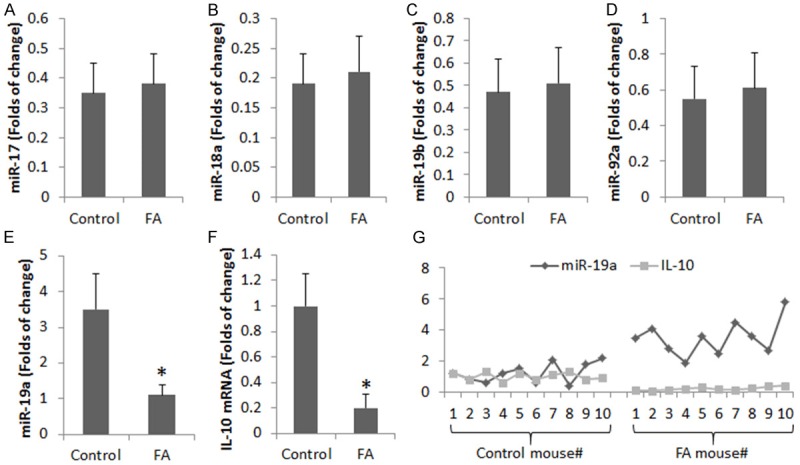
Assessment of miR-17-92 and IL-10 in B cells from the intestine. (A-F) The bars indicate the levels of miR-17-92 members (A-E) and IL-10 mRNA in B cells isolated from the mouse intestine (by RT-qPCR). Data are presented as mean ± SD. *p<0.01, compared with the control group. Each group consists of 10 mice. Samples from individual mice were processed separately. (G) The curves show the individual data of miR-19a and IL-10 mRNA in B cells isolated from the intestine.
miR-19a mediates the effect of IL-4 in suppression of IL-10 in B cells
We next observed the role of IL-4 in miR-19a expression in B cells. B cells were isolated from the mouse spleen and cultured in the presence of IL-4 at graded concentrations for 3 days. The B cells were analyzed by RT-qPCR. The results showed that exposure to IL-4 markedly increased the expression of miR-19a in B cells in an IL-4 dose-dependent manner (Figure 3A).
Figure 3.
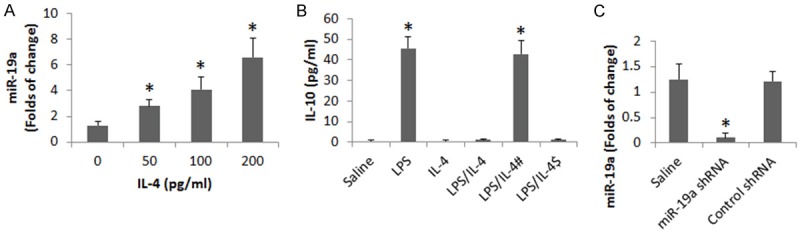
miR-19a mediates the IL-4-suppressed IL-10 expression in B cells. B cells were isolated from the peripheral blood samples of healthy subjects. The cells were treated with the conditions as denoted on the X axis for 3 days. (A) The bars indicate the miR-19a levels in the B cells (by RT-qPCR). (B) The bars indicate the IL-10 levels in the supernatant of B cell culture (by ELISA). (C) The bars indicate the miR-19a RNAi results. Data of bars are presented as mean ± SD. *p<0.01, compared with the dose 0 group (A) or the saline group (B). The data are summarized from 3 independent experiments.
With an established cell culture model [13], we up regulated the expression of IL-10 in B cells with LPS in the culture. Wild B cells and miR-deficient B cells were stimulated with LPS in the presence or absence of IL-4. The results showed that exposure to LPS increased the expression of IL-10 by B cells, which was suppressed by the presence of IL-4 in wild B cells, but not in miR-19a-deficient B cells (Figure 3B, 3C). The results suggest that miR-19a mediates the effect of IL-4 in the suppression of IL-10 expression in B cells.
