Abstract
Background: Human adenovirus type 7 (HAdV7) is globally attracting great concern as its high morbidity and severity in respiratory diseases, especially in Asia. Objective: To investigate the clinical and epidemiologic characteristics of HAdV7 infection outbreak in East China. Methods: The clinical samples were collected from the patients of an ARD outbreak in East Chinafor the detection of causative pathogens by multiplex PCR. The molecular type of human adenovirus isolates were identified by sequencing and homologous comparison based on their hexon genes. The spatiotemporal dynamics of global HAdV7 was investigated using the phylogenetic and phylogeographic analyses. Total 67 referenced HAdV7 hexon sequences (>800 bp) from GenBank were selected for constructing the maximum likelihood tree by MEGA 5.1.0, grouped according to the tree topology for the further migration analysis by PAUP* 4.0 and MigraPhyla 1.0 b to understand the transmission patterns of HAdV7 in global epidemics. Results: The results showed HAdV7 as the causative pathogen in this outbreak, and the outbreak strains had the hexon sequences highly identical with the isolates in Shaanxi (2012). The origin of HAdV7 was inferred as California, meanwhile a total of 21 migration routes were acquired. HAdV7 in this outbreak was statistically proven dispersed from Shaanxi province (2012). Conclusions: The analyses of epidemiology and transmission pattern of HAdV7 would not only enrich the molecular biological basic database but also provide theoretical basis for HAdV7 prevention and control strategy.
Keywords: Human adenovirus type 7, epidemiology, clinical characteristics, phylogeny, phylogeography
Introduction
Human adenovirus (HAdV) is a common virus found worldwide since it was first isolated from the tonsils and adenoids in children undergoing tonsillectomy and adenoidectomy in 1953 [1]. It is a non-enveloped icosahedral virus composed of double stranded linear DNA. Based on characteristics such as serology, DNA sequence homology, and phylogenomics, to date, more than 60 serotypes have been identified and classified into seven species (A-G, with B species further divided into B1 and B2 sub-species)[2].
Human adenovirus strains are widely distributed. HAdV infections are transmitted through respiratory route or the fecal-oral route, and cause a broad spectrum of clinical diseases, including respiratory, gastrointestinal, conjunctival illness, etc [3,4]. The children under the age of 5 years, close-quartered populations such as crowed communities, schools, military training camps, and immune-compromised individuals are generally the susceptible populations of the HAdV-infections according to the infectious route and pathogenicity [5,6].
HAdV infections have been considered for decades as being one of the predominant causes (other two viral agents are respiratory syncytial virus, parainfluenza virus) of respiratory diseases among military trainees worldwide with infection reports in USA [7,8], Asia [9] and Europe [10]. The HAdV serotypes most frequently associated with acute respiratory diseases (ARD) in both military and civilian communities have been reported as subspecies B1 HAdV3, HAdV7, HAdV21 and species E HAdV4 [7-9]. In the last 5 years, especially among the Asian area, a large scale population suffered from HAdV7 infections presented as ARD were reported continuously [11-13]. In mid-January 2014, another large scale of HAdV7 outbreak occurred in the military in east China. This apparent outbreak promoted us to use surveillance to describe the epidemiology, clinical features of HAdV7 infection. In the present study, we also investigated the transmission dynamics of HAdV7 from the origin to its dispersal in east China based on the phylogenetic and phylogeographic analyses [14,15]. We aimed to understand the evolutionary process of HAdV7 in China, which will aid in establishing more effective control strategies.
Materials and methods
Samples collection and etiology determination
Totally 119 double throat swabs and blood samples were collected from the hospitalized military trainees who had the ARD symptoms such as fever (over 37.7°C), cough, cough with sputum and sore throat in Zhejiang and Shanghai, East China. All specimens were preserved at -80°C for subsequent nucleic acid extraction, viral culture and isolation. The study was approved by the Hospital’s Human Use Committee and Institutional Review Board.
All the blood samples were subjected to microbe culture, routine blood examination including blood cell count, blood platelet count, hemoglobin test, and C-reactive protein (CRP), blood biochemical tests, for example, albumin (ALB), total bilirubin (TBIL), glutamic-pyruvic transaminase (GPT), creatinine (Cr), and glucose (GLU). Acute IgM/IgG serology was evaluated by using enzyme-linked immunosorbent assay (ELISA) to detect the common respiratory pathogens including adenovirus.
PCR detection, meanwhile, was performed to confirm the causative pathogens. Viral nucleic acids were extracted from 200 ul of the throat swab specimens using the QIAampMinElute Virus Spin Kit (QIAGEN, Shanghai, China) in accordance with the manufacturer’s instructions. For detection of Legionella pneumophila (L. pneumophila), Mycoplasma pneumoniae (M. pneumoniae) and Chlamydia pneumoniae (C. pneumoniae), previously published specific primers and probes targeted at specific regions of the mip gene, the P1 gene, and the 16S rRNA, respectively were used for real-time PCR assay [16-18]. Real-time PCR was performed on an ABI 7500 FAST Real-Time PCR System (Applied Biosystems, CA, USA). Amplification was according to the following parameters: 95°C for 10 min, followed by 50 cycles at 95°C for 15 s, 50-65°C for 1 min. Other 15 types of respiratory DNA/RNA virus were detected by using the Revert Aid First Strand cDNA synthesis kit (Fermentas, Ontario, Canada) and the Seeplex RV 15 ACE Detection Kit (Seegene, Seoul, Korea) in accordance with the manufacturer’s instruction.
