Abstract
Reverse transcription quantitative PCR (RT-qPCR) as the accurate and sensitive method is use for gene expression analysis, but the veracity and reliability result depends on whether select appropriate reference gene or not. To date, several reliable reference gene validations have been reported in fruits trees, but none have been done on preharvest and postharvest longan fruits. In this study, 12 candidate reference genes, namely, CYP, RPL, GAPDH, TUA, TUB, Fe-SOD, Mn-SOD, Cu/Zn-SOD, 18SrRNA, Actin, Histone H3, and EF-1a, were selected. Expression stability of these genes in 150 longan samples was evaluated and analyzed using geNorm and NormFinder algorithms. Preharvest samples consisted of seven experimental sets, including different developmental stages, organs, hormone stimuli (NAA, 2,4-D, and ethephon) and abiotic stresses (bagging and girdling with defoliation). Postharvest samples consisted of different temperature treatments (4 and 22°C) and varieties. Our findings indicate that appropriate reference gene(s) should be picked for each experimental condition. Our data further showed that the commonly used reference gene Actin does not exhibit stable expression across experimental conditions in longan. Expression levels of the DlACO gene, which is a key gene involved in regulating fruit abscission under girdling with defoliation treatment, was evaluated to validate our findings. In conclusion, our data provide a useful framework for choice of suitable reference genes across different experimental conditions for RT-qPCR analysis of preharvest and postharvest longan fruits.
Keywords: Longan (Dimocarpus longan Lour.), preharvest and postharvest fruits, gene expression, reference genes, RT-qPCR, normalization, ACO gene
Introduction
Reverse transcription quantitative PCR (RT-qPCR) as the sensitive and accurate method for gene expression analysis is employed in a wide range of applications (Bustin, 2002). However, the accuracy of RT-qPCR is affected by many factors, including differences in RNA sample quality and extraction method, and efficiencies of reverse transcription and PCR (Die et al., 2010; Schmidt and Delaney, 2010). Use of appropriate reference genes could help eliminate the variability introduced by differences across samples and ensure the veracity and reliability of RT-qPCR results.
A suitable reference gene should possess invariant expression across all samples and under different experimental conditions. Housekeeping genes are often selected as ideal reference genes because they are thought to exhibit such properties. Nevertheless, a few recent reports have indicated that even the widely used reference genes are not invariant under different experimental conditions (Nicot et al., 2005; Marino et al., 2008). GAPDH and Actin are reference genes that exhibit different expression levels in different plants, tissues and experimental conditions (Yperman et al., 2004; Barber et al., 2005). Gutierrez et al. (2008) also found that the gene expression of TUB, Actin, UBQ, and EF were varies across all samples. If the reference gene used for normalization in RT-qPCR was variant, it will led to inaccuracy conclusion (Czechowski et al., 2005; Gutierrez et al., 2008; Artico et al., 2010). Hence, reference genes must be systematically evaluated to ensure their stability of expression in each experimental system to obtain accurate and reliable results (Lee et al., 2002; Hong et al., 2008). Moreover, the optimal number of reference genes should be determined, as most gene expression analyses are expected to require multiple reference genes (Vandesompele et al., 2002).
To date, several reliable reference gene validations have been reported in fruits such as litchi (Zhong et al., 2011), banana (Chen et al., 2011), papaya (Zhu et al., 2012), apple (Tatiane et al., 2015), citrus (Mafra et al., 2012), berry (Reid et al., 2006), peach (Tong et al., 2009), pear (Wu et al., 2012; Xu et al., 2015), plum (Kim et al., 2015), melon (Carmen et al., 2015), watermelon (Kong et al., 2014), strawberry (Clancy et al., 2013), olive (Tjasa et al., 2013), and grapevine (Mauricio et al., 2013). Such reference gene validations have also been reported for other species such as Lilium regale (Liu et al., 2016), celery (Li et al., 2016), Lycoris aurea (Ma et al., 2016), tall fescue (Yang et al., 2015), moss (Li et al., 2015a), carrot leaves (Tian et al., 2015), Oxytropis ochrocephala Bunge (Zhuang et al., 2015), cotton (Tu et al., 2007; Artico et al., 2010), tobacco (Schmidt and Delaney, 2010), Petunia (Mallona et al., 2010), Arabidopsis (Czechowski et al., 2005), potato (Nicot et al., 2005), Lolium temulentum (Dombrowski and Martin, 2009), rice (Jain et al., 2006), zucchini (Obrero et al., 2011), and cucumber (Wan et al., 2010). However, there was still no report about selecting appropriate reference genes for RT-qPCR analysis in preharvest and postharvest longan fruits. Although Lin and Lai (2010, 2013) identified reference genes for longan, their results are only applicable for somatic embryogenesis of longan.
Longan (Dimocarpus longan Lour.), which belongs to the family of Sapindaceae, is an important tropical/subtropical fruit species. The main countries producing this fruit include China, Thailand and Vietnam. Several areas in India, Laos, Philippines, Myanmar, Malaysia and Indonesia also cultivate longan (Shi et al., 2015). Longan fruits are preferably eaten fresh, and they have a delicate and sweet-tasting flesh. This fruit can also be processed to make dried pulp, canned fruit, jam, drinks and wine. In China, longan fruit is mostly marketed locally but has been increasingly imported in recent years (Jiang et al., 2002).
Several preharvest and postharvest problems significantly reduce the yield and commercial value of longan (Jiang et al., 2002). These include fruit abscission during growth and development in preharvest, and fruit deterioration and pericarp browning, reducing the storage potential and marketability of longan, in postharvest periods. The cause of these problems may be at the physicochemical, biochemical and molecular levels (Lai et al., 2001; Jiang et al., 2002). RT-qPCR is a useful method to find the key genes relate to that processes, but there was still no report about selecting appropriate reference genes for RT-qPCR analysis in longan fruits. Although a few gene expression patterns have been reported in longan fruit using different references genes such as Actin (Zhao et al., 2011; Chen et al., 2012; You et al., 2014; Shuai et al., 2016), EF-1a (Shuai et al., 2016), and GAPDH (Shuai et al., 2015), stability of these reference genes has not been verified to date. Therefore, identification of appropriate references genes for RT-qPCR analyses in longan fruit is urgently needed.