miR-19a mediates the IL-4-induced DNA remolding at the promoter locus of IL-10 in B cells
Based on published data that HDAC11 is involved in the suppression of IL-10 [13], we wondered if HDAC11 was involved in the regulation of IL-10 expression in B cells regulated by IL4/LPS in the present experimental system. To this end, we assessed HDAC11 levels in B cells after exposure to LPS or/and IL-4. The results showed that, at the locus of IL-10 promoter, we detected higher levels of HDAC11 at the IL-10 promoter locus in B cells (Figure 4A). We further observed lower levels of H3K4, RNA polymerase II and c-Maf at the promoter locus of IL-10 in B cells after stimulated with IL-4 or IL-4/LPS as compared to LPS alone (Figure 4B-D); such a phenomenon did not occur in miR-19a-deficient B cells (Figure 4A-D).
Figure 4.
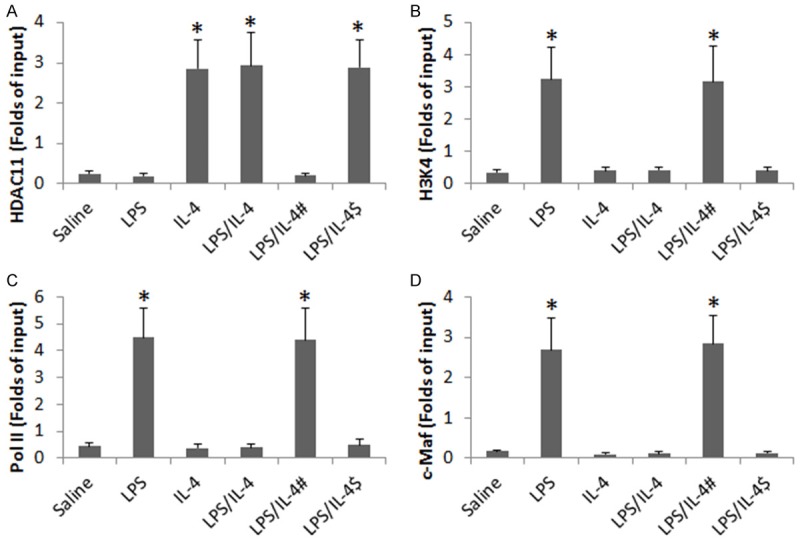
miR-19a mediates IL-4-induced DNA remolding at IL-10 promoter locus in B cells. B cells were isolated from the naive mouse spleen and stimulated with LPS (1 µg/ml) or/and IL-4 (200 pg/ml) in the culture for 48 h. The bars indicate the levels of HDAC11 (A), acetylated H3K4 (B), RNA polymerase II (Pol II; C) and c-Maf (D) at the IL-10 promoter locus. #, miR-19a was knocked down in the B cells. $, B cells were treated with control shRNA. Data are presented as mean ± SD. *p<0.01, compared with the saline group. The data were summarized from 3 independent experiments.
Deficiency of miR-19a prevents IL-4-inudced IL-10 suppression in B cells in vivo
To further test the role of miR-19a in the mediating the IL-4 induced IL-10 suppression in B cells, we treated wild and miR-19a-deficient mice with recombinant IL-4 or/and LPS via intraperitoneal injection daily for 5 days. B cells were isolated from the intestine and analyzed for IL-10 expression. The results showed that treatment with LPS increased IL-10 expression in B cells of both wild and miR-19a-deficient mice, which was blocked by IL-4 in wild mice but not in miR-19a-deficient mice (Figure 5).
Figure 5.
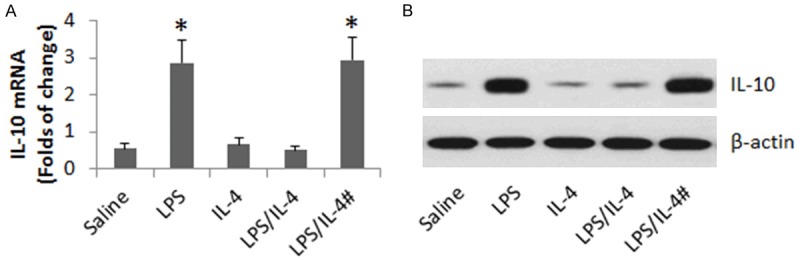
Assessment of miR-19a in mediating the role of IL-4 in suppressing IL-10 expression in B cells in vivo. C57BL/6 mice and miR-19a-deficient mice (#) were i.p. injected with LPS (10 mg/kg) or/and IL-4 (4 mg/kg) daily for 5 days. B cells were isolated from the intestine and analyzed by RT-qPCR and Western blotting. A. The bars indicate the mRNA levels of IL-10 in B cells. B. The immune blots indicate the protein levels of IL-10 in B cells. Each group consists of 6 mice. Samples from individual mice were processed separately.