Adenovirus culture, isolation and identification
From adenovirus-positive throat swab specimens identified by PCR analysis, 20 were randomly selected for incubating into HEp-2 cells and culturedin a maintenance medium (minimal essential mediumcontaining 2% FCS, 100 U/mL penicillin G, 100 mg/mL streptomycin) at 37°C in a closed system in a 5% CO2 incubator.Cultures exhibiting adenovirus-like CPE were passaged again to confirm the presence of the virus [13]. The genotype identification of HAdV was performed by sequencing and comparing the hexon gene sequence of the adenovirus positive isolate with that of other reference strains from NCBI GenBank database (http://www.ncbi.nlm.nih.gov/pubmed/). PCR amplification was optimized [19], and 3 pairs of specific primers were used for separate PCRs whose products had overlapping sequences of 250-300 bp and were sequenced in both directions (Table 1). These sequence segments were assembled using the SeqMan in Lasergene 8 (DNASTAR, Madison, WI, USA). Multiple nucleotide sequence alignments were conducted by Clustal W in BioEdit 5.0.9 (Ibis Therapeutics, Carlsbad, CA, USA) [20]. 17 reference sequences representing seven species of HAdVs were retrieved from GenBank database and neighbor-joining phylogenic tree with 1000 bootstrap pseudo-replicates was constructed based on the hexon gene sequences of the isolates from this outbreak by using MEGA 5 (CEMI, Tempe, AZ, USA) [21].
Table 1.
Primers used for amplification and sequencing of the complete hexon of HAdV7 isolates in this study
| Primer | Sequence (5’-3’) | Position* | Size (bp) |
|---|---|---|---|
| ADV-1F | GAGGAGAAAGGAAGAGGTCG | 18373-18392 | 1272 |
| ADV-1R | TATGCCATCCAGAGGAAAAC | 19626-19645 | |
| ADV-2F | CAGGGATAACTTTGTAGGCCTA | 19372-19393 | 946 |
| ADV-2R | CAGAGAGGTAGTCGTTGAATGA | 20297-20318 | |
| ADV-3F | TTTCCACATACAAGTGCCTCA | 20044-20064 | 1331 |
| ADV-3R | GGGAAACGCTTGTCAAAGGT | 21356-21375 |
Position in the complete genome of HAdV7 strain Gomen (AY594255).
Migration analysis of human adenovirus type 7
The HAdV7 global transmission characteristics in the human populations were investigated by phylogenetic and phylogeographic analyses that were firstly used by Wallace et al. [22]. A total of 66 HAdV7 sequences were acquired for the construction of the HAdV7 data set according to 3 selection criteria: 1) the sequences searchable in the NCBI were published before January 2014, with the genome lengths ranging from 800-35000 bp; 2) the strains collected from sporadic HAdV7 infectious cases or without clear isolated information would be excluded; 3) the replicates (100% identity) at the same epidemic would be excluded. The HAdV3 isolate AB330084 from Genbank database was added into the data set of the HAdV7 strains (n=67) as an outgroup in the phylogeny. The maximum likelihood tree was constructed using a Kimura 2-parameter model by the bootstrap method with the value of 1000 replicates in MEGA 5 (CEMI, Tempe, AZ, USA). Then 67 referenced sequences used in the migration analysis with MigraPhyla 1.0 b (http:pd.bio.uci.edu/ee/WallaceR/Migraphyla.html) [14] were grouped into 32 discrete modules, each of which was combined with the year of isolation and the isolated locality according to the tree topology [22]. The modules of the sequences in the tree were assigned to the tips as a single character with 31 states. And the modules of ancestral nodes were assigned by the method of maximum parsimony in PAUP* 4.0 (Sinauer Associates, Sunderland, MA, USA) when moving recursively up the tree to support the fewest possible migration events between modules consistent with the phylogenetic tree. Under the MigraPhyla protocol, a Monte Carlo test of 10000 trials was performed to calculate the P values that represented the probable frequencies of migration events between each pair of modules in the original migration tree more than that of migration events between each pair of modules randomly distributed across the tree tips. Furthermore, to test the significance of P values (P<0.05) across all localities, a corresponding sparse false discovery rate (sFDR) correction was calculated as the necessary for α, the normal type 1 error rate in the multiple tests across module pairs.
Inpatient analysis
Complete clinical, radiographic, and laboratory examinations were performed in all the hospitalized trainees with permission. Relevant data were recorded, surveillance of radiographic examinations including routine thoracic computed tomography (CT) scans, ultrasound and electrocardiographic examination on the patients were kept during the treatment. These analyses were aim for determining whether HAdV infection was associated with specific presenting or outcome variables.
Statistical analysis
Data were analyzed by SPSS version 11.02 (SPSS, IL, USA). Categorical variables were used for description of the clinical data, and continuous variables were signed as (x̅±SD). Monte Carlo test of 10000 trials were applied in Migration analysis to calculate the value of P. A value of P<0.05 was considered statistically significant.