In the present study, 12 reference genes, including cyclophilin (CYP), ribosomal protein L (RPL), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), alpha-tubulin (TUA), beta-tubulin (TUB), copper/zinc-superoxide dismutase (Cu/Zn-SOD), manganese superoxide dismutase (Mn-SOD), iron-superoxide dismutase (Fe-SOD), 18S ribosomal RNA (18SrRNA), Actin, Histone H3 and elongation factor 1-alpha (EF-1a) were selected based on evidence of stable expression in previous studies (Lin and Lai, 2010; Chen et al., 2011; Zhong et al., 2011; Zhu et al., 2012).
geNorm and NormFinder were useful programs to identify appropriate reference genes for RT-qPCR analysis, especially using together (Chen et al., 2011; Zhong et al., 2011). Base on the previous reports, this study selected the two programs to rank the stability of the candidate reference genes. Finally, identifying the suitable reference genes for RT-qPCR normalization in preharvest and postharvest longan fruits. The preharvest experiment consisted of seven sample sets, including different developmental stages, organs, hormone stimuli (NAA, 2,4-D, and ethephon) and abiotic stresses (bagging and girdling with defoliation). The postharvest experiment consisted of different temperature treatments (4 and 22°C) and longan varieties.
In addition, for verifying our results, the expression levels of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene under girdling with defoliation treatment was normalized to the most stable and unstable genes. Our findings provide important guidelines for selecting suitable reference genes under different experimental conditions in preharvest and postharvest longan fruits.
Materials and methods
Plant materials and treatments
Longan trees were selected in the South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China (2013). There were a total of 150 samples in this study. Pericarp and aril samples of six varieties: “Shixia,” “Chuliang,” “Linglong,” “Gushan,” “Caopushi,” and “Benzhan” were obtained at 110 days after anthesis (DAA). Pericarp and aril samples at six different developmental stages of “Shixia” (62, 76, 83, 89, 103, and 110 DAA) were collected. The tender root, shoot, young leaf, flower, fruitlets, and seed of “Shixia” were also sampled. The fruitlets and seeds were sampled at 40 and 110 DAA, respectively.
Three 10-year-old “Shixia” trees (2013) were used for treatment, and each tree was used as a biological replicate. The treatment was carried out at 90 DAA. Fifty similar size of the fruit-bearing shoots from each tree were tagged. 10 shoots were treated with 100 mg/L NAA. Meanwhile, 10 shoots were sprayed with 20 and 250 mg/L 2,4-D and ethephon, respectively, 10 shoots were bagged with adhesive-bonded fabric bags and the remaining 10 shoots were control that treated with distilled water. 10 fruits randomly collected from 10 shoots at 0, 2, and 3 d after treatment, and the pericarp was collected as sample.
For temperature samples, the fresh postharvest longan fruits (110 DAA in 2013) were put into unsealed plastic bags and then treated with 4°C (cold temperature) and 22°C (room temperature), respectively. Aril was collected at 2 and 3 d after treatment.
Three 10-year-old “Shixia” trees were selected for girdling plus defoliation treatment, and each tree was a biological replicate. The treatment was performed at 40 DAA. Twenty fruit-bearing shoots were tagged with a uniform diameter from each tree. 10 shoots were girdled at a width of 0.5 cm and defoliated all leaves above the girdle, and the remaining untreated 10 shoots were used as control. The fruitlets and fruit abscission zones of 10 fruits randomly harvested from 10 fruit-bearing shoots were sampled at 4 and 5 d after treatment. Fruit abscission zone samples were collected by cutting ~1 mm piece at each side of the abscission fracture plane.
Table 1 summarizes the data of all sample sets mentioned above. All tissues were immediately frozen in liquid nitrogen after separation and stored at −80°C for future use.
Table 1.
Sample sets considered in the present study.
| Experimental sample sets | Tissue type | Sampling dates | Cultivars | Biological replicates | Total number of samples |
|---|---|---|---|---|---|
| Different varieties | Pericarp, aril | 110 DAA | “Shixia,” “Chuliang,” “Linglong,” “Gushan,” “Caopushi,” and “Benzhan” | 3 | 36 |
| Different developmental stages | Pericarp, aril | 62, 76, 83, 89, 103, 110 DAA | “Shixia” | 3 | 36 |
| Different organs | Root, shoot, leaf, flower, fruitlets seed | 40, 110 DAA | “Shixia” | 3 | 18 |
| NAA | Pericarp | 0, 2, 3 DAT | “Shixia” | 3 | 9 |
| 2,4-D | Pericarp | 0, 2, 3 DAT | “Shixia” | 3 | 9 |
| Ethephon | Pericarp | 0, 2, 3 DAT | “Shixia” | 3 | 9 |
| Bagging | Pericarp | 0, 2, 3 DAT | “Shixia” | 3 | 9 |
| Temperature (4°C, 22°C) | Aril | 2, 3 DAT | “Shixia” | 3 | 12 |
| Girdling with defoliation | Fruitlets, fruit abscission zone | 4, 5 DAT | “Shixia” | 3 | 12 |
DAA, days after anthesis; DAT, days after treatment. Total number of samples: 150.
RNA isolation, quality control, and cDNA synthesis
According to the protocol of Wu et al. (2011), total RNA was extracted from the frozen samples. DNase I (TaKaRa, Otsu, Japan) and RNAse-free columns (Huayueyang, Beijing, China) were used to remove potentially contaminating DNA and purify total RNA, respectively. RNA integrity was confirmed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). For reverse transcription, 2 μg of total RNA and oligo-dT primers were used with the M-MLV cDNA synthesis kit following the manufacturer's instructions (Promega).
Selection of candidate reference genes for preharvest and postharvest longan fruits
Based on previous studies, 12 reference genes were selected to compare the stability of their expression across samples. CYP and RPL were selected from our transcriptome database (Zhang et al., 2016; http://www.ncbi.nlm.nih.gov/sra/SRX1395356/). GAPDH (GeneBank number FJ694012), TUA (GeneBank number FJ479617), TUB (GeneBank number KC921220), Fe-SOD (GeneBank number EU330204), Mn-SOD (GeneBank number EU563945), and Cu/Zn-SOD (GeneBank number EU157910) were obtained from NCBI. 18SrRNA, Actin, Histone H3 and EF-1a were picked based on the report by Lin and Lai (2010).
Design and validation of RT-qPCR primers
Primer pairs of CYP, RPL, GAPDH, TUA, TUB, Fe-SOD, Mn-SOD, and Cu/Zn-SOD were designed using Primer Premier 5.0 software, while primer pairs of 18SrRNA, Actin, Histone H3, and EF-1a were selected based on the report by Lin and Lai (2010). All primers were obtained from a commercial supplier (Sangon, Guangzhou, China). Prior to RT-qPCR, each primer pair was tested via melting curve, the good melting curve should meet the requirement that there was a single peak in the template and no amplicon was observed in no template control (NTC). The size specificity of the amplicon were confirmed by using 2.0% agarose gel electrophoresis. In orded to verify the cDNA sequence of the PCR products the target amplicons were sequenced. Standard curve was used to calculate the PCR efficiency for each gene. Table 2 shows the selected candidate reference genes, primers, and amplicon characteristics.