Discussion
The present data show that the expression of IL-10 was lower in B cells from intestine of FA mice. The expression of IL-10 was negatively correlated with the miR-19a levels in the B cells. We also found that the serum levels of IL-4 were positively correlated with the miR-19a levels of B cells. Exposure of B cells to IL-4 in the culture markedly suppressed the expression of IL-10, which was mediated by miR-19a. This study has identified a previously unknown phenomenon that miR-19a interferes with the expression of IL-10 in B cells.
Cumulative reports indicate that miR-17-92 cluster is a group of important molecules in the immune regulation. They are involved in a large number of immune activities, such as in allergy [11], Cancer [14] and inflammatory disorders [15]. There are 6 members in the miR-17-92 cluster. Our data show that the miR-19a, one of the members of miR-17-92 cluster, was up regulated in FA mice. This is in line with the data reported by Simpson et al, in which miR-19a was found higher in asthma patients as compared with healthy subjects [11]. Both of our data indicate that miR-19a plays a role in the promotion of Th2 polarization. Simpson’s data indicate the increase in miR-19a was in CD4+ T cells, which facilitated the production of Th2 cytokines, while our data indicate that miR-19a was increased in B cells in the intestine to suppress the production of IL-10 in B cells.
The expression of IL-10 by B cells is of significance in immune regulation. Cumulative evidence indicates that IL-10+ B cells have immune regulatory function. Our previous studies show that IL-10+ B cells of the intestine are less in FA mice than in naive control mice [16,17]. Lee et al indicate that peripheral IL-10+ B cells are less in patients with food allergy than healthy subjects [5]. Liao et al found that in asthma patients, the peripheral IL-10+ B cells were also less as compared with healthy controls [18]. Therefore, to identify the causative factors in the suppression of IL-10 expression in B cells is beneficial for innovating novel therapeutic remedies for allergic disorders. The present data indicate that miR-19a is one of the factors that repress the expression of IL-10 in B cells, indicating that miR-19a may be a novel target to regulate the development of IL-10+ B cells.
The data show that exposure of B cells to IL-4 represses the expression of IL-10. IL-4 is the signature cytokine of Th2 response. It is well documented that IL-4 is increased in allergic diseases [19]. It is also observed that IL-10+ B cells are less frequent in the subjects with allergic disorders [18]. These two phenomena are in parallel in subjects with allergic diseases. The present data identified a negative correlation between serum IL-4 levels and the expression of IL-10 in B cells, indicating high levels of IL-4 are one of the factors to compromise the expression of IL-10 in B cells.
In summary, the present data indicate that miR-19a plays a critical role in the suppression of IL-10 in B cells; the data suggest that to regulate the expression of miR-19 may be novel approach in the improvement of regulatory B cell development.
Acknowledgements
This study was supported by grants from the innovation of science and Technology Commission of Shenzhen Municipality (JCYJ20150402090413008, JCYJ20140418095735611 and ZDSYS201506050935272), the Natural Science Foundation of China (81373176, 31570932, 31400856, 81571790 and 81501573).
Disclosure of conflict of interest
None.
Authors’ contribution
ZQL, GY, XRG, JQL and LHM performed experiments, analyzed data and reviewed the manuscript. ZGL and PCY organized the study and supervised the experiments. PCY designed the project and wrote the manuscript.