Results
Epidemiology and clinical features of ARD infection
In mid-January 2014, an ARD outbreak occurred in the military training bases where a total of 1200 soldiers were taking the training in Zhejiang and Shanghai, China. Centralized teaching courses and training activities were carried out for recruit trainees. The index cases were two recruits aged both 20 years who lived in the same dormitory presented with fever (oral temperature of 38.1°C, 38.5°C, respectively), and a series of ARD symptoms, including cough, sputum, sore throat, and headache. The disease spread rapidly and peaked 4 days after the index cases. Medical staffs from three military hospitals were sent to aid the base medics in treating the sick trainees. During this outbreak, 342 trainees aged 17-23 years developed disease symptoms. Infectious patients who had febrile respiratory symptoms with severe moist rales heard or shadows in the lung by fluoroscopy were suggested to be hospitalized. Remaining mild cases were reviewed by physicians, quarantined and treated in the training base. In this study, 119 ill trainees were hospitalized and their clinical features were summarized in Table 2. Among these hospitalized trainees, the most common symptom was fever (85, 71.4%). Neither gastrointestinal symptoms nor conjunctivitis was observed. Skin rash (1, 0.9%) and anorexia (3, 2.7%) were less common. In addition, cervical lymphadenopathy was detected in 10 (8.4%) patients.
Table 2.
Clinical data of 119 military trainees hospitalized for adenovirus infection in east China
| Parameter | Value |
|---|---|
| Age, years (Mean ± SD) | 19.23 ± 2.89 |
| Median hospitalization, days (range) | 22 (8-38) |
| Fever | 71.43 (85/119) |
| Duration, days (range) | 5 (1-17) |
| Median peak temperature, (range) | 38.7 (37.7-40.2) |
| Sore throat/Pharyngeal hyperemia and edema | 49.58 (59/119) |
| Bilateral tonsil I-II enlargement | 47.06 (56/119) |
| Cough | 66.39 (79/119) |
| Acratia/Muscular Soreness | 21.01 (25/119) |
| Expectoration | 60.50 (72/119) |
| White sputum | 33.61 (40/119) |
| Yellow sputum | 26.89 (32/119) |
| Pneumonia | 48.74 (58/119) |
| Right upper lobe infiltration | 1.68 (2/119) |
| Right middle lobe infiltration | 6.72 (8/119) |
| Right lower lobe infiltration | 6.72 (8/119) |
| Left upper lobe infiltration | 1.68 (2/119) |
| Left lower lobe infiltration | 21.85 (26/119) |
| Lateral lung infiltration | 10.08 (12/119) |
Notes: Values are % (no. abnormal inpatients/total inpatients), except as indicated.
Clinical laboratory parameters and radiographic findings
The results of blood culture showed all blood samples were negative for microbe. The routine blood examinationexhibited abnormal white blood cell (WBC) levels including Leukocytosis (>10.0×109/L, 12.61%) and leucopenia (<4.0×109/L, 31.93%), and elevated C-reactive protein (CRP, >5.0 mg/L). No obviously abnormal biochemical indexes were observed. Six kinds of pathogen-specific IgM antibodies were detected. All samples were positive for HAdV IgM, 4 were simultaneously positive for M. pneumoniae IgM, and one each positive for IgM specific to L. pneumophila, C. pneumoniae, influenza A virus and human parainfluenza virus, respectively. Further chest radiography that performed in all inpatients was reviewed. 