Table 2.
Selected candidate reference genes, primers, and amplicon characteristics.
| Gene | Primer sequences (forward/reverse) | Amplicon length (bp) | Tm (°C) | Amplification efficiency |
|---|---|---|---|---|
| CYP | AAGGAGATGGAGAAGGTTGGTT | 213 | 83.37 | 2.072 |
| CCCTTTCTGACTTTGGGTAGACATA | ||||
| RPL | TCAGACAGTGATTACACCGAGTTC | 246 | 80.87 | 1.914 |
| GCCAGTAGAGACAAAAAGGCAAGA | ||||
| GAPDH | GTCGCTTGGCTGCCTATAATCT | 204 | 81.79 | 1.962 |
| AGACCGTTGACTGAACCATCC | ||||
| TUA | GATTATGAGGAAGTTGGGGCTG | 175 | 81.09 | 1.986 |
| AAGCAACCTAGCACATAGTGAAAGT | ||||
| TUB | AGATGTTCCGTCGTGTGAGTGA | 86 | 83.43 | 1.977 |
| CCTCCTCATATTCATCCTCATCAG | ||||
| Fe-SOD | AAGAGGAGAAAGAGCAAGAGTCAGA | 114 | 80.80 | 1.917 |
| CCGATACAACAAACCCTGAAATG | ||||
| Mn-SOD | ACTACCTACAGTACAAGAATGTCAGACC | 184 | 81.44 | 1.949 |
| GGGCTCCTCATCCTATATCGTT | ||||
| Cu/Zn-SOD | TTGTAGGAAGGGCTGTCGTTG | 207 | 82.46 | 1.992 |
| TCTCGTCAAGTCACTCTCAAGCAT | ||||
| 18SrRNA | CCTGAGAAACGGCTACCACAT | 171 | 83.93 | 1.931 |
| CACCAGACTTGCCCTCCA | ||||
| Actin | TGCTATCCTTCGGTTGGACC | 93 | 81.77 | 1.984 |
| CGGACGATTTCCCGTTCAG | ||||
| Histone H3 | ATCCGCAAGTACCAGAAGAGCA | 155 | 85.71 | 2.002 |
| CCCACCAAGTAAGCCTCAG | ||||
| EF-1a | GATGATTCCCACCAAGCCCAT | 129 | 84.57 | 2.072 |
| GGGTCCTTCT TCTCAACACTCT |
RT-qPCR conditions
RT-qPCR was conducted on a LightCycler480 II System (Roche, Switzerland) using DyNAmo Flash SYBR Green q-PCR kit (Thermo, USA). Each reaction mix contained 1 and 5 μL of diluted cDNAs and SYBR Green PCR Master Mix (Thermo, USA), respectively, and 250 nM of each primer to a final volume of 10 μL. The amplification conditions were as follows: 50°C for 2 min; 95°C for 10 min; 40 cycles at 95°C for 15 s; 55°C for 30 s; and 72°C for 30 s in 384-well optical reaction plates (Roche, Switzerland). The dissociation curve was analyzed at 60–95°C over 40 cycles to determine primer specificity. Each assay included three technical and biological replicates.
Data analysis
The stability of the 12 selected reference genes were ranked by geNorm and NormFinder algorithms, and the gene expression levels were determined using crossing point (Cp) values. The raw Cp values should transform to relative quantities called the Q values, when the data input into the two algorithms. The Q values were calculated using the equation: Q = 2(minCp−sampleCp). After the geNorm algorithm was applied, the stability value (M) for each gene was generated, and pairwise variation (V) between genes was obtained. The M and V values indicate the most appropriate reference genes and the optimal number of reference genes, respectively (Vandesompele et al., 2002). The NormFinder algorithm calculates the stability for each gene, with lower stability value indicating more stable reference genes (Andersen et al., 2004).
Normalization of DlACO
The ACO gene converting ACC into ethylene. ACO gene expression is positively correlated with ethylene production rates and upregulated during fruit abscission (Li and Yuan, 2008a,b; Li et al., 2015b). One DlACO gene (GeneBank number GU123929.1) was used to verify the results of reference gene stability analysis in RT-qPCR. Gene expression level of DlACO in the fruit abscission zone was quantified during girdling with defoliation using one or two most suitable reference genes and the most unsuitable gene that determined by geNorm. The primer pairs (forward: 5′-CAACTTGAGGTTATCACAAATGGG -3′ and reverse: 5′-ACAATGCTGGAGCTGGGTAGA -3′) for DlACO were also confirmed with the same methods described above.
Results
Verification of amplicons, primer specificity, and PCR amplification efficiency
The agarose gel electrophoresis showed there were only one amplicon with the designed size after PCR amplified in all candidate reference genes, and the same results were observed in melting curve. The melting curve showed a single peak in the template and no amplicon was observed in NTC for each selected reference gene (Figure S1). Furthermore, sequence analysis showed that all of the sequenced amplified fragments were matched to the sequences used for primer design. The PCR efficiency for the 12 reference genes between 1.914 (RPL) and 2.072 (CYP; Table 2).
Cq values of candidate reference genes in preharvest and postharvest longan fruits
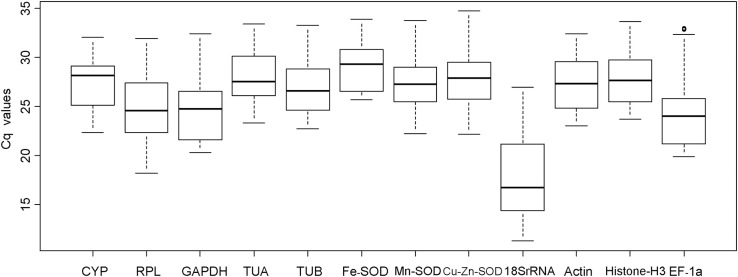
All the Cq values of the reference genes are shown in Table S1. The Cq values of the 12 reference genes were variable (Figure 1), ranging between 11.25 (18SrRNA) and 34.72 (Cu/Zn-SOD), while the mean Cq values ranged between 17.43 (18SrRNA) and 29.02 (Fe-SOD; Table S2). Moreover, 18SrRNA levels were the most variable (15.73 Cq, the maximun and minimum values were 26.98 and 11.25, respectively) while Fe-SOD was the least variable (8.21 Cq, the maximun and minimum values were 33.88 and 25.67, respectively). Since Cq values are negatively correlated to expression levels, our data indicate that 18SrRNA had high expression while Fe-SOD had low expression (Table S2).