References
- 1.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–81. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 2.Fillatreau S, Gray D, Anderton SM. Not always the bad guys: B cells as regulators of autoimmune pathology. Nat Rev Immunol. 2008;8:391–7. doi: 10.1038/nri2315. [DOI] [PubMed] [Google Scholar]
- 3.Mauri C, Blair PA. Regulatory B cells in autoimmunity: developments and controversies. Nat Rev Rheumatol. 2010;6:636–43. doi: 10.1038/nrrheum.2010.140. [DOI] [PubMed] [Google Scholar]
- 4.Li R, Rezk A, Healy LM, Muirhead G, Prat A, Gommerman JL, Bar-Or A MSSRF Canadian B cells in MS Team. Cytokine-Defined B Cell Responses as Therapeutic Targets in Multiple Sclerosis. Front Immunol. 2015;6:626. doi: 10.3389/fimmu.2015.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SJ, Noh G, Lee JH. In Vitro Induction of Allergen-Specific Interleukin-10-Producing Regulatory B Cell Responses by Interferon-gamma in Non-Immunoglobulin E-Mediated Milk Allergy. Allergy Asthma Immunol Res. 2013;5:48–54. doi: 10.4168/aair.2013.5.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noh J, Noh G, Lee SJ, Lee JH, Kim A, Kim HS, Choi WS. Tolerogenic effects of interferon-gamma with induction of allergen-specific interleukin-10-producing regulatory B cell (Br1) changes in non-IgE-mediated food allergy. Cell Immunol. 2012;273:140–9. doi: 10.1016/j.cellimm.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 7.de Candia P, Torri A, Pagani M, Abrignani S. Serum microRNAs as Biomarkers of Human Lymphocyte Activation in Health and Disease. Front Immunol. 2014;5:43. doi: 10.3389/fimmu.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mihailovich M, Bremang M, Spadotto V, Musiani D, Vitale E, Varano G, Zambelli F, Mancuso FM, Cairns DA, Pavesi G, Casola S, Bonaldi T. miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nat Commun. 2015;6:8725. doi: 10.1038/ncomms9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 10.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, Wang Y, Pua HH, Baumjohann D, Montoya MM, Panduro M, Remedios KA, Huang X, Fahy JV, Arron JR, Woodruff PG, Ansel KM. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. 2014;15:1162–70. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Xu LZ, Peng K, Wu W, Wu R, Liu ZQ, Yang G, Geng XR, Liu J, Liu ZG, Liu Z, Yang PC. Specific immunotherapy in combination with Clostridium butyricum inhibits allergic inflammation in the mouse intestine. Sci Rep. 2015;5:17651. doi: 10.1038/srep17651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, Pinilla-Ibarz J, Seto E, Sotomayor EM. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conev NV, Donev IS, Konsoulova-Kirova AA, Chervenkov TG, Kashlov JK, Ivanov KD. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci Trends. 2015;9:393–401. doi: 10.5582/bst.2015.01170. [DOI] [PubMed] [Google Scholar]
- 15.Kara M, Kirkil G, Kalemci S. Differential Expression of MicroRNAs in Chronic Obstructive Pulmonary Disease. Adv Clin Exp Med. 2016;25:21–6. doi: 10.17219/acem/28343. [DOI] [PubMed] [Google Scholar]
- 16.Yang G, Geng XR, Liu ZQ, Liu JQ, Liu XY, Xu LZ, Zhang HP, Sun YX, Liu ZG, Yang PC. Thrombospondin-1 (TSP1)-producing B cells restore antigen (Ag)-specific immune tolerance in an allergic environment. J Biol Chem. 2015;290:12858–67. doi: 10.1074/jbc.M114.623421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZQ, Wu Y, Song JP, Liu X, Liu Z, Zheng PY, Yang PC. Tolerogenic CX3CR1+ B cells suppress food allergy-induced intestinal inflammation in mice. Allergy. 2013;68:1241–8. doi: 10.1111/all.12218. [DOI] [PubMed] [Google Scholar]
- 18.Liao HY, Tao L, Zhao J, Qin J, Zeng GC, Cai SW, Li Y, Zhang J, Chen HG. Clostridium butyricum in combination with specific immunotherapy converts antigen-specific B cells to regulatory B cells in asthmatic patients. Sci Rep. 2016;6:20481. doi: 10.1038/srep20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol. 2010;10:838–48. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]