53 chest abnormal cases were observed by chest CT scan at admission. Besides, another 5 patients hospitalized for 2 days with worsen conditions were found chest abnormal. In summary, 48.7% (58/119) patients signed as pneumonia. By reading the film results of chest CT scan, high density mottling or patchy shadows were predominantly existed rather than homogenous lobar or segmental shadows, which were mainly distributed as a unilateral lung form, especially in the left lung (26, 21.8%) (Figure 1A and Table 2). Besides, similar patchy shadows were observed in the right lung (18, 15.1%) (Figure 1C and Table 2), and in bilateral lungs (12, 10.1%) (Figure 1E and Table 2).
Figure 1.
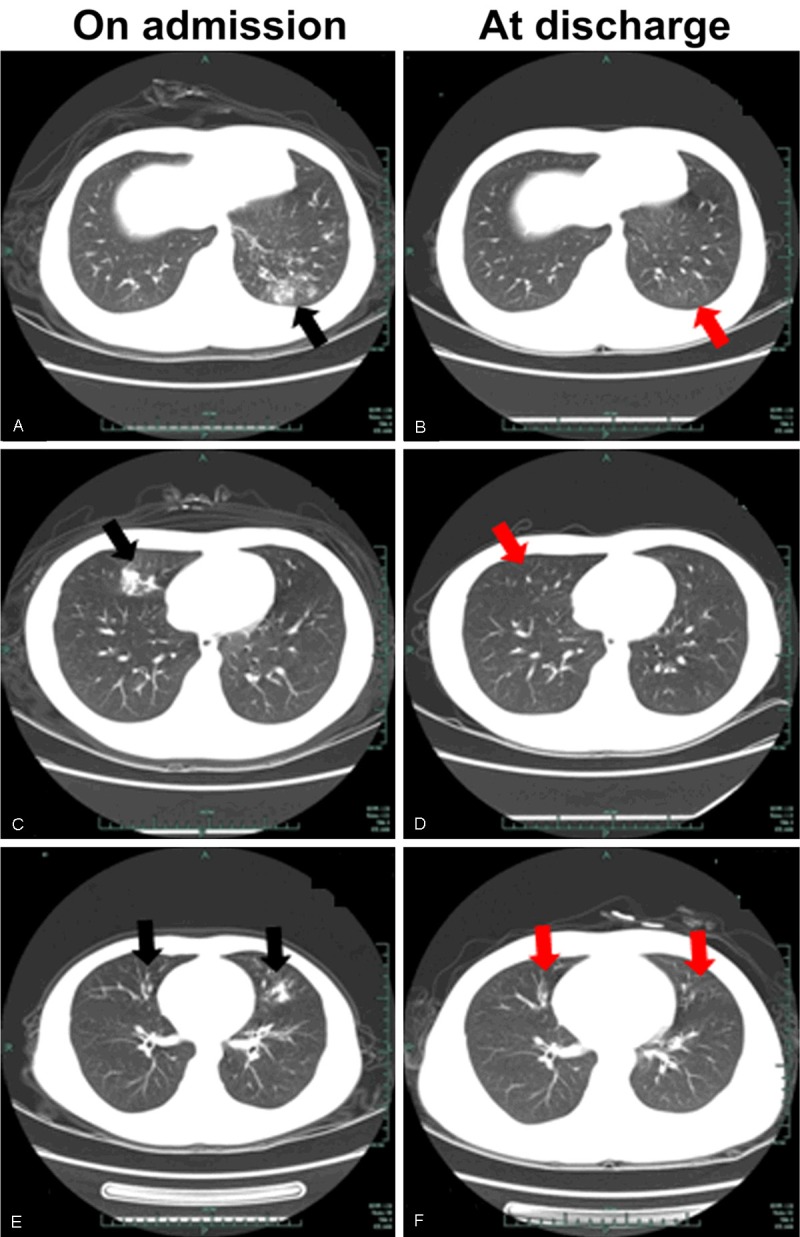
Chest computer tomography (CT) scans in 3 hospitalized patients with HAdV7 infection on admission (A, C, E) and at discharge (B, D, F). Three pulmonary infection pattern were showed: high density mottling or patchy shadows (black arrows) in the left lobe (A), in the right lobe (C), and in bilateral lungs (E).
PCR determination of the outbreak
By detecting the causative pathogens using extracted nucleic acid from 119 throat swab samples under PCR, positive adenovirus DNA were observed in all samples. However, co-infection with other pathogens were not common in this outbreak, in the adenovirus infectious patients, 8 (6.7%) were found to be concomitantly infected with M. pneumonia (4), C. pneumoniae (1), influenza A virus (1), human parainfluenza virus 1 (1), and L. pneumophila (1) (Table 3).
Table 3.
The major laboratory data of 119 patients hospitalized for adenovirus infection in east China
| Test | Laboratory data | Mean ± SD* | % (no. corresponding inpatients/totalinpatients) |
|---|---|---|---|
| Blood | WBCcount†, 1000 cells/uL | ||
| <4.0 | 3.37 ± 0.54 | 31.93 (38/119) | |
| 4.0-1.0 | 5.61 ± 1.38 | 55.46 (66/119) | |
| >10.0 | 10.8 ± 1.85 | 12.61 (15/119) | |
| Neutrophils, % | 60.7 ± 12.8 | 100 | |
| Lymphocytes, % | 30.9 ± 11.7 | 100 | |
| C-reactive protein, mg/L | |||
| >5.0 | 26.6 ± 19.1 | 72.27 (86/119) | |
| <5.0 | 2.57 ± 2.1 | 27.73 (33/119) | |
| Serum | Legionella pneumophila-IgM# | + | 0.84 (1/119) |
| Mycoplasma pneumonia-IgM# | + | 3.36 (4/119) | |
| Chlamydia pneumonia-IgM# | + | 0.84 (1/119) | |
| PCR | Influenza A virus | + | 0.84 (1/119) |
| Human parainfluenza virus | + | 0.84 (1/119) | |
| Human adenovirus | + | 100 |
SD, standard deviation;
WBC, white blood cell;
+detection as positive;
PCR, reconfirmed as positive.
Molecular analysis of human adenovirus
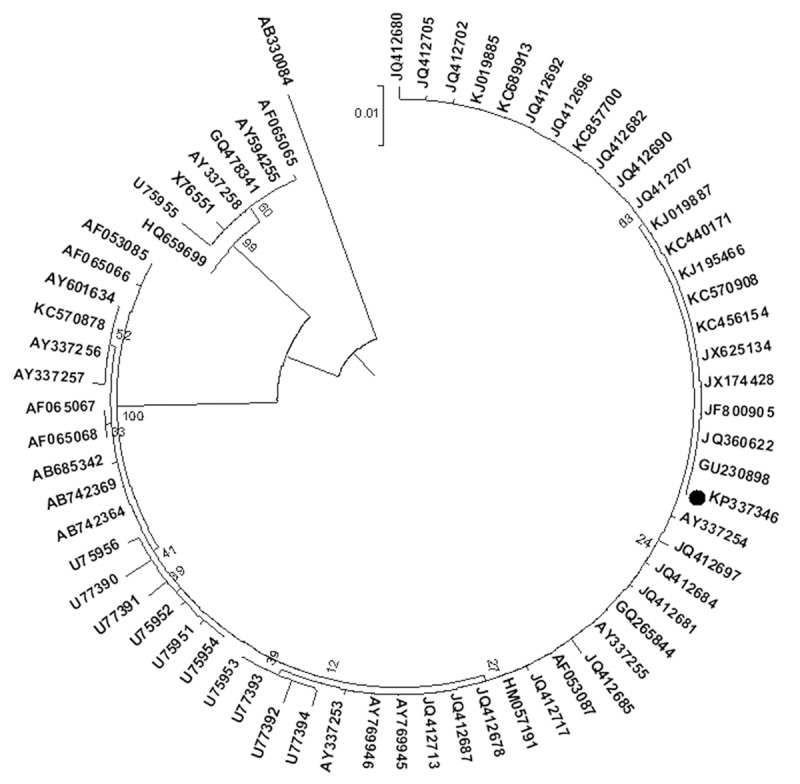
From 20 randomly selected adenovirus-positive throat swab specimens identified by PCR analysis, 18 isolates were observed to produce visible adenovirus-like CPE upon culturing in A549 cells. Comparative analysis and online Nucleotide BLAST analysis of the entire hexon sequences showed that all these 18 outbreak isolates shared 100% similarities with each other and were identified as the same genotype HAdV7, of which 3 representative isolates from different time points (16 January, 20 January and 27 January) were archived as HZU2014-07, HZU2014-23 and HZU2014-81, and deposited in GenBank with the corresponding accession number KP337345, KP337346, and KP337347. Further, phylogenetic analysis results showed that the isolates from this study belonged to HAdV7 and had high identity with the hexon gene of HAdV SXWN1203 strain KC689913 [13], HAdV CHN/DG01/2011/7 [P7H7F7] strain KC440171 (Figure 2) [23].