Figure 1.
Cq values of candidate reference genes in all longan samples. Lines across boxes indicate median values. Boxes indicate 25/75 percentiles. Whisker caps indicate the minimum and maximum values. Circles represent outliers.
Determination of the optimal number of reference genes in preharvest and postharvest longan fruits
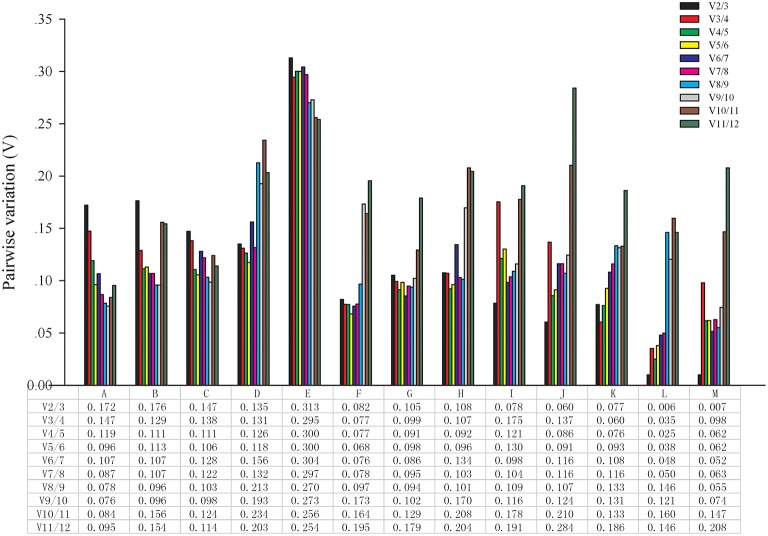
The optimal number of reference genes was determined using geNorm. The software can calculate the pairwise variation (V) values of normalization factor by introducing new reference genes, and the most appropriate number is determined according to the ratio Vn/Vn+1. As shown in Figure 2, two reference genes were sufficient for normalizing gene expression at different developmental stages, NAA, 2,4-D, ethephon, bagging, temperature (4 and 22°C) and girdling with defoliation treatments because the V2/3 value was lower than 0.15. Three reference genes were sufficient for RT-qPCR analysis in different varieties because the V2/3 value (0.172 and 0.176) was higher than 0.15, and the V3/4 value (0.147 and 0.129) was lower than 0.15. However, none of the genes selected was appropriate for different organ samples because all Vn/n +1 values were higher than 0.15.
Figure 2.
Determination of the optimal number of reference genes. Pairwise variation (Vn/n +1) analysis between the normalization factors (NFn and NFn +1) was performed using the geNorm program in all samples. A, different varieties (pericarp); B, different varieties (aril); C, different developmental stages (pericarp); D, different developmental stages (aril); E, different organs; F, NAA stimuli; G, 2,4-D stimuli; H, ethephon stimuli; I, bagging; J, cold temperature treatment (4°C); K, room temperature treatment (22°C); L, girdling with defoliation (fruit abscission zone); M, girdling with defoliation (fruitlets).
Expression stability of candidate reference genes in preharvest and postharvest longan fruits
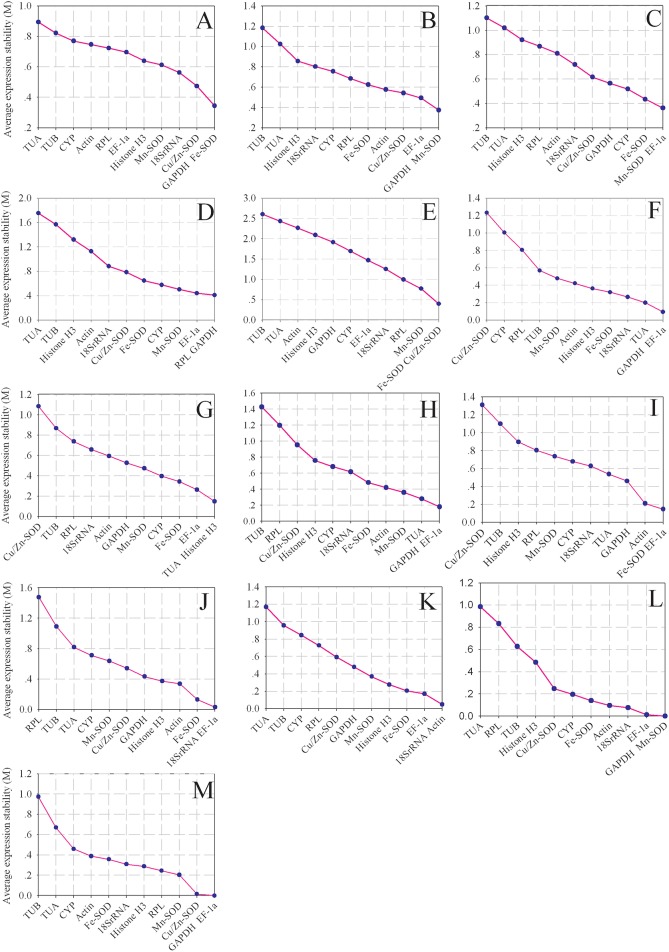
The results were shown in Figure 3 and summarized in Table S3 by geNorm algorithm. The GAPDH and Fe-SOD genes were the most suitable candidates for normalization in the pericarp of longan fruit in different varieties with an M value of 0.343 (Figure 3A), whereas GAPDH and Mn-SOD genes were the most suitable genes in the aril of longan fruit in different varieties with an M value of 0.374 (Figure 3B). For the pericarp or aril of longan fruit samples at different developmental stages, the most stable genes were Mn-SOD and EF-1a, and GAPDH and RPL with M values of 0.362 and 0.409, respectively (Figures 3C,D). Fe-SOD and Cu/Zn-SOD were the most stable genes across different organ samples with an M value of 0.398 (Figure 3E). The GAPDH and EF-1a genes ranked high in the pericarp of longan fruit under NAA and ethephon conditions, with M values of 0.093 and 0.179, respectively (Figures 3F,H). TUA and Histone H3 genes were the most stable genes in the pericarp of longan fruit under 2,4-D conditions with an M value of 0.147 (Figure 3G). Fe-SOD and EF-1a were the most appropriate genes with an M value of 0.145 in the bagging condition (Figure 3I). For temperature treatments at 4 and 22°C, the gene pairs with the highest expression stability were 18SrRNA and EF-1a, and 18SrRNA and Actin with M values of 0.031 and 0.050, respectively (Figures 3J,K). For girdling with defoliation treatment, the suitable genes were GAPDH and Mn-SOD genes with an M value of 0.000 in the fruit abscission zone, and GAPDH and EF-1a genes with an M value of 0.000 in fruitlets (Figures 3L,M). These results indicate that different reference genes should be selected depending on experimental conditions.
Figure 3.
Average expression stability values (M) of candidate reference genes. The average M values of the reference genes were measured by stepwise exclusion of the least stable reference genes. A lower M value indicates more stable expression, as analyzed by the geNorm software in longan sample sets under different experimental conditions. (A) different varieties (pericarp), (B) different varieties (aril), (C) different developmental stages (pericarp), (D) different developmental stages (aril), (E) different organs, (F) NAA stimuli, (G) 2,4-D stimuli, (H) ethephon stimuli, (I) bagging, (J) cold temperature treatment (4°C), (K) room temperature treatment (22°C), (L) girdling with defoliation (fruit abscission zone), (M) girdling with defoliation (fruitlets).
According to the optimal number of reference genes in longan fruit obtained from geNorm analysis, the most suitable combinations for 12 sample sets were GAPDH + Fe-SOD + Cu/Zn-SOD (pericarp of different varieties), GAPDH + Mn-SOD + EF-1a (aril of different varieties), Mn-SOD + EF-1a (pericarp of different developmental stages), GAPDH + RPL (aril of different developmental stages), GAPDH + EF-1a (pericarp of NAA and ethephon treatment), TUA + Histone H3 (pericarp of 2,4-D treatment), Fe-SOD + EF-1a (pericarp of bagging treatment), 18SrRNA + EF-1a (4°C temperature treatment), 18SrRNA + Actin (22°C temperature treatment), GAPDH + Mn-SOD (fruit abscission zone of girdling with defoliation treatment), and GAPDH + EF-1a (fruitlets of girdling with defoliation treatment). However, none of the possible gene combinations was deemed suitable for the different organ samples since all of the Vn/n +1 values were higher than 0.15.
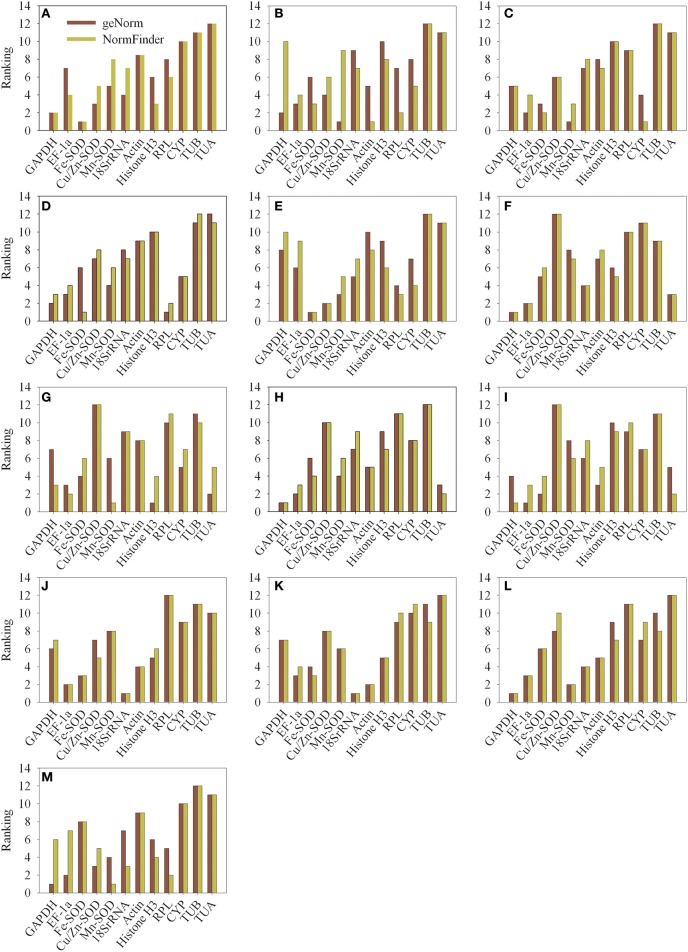
The NormFinder program was used to further verify the results obtained from geNorm analysis. The results were shown in Table S4 by NormFinder analysis. Some differences were observed in the results obtained from geNorm and NormFinder analyses. The six most stable reference genes (half of the total) were almost similar, but the ranking orders were different in the results yielded by the two algorithms (Figure 4 and Table S5). Table 3 summarizes the same results got from geNorm and NormFinder programs. In all experimental conditions, the most appropriate gene outcome by NormFinder analysis was in the top six appropriate genes obtain by geNorm analysis. In addition, the two most unstable genes were the same in both geNorm and NormFinder analyses.
Figure 4.
Comparison of the ranking of the candidate reference genes according to their stability values calculated using geNorm and NormFinder. (A) different varieties (pericarp), (B) different varieties (aril), (C) different developmental stages (pericarp), (D) different developmental stages (aril), (E) different organs, (F) NAA stimuli, (G) 2,4-D stimuli, (H) ethephon stimuli, (I) bagging, (J) cold temperature treatment (4°C), (K) room temperature treatment (22°C), (L) girdling with defoliation (fruit abscission zone), (M) girdling with defoliation (fruitlets).
Table 3.
Consensus of stability ranking of the reference genes estimated by geNorm and NormFinder.