Figure 2.

Neighbor joining phylogeny of hexon sequences for HAdV7 isolates described in this study and other reference hexon sequences representing seven species of HAdV from GenBank.
Clinical treatment and prognosis
According to the clinical symptoms and signs, HAdV7 was determined as the causative pathogen for this outbreak in the military training bases combining with the laboratory results. Following the diagnosis and treatment guideline for adenovirus infection issued by Chinese Military Commission for Infectious Diseases [24], we used the treatments such as, methylprednisolone sodium succinate (methylprednisolone) for anti-inflammatory, ribavirin for antiviral, levofloxacin lactate (lailixin) for preventing infection, tanreqing (a Chinese patent medicine) treatment, vitamin C for heat clearing and detoxifying, rebamipide tablets for protecting gastric mucosa, omeprazole enteric coated capsules for inhibition of gastric acid. All HAdV7 infectious patients received the same antibiotic treatment exclude the ones who were co-infected with M. pneumonia or C. pneumonia or L.pneumophila were added the extra azithromycin to the antibiotic treatment.
Migration analysis of human adenovirus type 7
As described, MigraPhyla was used for tracking the migration of HAdV7 during its evolutionary history [14,22]. Total 67 sequences (excluding the replicates in the same epidemic and individual infectious cases) in 31 modules across 19 localities worldwide from its first discovery to January 2014 were archived for constructing the maximum likelihood phylogenetic tree (Figure 3).
Figure 3.
Maximum likelihood phylogeny of hexon sequences of HAdV7 viruses from this study in Hangzhou, China and other global locations. 66 HAdV7 hexon sequences across the world in different years from Genbank were analyzed.
MigraPhyla results showed that among the migration events detectable cells, California was considered as the primary source for multiple regional and international HAdV7 migration events. Monte Carlo test results showed that California was reconfirmed as the primary source for HAdV7 migration events, and total 22 migration routes were statistically significant (P<0.05). Washington D.C., Great Lakes regions, and Beijing, Guangzhou, the countries of Mongolia, the Philippine city of Tacloban, were inferred as the secondary sources for HAdV7 migration events from where HAdV7 was spread to the tertiary localities. In this outbreak, HAdV7 migration route was from the initial source of California to the Great Lakes regions, next to the southern Chinese city of Guangzhou, and back north to Shaanxi, finally to Hangzhou (Figure 4).
Figure 4.
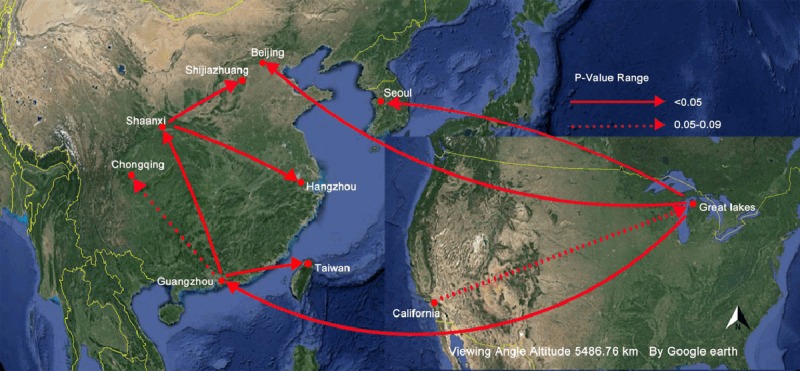
Global migration maps of HAdV7 hexon sequences that related to this outbreak. The node denotes the locality, the red solid arrow lines indicate the statistically significant routes between two nodes (P<0.05) under 10000 Monte Carlo tests and a sparse false discovery rate (sFDR) correction.