| Experimental sample sets | Six most stable genes by geNorm | Most stable genes by NormFinder | Two least stable genes by geNorm and NormFinder |
|---|---|---|---|
| Different varieties (pericarp) | Fe-SOD, GAPDH, Cu/Zn-SOD, 18SrRNA, Mn-SOD, Histone H3 | Fe-SOD | TUA, TUB |
| Different varieties (aril) | GAPDH, Mn-SOD, EF-1a, Cu/Zn-SOD, Actin, Fe-SOD | Actin | TUB, TUA |
| Different developmental stages (pericarp) | Mn-SOD, EF-1a, Fe-SOD, CYP, GAPDH, Cu/Zn-SOD | CYP | TUB, TUA |
| Different developmental stages (aril) | GAPDH, RPL, EF-1a, Mn-SOD, CYP, Fe-SOD | Fe-SOD | TUB, TUA |
| Different organs | Fe-SOD, Cu/Zn-SOD, Mn-SOD, RPL, 18SrRNA, EF-1a | Fe-SOD | TUB, TUA |
| NAA treatment (pericarp) | GAPDH, EF-1a, TUA, 18SrRNA, Fe-SOD, Histone H3 | GAPDH | Cu/Zn-SOD, CYP |
| 2,4-D treatment (pericarp) | TUA, Histone H3, EF-1a, Fe-SOD, CYP, Mn-SOD | Mn-SOD | Cu/Zn-SOD, TUB/RPL |
| Ethephon treatment (pericarp) | GAPDH, EF-1a, TUA, Mn-SOD, Actin, Fe-SOD | GAPDH | TUB, RPL |
| Bagging treatment (pericarp) | Fe-SOD, EF-1a, Actin, GAPDH, TUA, 18SrRNA | GAPDH | Cu/Zn-SOD, TUB |
| Cold temperature treatment (4°C aril) | 18SrRNA, EF-1a, Fe-SOD, Actin, Histone H3, GAPDH | 18SrRNA | RPL, TUB |
| Room temperature treatment (22°C aril) | 18SrRNA, Actin, EF-1a, Fe-SOD, Histone H3, Mn-SOD | 18SrRNA | TUA, TUB/ CYP |
| Girdling with defoliation (fruit abscission zone) | GAPDH, Mn-SOD, EF-1a, 18SrRNA, Actin, Fe-SOD | GAPDH | TUA, RPL |
| Girdling with defoliation (fruitlets) | GAPDH, EF-1a, Cu/Zn-SOD, Mn-SOD, RPL, Histone H3 | Mn-SOD | TUB, TUA |
Reference gene validation
The expression level of DlACO under girdling with defoliation treatment was evaluated to validate our findings. The results were shown in Figure 5. The analysis revealed that the expression level of DlACO in fruit abscission zone increased progressively after girdling with defoliation treatment. Similar change patterns with minor differences were observed when using GAPDH alone or combination of GAPDH + Mn-SOD as reference gene(s). While, when using TUA gene (the least stable) as reference gene resulted in overestimation of DlACO expression (Figure 5). The effect was clearly caused by low stability of TUA gene expression in the sample and indicated that an appropriate reference gene was urgently needed in RT-qPCR analysis.
Figure 5.
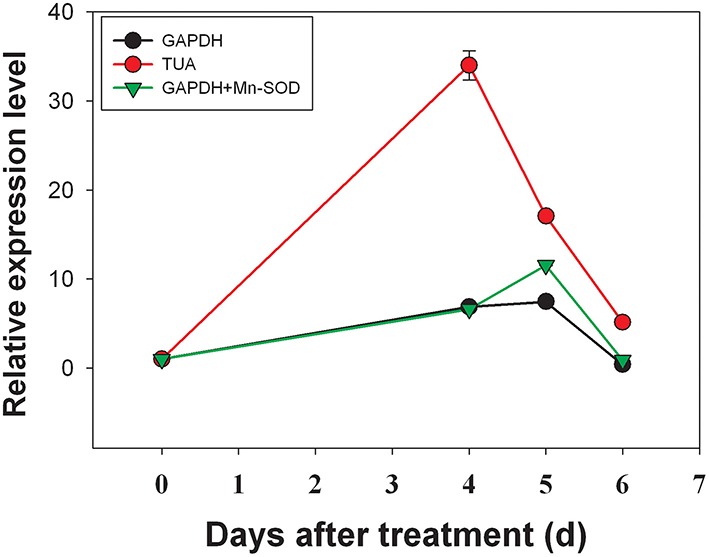
Relative quantification of DlACO expression using validated reference genes for normalization under girdling with defoliation conditions.
Discussion
RT-qPCR as the accurate and sensitive method for gene expression analysis, but the veracity and reliability result depends on whether select appropriate reference gene or not (Nicot et al., 2005). A suitable reference gene should possess invariant expression across all samples and in different experimental conditions. However, no gene has been found to meet this requirement (Artico et al., 2010). Therefore, it is important to find the appropriate reference gene(s) suitable for RT-qPCR normalization in the tissue or experimental condition of interest (Czechowski et al., 2005; Narsai et al., 2010). As far as we know, this study was the first to systematically select appropriate reference genes for RT-qPCR analysis in preharvest and postharvest longan fruits.
In this study, twelve candidate reference genes that have been validated previously in tropical/subtropical fruit trees were selected. Actin and EF-1a have been reported previously in somatic embryogenesis of longan (Lin and Lai, 2010), litchi (Zhong et al., 2011), banana (Chen et al., 2011), and papaya (Zhu et al., 2012); GAPDH and TUA have been validated in litchi (Zhong et al., 2011), banana (Chen et al., 2011), and papaya (Zhu et al., 2012); CYP has been studied in banana (Chen et al., 2011) and papaya (Zhu et al., 2012); 18SrRNA has been used to study somatic embryogenesis of longan (Lin and Lai, 2010) and papaya (Zhu et al., 2012); TUB has been reported in somatic embryogenesis of longan (Lin and Lai, 2010) and litchi (Zhong et al., 2011); Fe-SOD, Mn-SOD, Cn/Zn-SOD, and Histone H3 have been validated in somatic embryogenesis of longan (Lin and Lai, 2010); RPL has been studied in banana (Chen et al., 2011). ACO as the target gene was valuated to validate our results. Agarose gel electrophoresis, amplicon sequencing and dissociation curve analysis were used to verify the specificity of the RT-qPCR primer pairs (Figure S1). Standard curves were generated to calculate PCR amplification efficiency.
The appropriate reference gene should exhibit stable expression in different experimental conditions and in all samples (Chen et al., 2011; Zhong et al., 2011). However, no reference gene was found to have invariant expression in our experiments (Figure 1), which is consistent with the results obtained by Anna et al. (2009) and Jain et al. (2006), both of whom highlighted the importance of determining the appropriate reference genes under given conditions. In previous studies, the Cq values and mean Cq values ranged from 11 to 35 and 16 to 33, respectively, across all tested samples (Jain et al., 2006). In the current study, the Cq values ranged from 11.25 (18SrRNA) to 34.72 (Cu/Zn-SOD), and the mean Cq values lied between 17.43 (18SrRNA) and 29.02 (Fe-SOD). Variation (Cqmax–Cqmin) within an experiment was always less than 3, but the variation values across the tested samples were within the range of 8–15 (Table S1), which were higher than those reported by Zhong et al. (2011) and Chen et al. (2011) for all tested samples. The differences between the studies may be explained by use of different species and treatment conditions. Furthermore, these results showed it was urgently needed to compare the stability of the reference gene before using them for normalization in RT-qPCR assays (Chandna et al., 2012; Marum et al., 2012).
There was only one reference gene for normalization in RT-qPCR analysis in longan fruit (Zhao et al., 2011; Chen et al., 2012). While, many reports indicated that more than one reference genes should be used for normalization in RT-qPCR analysis (Vandesompele et al., 2002; Die and Rowland, 2013). The geNorm result showed that two reference genes were sufficient to normalization in RT-qPCR analysis in different fruit developmental stages, NAA, 2,4-D, ethephon, bagging, temperature and girdling with defoliation treatments, except in different varieties and organs. Three genes were sufficient for normalizing gene expression in different varieties of samples. When 0.15 as the cut-off value, none of the gene selected was appropriate across different organ samples. However, this cut-off value can be adjusted depending on experimental conditions. Based on our findings and previous reports (Vandesompele et al., 2002), we conclude that at least three most stable reference genes are needed for normalization in RT-qPCR under different organ conditions.
Previous reports showed that different sets of samples have their own most suitable reference genes (Chen et al., 2011; Zhong et al., 2011), and the same results were obtained in the present study (Figure 3). For example, Fe-SOD and Cu/Zn-SOD were the most stable genes under different organs. GAPDH and Mn-SOD genes ranked higher in different varieties of longan aril and fruit abscission zone in girdling with defoliation treatment, whereas Fe-SOD and GAPDH genes did better than GAPDH and Mn-SOD genes in the pericarp of longan fruit in different varieties. GAPDH and EF-1a were the most stable genes under NAA, ethephon treatment and fruitlets in girdling with defoliation conditions. GAPDH and RPL were the most suitable reference genes in aril samples at different developmental stages. GAPDH gene was one of the best reference genes in the seven experiments in this study, and the same result was found in litchi (Zhong et al., 2011), pear (Wu et al., 2012), plum (Kim et al., 2015), and Chinese wolfberry (Wang et al., 2013). Interestingly, GAPDH is one of the least stable genes in watermelon (Kong et al., 2014).
Mn-SOD and EF-1a genes were ranked higher in pericarp at different developmental stages. Fe-SOD and EF-1a, or 18SrRNA, and EF-1a were the most suitable reference genes under bagging or cold temperature (4°C) treatment. EF-1a was one of the most suitable reference genes across six experiments, which is consistent with previous findings in litchi (Zhong et al., 2011), pear (Wu et al., 2012), berry (Reid et al., 2006), plum (Kim et al., 2015), rice (Jain et al., 2006), and Lolium temulentum (Dombrowski and Martin, 2009). However, EF-1a is one of the least stable genes in banana (Chen et al., 2011).
18SrRNA and Actin were the most stable genes in room temperature (22°C) treatment. Actin has been used as reference gene for normalization in many RT-qPCR studies (Kandasamy and Meagher, 1999; Reid et al., 2006), including those on longan fruit (Zhao et al., 2011; Chen et al., 2012). The present study showed that Actin was one of the most suitable reference genes but only at room temperature (22°C) and not in most other conditions. Therefore, more suitable reference genes should be considered in RT-qPCR analysis in longan fruit in future studies. Furthermore, previous studies demonstrated that Actin transcription in tissues subjected to cold treatment is less stable (Lopez-Pardo et al., 2012; Saha and Vandemark, 2012), suggesting that low temperatures influence its transcription and/or turnover rates. Consistent with this, Actin was previously reported as one of the least stable genes by Tatiane et al. (2015) and Wu et al. (2012). The 18SrRNA was not the stable reference gene in Libault et al. (2008) report. In this research, 18SrRNA was one of the most suitable reference genes under room temperature (22°C) treatment; however, in most other conditions, 18SrRNA was either one of the most or least stable genes. Previous studies have also reported that this gene is unstable in peach (Tong et al., 2009), pear (Wu et al., 2012), melon (Carmen et al., 2015), zucchini (Obrero et al., 2011), and cucumber (Wan et al., 2010). In addition, 18SrRNA gene expression was dramatically higher than those of other reference genes (Table S1), so 18SrRNA is not an appropriate reference gene for normalization in RT-qPCR studies involving target genes with mid- or low-level expression.
TUA and Histone H3 were the suitable reference genes under 2,4-D treatment. Histone H3 was found to be one of the best reference genes in the 2,4-D treatment group, but it did not rank high in other treatment conditions. Notably, this gene was previously reported to be the least stable reference gene in citrus (Mafra et al., 2012). Interestingly, while TUA was found to be one of the most suitable reference genes under 2,4-D treatment, it was one of the least stable reference genes in most samples used in this study. Consistent with this, TUA was previously reported to be the least stable gene in litchi (Zhong et al., 2011), pear (Xu et al., 2015), zucchini (Obrero et al., 2011), and cucumber (Wan et al., 2010). Thus, a gene that is invariant in one experimental condition may have varying expression in other experiments. Therefore, verifying the stability of reference genes for each experimental condition is critical before using them for normalization in RT-qPCR assays.
The stability of the candidate reference genes was evaluated using geNorm and NormFinder algorithms in this study. While some studies have reported small differences in gene stability ranking generated by the two algorithms (Cruz et al., 2009; Lee et al., 2010), some have found large differences (Anna et al., 2009; Lin and Lai, 2010). In this study, slight differences were found between the geNorm and NormFinder results. The six most stable reference genes (half of the total) were similar, but the ranking order was different in the two algorithms (Figure 4 and Table S5). Moreover, the two most unstable reference genes identified by the two programs were the same (Table 3). These results are consistent with those reported by other groups (Marino et al., 2008; Artico et al., 2010; Lin and Lai, 2010; Wan et al., 2010). Given the differences in statistical algorithm used by geNorm and NormFinder programs, it is not surprising that there are some differences in their results. Nevertheless, there is substantial overlap in the results generated by the two programs, so use of both methods allows affirmation of candidate reference genes.
In orded to validate our results, relative mRNA levels of DlACO in the fruit abscission zone after girdling with defoliation treatment were calculated using different reference genes. Results showed that when used the most unstable reference gene result in overestimation expression (Figure 5), which indicated that an appropriate reference gene was urgently needed in RT-qPCR analysis.