Discussion
In the present study, we reported a large-scale outbreak of HAdV7 infections associated with acute respiratory disease in a military training base in Zhejiang Province and Shanghai, East China, in mid-January, 2014. Compared with the clinical symptoms that published previously, symptoms in this epidemic showed no apparent difference, such as fever, cough, expectoration, and pneumonia [5,25,26]. Positive HAdV-IgM in serum samples from patients were determined, however could not be used as an early diagnosis for infection. As described by Julkunen et al., IgM responses were faint and inconsistent comparing with IgA and IgG responses [27]. Therefore, the IgM antibodies test by ELISAwas not appropriate for the early diagnosis of HAdV infection. We took the multiplex PCR approach by using the Dual priming oligonucleotide system technology for detecting the adenovirus [28], and identified the molecular type of HAdV by using direct sequencing of the hexon gene where the hypervariable regions (HVRs) were existed. Up to now, this hexon gene sequencing and homologous analysis is the most traditional but highest accurate method for identification of HAdV [29]. However, compared to other type-specific PCR or nested PCR methods, it also costs much and is time-consuming on the cases of numerous clinical specimens. Therefore, the need to establish and improve the efficient epidemiological and common etiological detection for HAdV infection is emphasized.
Among HAdV serotypes, HAdV3, HAdV7, HAdV21 and HAdV4 have been reported to cause outbreaks of respiratory infection in close-quartered populations including civilian communities, hospitals, schools, and military who mostly recovered with no residual sequelae [8,30,31]. HAdV7 is of particular concern as it is often with more severe or higher levels of morbidity in the related respiratory diseases than other HAdV serotypes [23]. In recent 5 years, relevant reports of the outbreaks of HAdV7 associated ARD that affect large population trend to be increased [11-13]. In mainland China, epidemic outbreaks of HAdV7 that affected numerous of military recruits, populations in schools or hospitals consecutively occurred in Chongqing (2010) [23], Guangzhou (2011) [23], Shijiazhuang (2011) [32], Beijing (2012-2013) [33], Shaanxi (2012) [13], Hangzhou (2014) (In this study). In this outbreak, the genotype of HAdV isolates was identified as HAdV7 through phylogenetic analysis, and each hexon gene of the isolates were identical, these isolates also had high identity (100%) with the HAdV SXWN1203 strain KC689913.1 [13] and the HAdV CHN/DG01/2011/7 [P7H7F7] strain KC440171 (Figure 2) [23]. Homologous analysis also performed on the sequence data set of global HAdV7 hexon genes, these results indicated not only the very low mutation frequency of HAdV7 isolates during the epidemic, but also the relative low mutation rate in the evolutionary history.
It has been known that viral infection has close relationships with the body immune competence [34]. Once the virus invades the body, human adaptive immunity is activated, mainly through 2 pathways including the re-infection and vaccination. During the re-infection of human adenovirus, it has been reported that most serotypes of the human adenovirus cause multi-system infections including respiratory, gastrointestinal, and central nervous system infections that are commonly mild or recessive infections. These are consistent with the neutralizing antibody responses to HAdV. Neutralizing antibodies that are produced after viral infection indicate the nature of persistent protective immunity in humans to HAdV infection [34]. As described, HAdV7 vaccines played a significant role in defense and decrease of adenovirus infection after the first application in the USA military recruits in 1971 and the reuse of HAdV7 vaccines in late 2011 [35,36]. According to the Naval Health Research Center (NHRC) ongoing adenovirus surveillance, the resumption of the vaccines in recruits made adenovirus rate plummeting from 5.8 to 0.02 cases per 1000 person-weeks in 2012-2013 [36]. This finding confirmed the effectiveness of the vaccines in prevention of HAdV7 infection. The relatively low mutation rates of HAdV7 were proven in the evolutionary history. To date, the vaccines have not been deployed in China. It has been one of the most causes that lead to high frequent outbreaks of HAdV7 in China. Thus the urgency of vaccination in vulnerable groups including students and military personnel in China should be considered.
By now, there have been no relevant reports or experimental data from the perspective of either the virus’s victims or the virus alone inferring the transmission patterns of HAdV7 in the epidemic all over the world. And this is the first time temporal and spatial transmission dynamics of HAdV7 across the world has been investigated by phylogeographic analysis. MigraPhyla results showed that among 56 observed HAdV7 migration routes, 22 migration routes were statistically significant (P<0.05) with sFDR supported. HAdV7 in this outbreak was statistically proven dispersed from Shaanxi province (2011), and this migration route belonged to the branch of initial source California (1953). As all these large population outbreaks of HAdV7 have been concentrated in the economically developed cities and the surrounding regions, accelerated integration of the global economy and developed traffic may act as an epidemiological attractant of HAdV7 [37]. In this study, too short HAdV7 hexon gene sequences or unclear HAdV7 isolates in some epidemics were exclude to the migration analysis of HAdV7 globally, the possibility of other transmission routes are needed for further research and confirmation.
Conclusions
Our study reported a large scale HAdV7 outbreak in Chinese military and reviewed the high frequency of HAdV7 infections in Asia (especially in China) in the last 5years. Phylogenetic and phylogeographic analyses of HAdV7 globally play an important role in learning the human adenovirus world and create a foundation towards exploitation of the more effective HAdV7 molecular diagnosis reagents and the relevant vaccines, also offered valuable accordance for HAdV7 prevention and control strategy. By recognizing the benefit for the application of the effective vaccinesin preventing the transmission of HAdV in USA, the development of AdVvaccines and vaccination application in the most vulnerable groups including students, recruits and community population in Asia (especially in China) should be emphasized.
Acknowledgements
This work was supported in part by a grant from the Medical Science Foundation of Nanjing Military Command (NO. 12MA117) and the Natural Science Foundation of Zhejiang Province (NO. Y15H200001). We are grateful to Pengfei Li at Beijing Genomics Institute in Hangzhou, Chinafor his collaboration. We also thank Prof. Jicheng Li (Department of Anatomy and Cell Biology, Institute of Basic Medicine, Zhejiang University, Hangzhou, China) for her editorial assistance and revision of the paper.
Disclosure of conflict of interest
None.
References
- 1.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 2.Han G, Niu H, Zhao S, Zhu B, Wang C, Liu Y, Zhang M, Yang S, Liu F, Wan C, Zhang Q. Identification and typing of respiratory adenoviruses in Guangzhou, Southern China using a rapid and simple method. Virol Sin. 2013;28:103–108. doi: 10.1007/s12250-013-3308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones MS 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushima Y, Shimizu H, Kano A, Nakajima E, Ishimaru Y, Dey SK, Watanabe Y, Adachi F, Suzuki K, Mitani K, Fujimoto T, Phan TG, Ushijima H. Novel human adenovirus strain, Bangladesh. Emerg Infect Dis. 2012;18:846–848. doi: 10.3201/eid1805.111584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon HJ, Rhie YJ, Seo WH, Jang GY, Choi BM, Lee JH, Lee CK, Kim YK. Clinical manifestations of respiratory adenoviral infection among hospitalized children in Korea. Pediatr Int. 2013;55:450–454. doi: 10.1111/ped.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Kong M, Su X, Zou M, Guo L, Dong X, Li L, Gu Q. An outbreak of acute respiratory disease in China caused by human adenovirus type B55 in a physical training facility. Int J Infect Dis. 2014;28:117–122. doi: 10.1016/j.ijid.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrix RM, Lindner JL, Benton FR, Monteith SC, Tuchscherer MA, Gray GC, Gaydos JC. Large, persistent epidemic of adenovirus type 4-associated acute respiratory disease in U. S. army trainees. Emerg Infect Dis. 1999;5:798–801. doi: 10.3201/eid0506.990609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajon AE, Hang J, Hawksworth A, Metzgar D, Hage E, Hansen CJ, Kuschner RA, Blair P, Russell KL, Jarman RG. Molecular Epidemiology of Adenovirus Type 21 Respiratory Strains Isolated From US Military Trainees (1996-2014) J Infect Dis. 2015;212:871–880. doi: 10.1093/infdis/jiv141. [DOI] [PubMed] [Google Scholar]
- 9.Lai CY, Lee CJ, Lu CY, Lee PI, Shao PL, Wu ET, Wang CC, Tan BF, Chang HY, Hsia SH, Lin JJ, Chang LY, Huang YC, Huang LM Taiwan Pediatric Infectious Disease Alliance. Adenovirus serotype 3 and 7 infection with acute respiratory failure in children in Taiwan, 2010-2011. PLoS One. 2013;8:e53614. doi: 10.1371/journal.pone.0053614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Veen J, Oei KG, Abarbanel MF. Patterns of infections with adenovirus types 4, 7 and 21 in military recruits during a 9-year survey. J Hyg (Lond) 1969;67:255–268. doi: 10.1017/s0022172400041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yusof MA, Rashid TR, Thayan R, Othman KA, Hasan NA, Adnan N, Saat Z. Human adenovirus type 7 outbreak in Police Training Center, Malaysia, 2011. Emerg Infect Dis. 2012;18:852–854. doi: 10.3201/eid1805.110865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsou TP, Tan BF, Chang HY, Chen WC, Huang YP, Lai CY, Chao YN, Wei SH, Hung MN, Hsu LC, Lu CY, Shao PL, Mu JJ, Chang LY, Liu MT, Huang LM. Community outbreak of adenovirus, Taiwan, 2011. Emerg Infect Dis. 2012;18:1825–1832. doi: 10.3201/eid1811.120629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu P, Ma C, Nawaz M, Han L, Zhang J, Du Q, Zhang L, Feng Q, Wang J, Xu J. Outbreak of acute respiratory disease caused by human adenovirus type 7 in a military training camp in Shaanxi, China. Microbiol Immunol. 2013;57:553–560. doi: 10.1111/1348-0421.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace RG, Fitch WM. Influenza A H5N1 immigration is filtered out at some international borders. PLoS One. 2008;3:e1697. doi: 10.1371/journal.pone.0001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito M, Oshitani H, Orbina JR, Tohma K, de Guzman AS, Kamigaki T, Demetria CS, Manalo DL, Miranda ME, Noguchi A, Inoue S, Quiambao BP. Genetic diversity and geographic distribution of genetically distinct rabies viruses in the Philippines. PLoS Negl Trop Dis. 2013;7:e2144. doi: 10.1371/journal.pntd.0002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reischl U, Lehn N, Simnacher U, Marre R, Essig A. Rapid and standardized detection of Chlamydia pneumoniae using LightCycler real-time fluorescence PCR. Eur J Clin Microbiol Infect Dis. 2003;22:54–57. doi: 10.1007/s10096-002-0858-2. [DOI] [PubMed] [Google Scholar]
- 17.Ursi D, Dirven K, Loens K, Ieven M, Goossens H. Detection of Mycoplasma pneumoniae in respiratory samples by real-time PCR using an inhibition control. J Microbiol Methods. 2003;55:149–153. doi: 10.1016/s0167-7012(03)00131-3. [DOI] [PubMed] [Google Scholar]
- 18.Diederen BM, de Jong CM, Marmouk F, Kluytmans JA, Peeters MF, Van der Zee A. Evaluation of real-time PCR for the early detection of Legionella pneumophila DNA in serum samples. J Med Microbiol. 2007;56:94–101. doi: 10.1099/jmm.0.46714-0. [DOI] [PubMed] [Google Scholar]
- 19.Huang G, Yu D, Zhu Z, Zhao H, Wang P, Gray GC, Meng L, Xu W. Outbreak of febrile respiratory illness associated with human adenovirus type 14p1 in Gansu Province, China. Influenza Other Respir Viruses. 2013;7:1048–1054. doi: 10.1111/irv.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tippmann HF. Analysis for free: comparing programs for sequence analysis. Brief Bioinform. 2004;5:82–87. doi: 10.1093/bib/5.1.82. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci U S A. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao S, Wan C, Ke C, Seto J, Dehghan S, Zou L, Zhou J, Cheng Z, Jing S, Zeng Z, Zhang J, Wan X, Wu X, Zhao W, Zhu L, Seto D, Zhang Q. Re-emergent human adenovirus genome type 7d caused an acute respiratory disease outbreak in Southern China after a twenty-one year absence. Sci Rep. 2014;4:7365. doi: 10.1038/srep07365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diseases CMCfI. Diagnosis and treatment guideline for adenovirus infection (in Chinese) Jie Fang Jun Yi Xue Za Zhi. 2013;38:529–534. [Google Scholar]
- 25.Cui X, Wen L, Wu Z, Liu N, Yang C, Liu W, Ba Z, Wang J, Yi S, Li H, Liang B, Li P, Jia L, Hao R, Wang L, Hua Y, Wang Y, Qiu S, Song H. Human adenovirus type 7 infection associated with severe and fatal acute lower respiratory illness and nosocomial transmission. J Clin Microbiol. 2015;53:746–749. doi: 10.1128/JCM.02517-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q, Jiang W, Chen Z, Huang L, Wang Y, Huang F, Ji W, Zhang X, Shao X, Yan Y. Epidemiology and clinical features of respiratory adenoviral infections in children. Eur J Pediatr. 2014;173:441–444. doi: 10.1007/s00431-013-2188-z. [DOI] [PubMed] [Google Scholar]
- 27.Julkunen I, Lehtomaki K, Hovi T. Immunoglobulin class-specific serological responses to adenovirus in respiratory infections of young adult men. J Clin Microbiol. 1986;24:112–115. doi: 10.1128/jcm.24.1.112-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007;35:e40. doi: 10.1093/nar/gkm051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol. 2004;42:3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong JY, Lee HJ, Piedra PA, Choi EH, Park KH, Koh YY, Kim WS. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis. 2001;32:1423–1429. doi: 10.1086/320146. [DOI] [PubMed] [Google Scholar]
- 31.Lin KH, Lin YC, Chen HL, Ke GM, Chiang CJ, Hwang KP, Chu PY, Lin JH, Liu DP, Chen HY. A two decade survey of respiratory adenovirus in Taiwan: the reemergence of adenovirus types 7 and 4. J Med Virol. 2004;73:274–279. doi: 10.1002/jmv.20087. [DOI] [PubMed] [Google Scholar]
- 32.Guo JH, Zhang SY, Tian HF, Xu M, Zhou JK. Survey on pharyngoconjunctival fever outbreak caused by type 7 adenovirus (in Chinese) Huan Jing Wei Sheng Za Zhi. 2015;5:369–372. [Google Scholar]
- 33.Qiu S, Li P, Liu H, Wang Y, Liu N, Li C, Li S, Li M, Jiang Z, Sun H, Li Y, Xie J, Yang C, Wang J, Li H, Yi S, Wu Z, Jia L, Wang L, Hao R, Sun Y, Huang L, Ma H, Yuan Z, Song H. Whole-genome Sequencing for Tracing the Transmission Link between Two ARD Outbreaks Caused by a Novel HAdV Serotype 7 Variant, China. Sci Rep. 2015;5:13617. doi: 10.1038/srep13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Xing M, Zhang C, Yang Y, Chi Y, Tang X, Zhang H, Xiong S, Yu L, Zhou D. Neutralizing antibody responses to enterovirus and adenovirus in healthy adults in China. Emerg Microbes Infect. 2014;3:e30. doi: 10.1038/emi.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blasiole DA, Metzgar D, Daum LT, Ryan MA, Wu J, Wills C, Le CT, Freed NE, Hansen CJ, Gray GC, Russell KL. Molecular analysis of adenovirus isolates from vaccinated and unvaccinated young adults. J Clin Microbiol. 2004;42:1686–1693. doi: 10.1128/JCM.42.4.1686-1693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radin JM, Hawksworth AW, Blair PJ, Faix DJ, Raman R, Russell KL, Gray GC. Dramatic decline of respiratory illness among US military recruits after the renewed use of adenovirus vaccines. Clin Infect Dis. 2014;59:962–968. doi: 10.1093/cid/ciu507. [DOI] [PubMed] [Google Scholar]
- 37.Kim JS, Kim JJ, Kim SJ, Jeon SE, Seo KY, Choi JK, Kim NO, Hong S, Chung GT, Yoo CK, Kim YT, Cheun HI, Bae GR, Yeo YH, Ha GJ, Choi MS, Kang SJ, Kim J. Outbreak of Ciprofloxacin-Resistant Shigella sonnei Associated with Travel to Vietnam, Republic of Korea. Emerg Infect Dis. 2015;21:1247–1250. doi: 10.3201/eid2107.150363. [DOI] [PMC free article] [PubMed] [Google Scholar]