Conclusion
As far as we know, this study was the first to systematically select appropriate reference genes for RT-qPCR in preharvest and postharvest longan fruits. Accurate results could be obtained with the use of two or three appropriate reference genes for normalization by RT-qPCR in preharvest and postharvest longan fruits. The most suitable combinations used to normalize the expression profiles in preharvest longan fruit are as follows: GAPDH + EF-1a for NAA, ethephon treatment and fruitlets in girdling with defoliation conditions; GAPDH + Mn-SOD for fruit abscission zone in girdling with defoliation conditions; GAPDH + RPL or Mn-SOD + EF-1a for aril or pericarp at different developmental stages, respectively; Fe-SOD + EF-1a for bagging treatment; TUA + Histone H3 for 2,4-D treatment; and Fe-SOD + Cu/Zn-SOD + Mn-SOD for different organs. In postharvest longan fruit, 18SrRNA + EF-1a and 18SrRNA + Actin were the most suitable combinations for 4 and 22°C temperature conditions, respectively, while Fe-SOD + GAPDH + Cu/Zn-SOD and GAPDH + Mn-SOD+ EF-1a were suitable for pericarp and aril of longan fruit, respectively. Analysis of DlACO gene expression indicated that use of suitable reference genes was crucial for getting accurate results in RT-qPCR analysis. Taken together, results from the present work provide a reference for choosing suitable reference genes for more accurate RT-qPCR analysis in preharvest and postharvest longan fruits.
Author contributions
JW and HZ conceived and designed the experiments. YW and HZ performed the experiments. WL analyzed the data. SS and LL contributed reagents/materials. JW and YW wrote the paper. All authors read and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by National Natural Science Foundation of China (No. 31572087), China Agricultural Research Service (No. CARS-33), Natural Science Foundation of Guangdong Province (No. 2014A030307031), Natural Science Foundation of Lingnan Normal University (No. QL1514), and The Science and Technology Foundation of Zhanjiang (No. 2016B01106).
Glossary
Abbreviations
- ACO
1-aminocyclopropane-1-carboxylate oxidase
- Actin
Actin
- Cu/Zn-SOD
Copper/zinc-superoxide dismutase
- CYP
Cyclophilin
- EF-1a
Elongation factor 1-alpha
- Fe-SOD
Iron-superoxide dismutase
- GAPDH
Glyceraldehyde-3- phosphate dehydrogenase
- Histone H3
Histone H3
- Mn-SOD
Manganese superoxide dismutase
- NAA
Naphthaleneacidic acid
- PCR
Polymerase chain reaction
- RPL
Ribosomal protein L
- RT-qPCR
Reverse transcription quantitative PCR
- TUA
Alpha-tubulin
- TUB
Beta-tubulin
- UBQ
Ubiquitin
- 18SrRNA
18S ribosomal RNA
- 2,4-D
2,4-(dichlorophenoxy)acetic acid.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00780
Single-peaked melt curves for all reference genes from three technical replicates of one set of cDNA pools with non-template control.
Cq values of all samples in longan.
Data statistics of Cq values of candidate reference genes in longan. Mean, median, minimum, and maximun of Cq values were determined by statistical analysis.
Candidate genes ranked according to their expression stability values (M) estimated using the geNorm algorithm.
Ranking of the candidate reference genes in different samples according to their stability value calculated by NormFinder analysis.
Comparison of the ranking of candidate reference genes according to their stability value calculated by geNorm and NormFinder analysis.
References
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Anna R. P., Oronzo A. T., Enrico P., Mario C. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. 10.1186/1471-2199-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artico S., Nardeli S. M., Brilhante O., Grossi-de-Sa M. F., Alves-Ferreira M. (2010). Identification and evaluation of new reference genes in gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 10:49. 10.1186/1471-2229-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber R. D., Harmer D. W., Coleman R. A., Clark B. J. (2005). GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 21, 389–395. 10.1152/physiolgenomics.00025.2005 [DOI] [PubMed] [Google Scholar]
- Bustin S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Mol. Endocrinol. 29, 23–39. 10.1677/jme.0.0290023 [DOI] [PubMed] [Google Scholar]
- Carmen L., Claudio M., Cristina E., Ronan S., John E. L., Frank L., et al. (2015). Variability of candidate genes, genetic structure and association with sugar accumulation and climacteric behavior in a broad germplasm collection of melon (Cucumis melo L.). BMC Genet. 16:28 10.1186/s12863-015-0183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandna R., Augustine R., Bisht N. C. (2012). Evaluation of candidate reference genes for gene expression normalization in brassica juncea using real time quantitative RT-PCR. PLoS ONE 7:e36918. 10.1371/journal.pone.0036918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., He X. H., Luo C., Yang L. T., Huang X., Hu Y. (2012). Cloning and expression of longan carbonic anhydrase gene under low temperature stress. Acta Hortic. Sin. 39, 243–252. 10.16420/j.issn.0513-353x.2012.02.011 [DOI] [Google Scholar]
- Chen L., Zhong H. Y., Kuang J. F., Li J. G., Lu W. J., Chen J. Y. (2011). Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234, 377–390. 10.1007/s00425-011-1410-3 [DOI] [PubMed] [Google Scholar]
- Clancy M. A., Rosli H. G., Chamala S., Barbazuk W. B., Civello P. M., Folta K. M. (2013). Validation of reference transcripts in strawberry (Fragaria spp.). Mol. Genet. Genomics 288, 671–681. 10.1007/s00438-013-0780-6 [DOI] [PubMed] [Google Scholar]
- Cruz F., Kalaoun S., Nobile P., Colombo C., Almeida J., Barros L., et al. (2009). Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 23, 607–616. 10.1007/s11032-009-9259-x [DOI] [Google Scholar]
- Czechowski T. M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Die J. V., Roman B., Nadal S., Gonzalez-Verdejo C. I. (2010). Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232, 145–153. 10.1007/s00425-010-1158-1 [DOI] [PubMed] [Google Scholar]
- Die J. V., Rowland L. J. (2013). Superior cross-species reference genes: a blueberry case study. PLoS ONE 8:e73354. 10.1371/journal.pone.0073354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski J. E., Martin R. C. (2009). Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Sci. 176, 390–396. 10.1016/j.plantsci.2008.12.005 [DOI] [Google Scholar]
- Gutierrez L., Mauriat M., Guenin S., Pelloux J., Lefebvre J. F., Louvet R., et al. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. 10.1111/j.1467-7652.2008.00346.x [DOI] [PubMed] [Google Scholar]
- Hong S. Y., Seo P. J., Yang M. S., Xiang F., Park C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112. 10.1186/1471-2229-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A. K., Khurana J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. 10.1016/j.bbrc.2006.04.140 [DOI] [PubMed] [Google Scholar]
- Jiang Y. M., Zhang Z. Q., Dary C., Joyce C., Saichol K. (2002). Postharvest biology and handling of longan fruit (Dimocarpus longan Lour.). Postharvest Biol. Technol. 26, 241–252. 10.1016/S0925-5214(02)00047-9 [DOI] [Google Scholar]
- Kandasamy M. K., Meagher R. B. (1999). Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil. Cytoskel. 44, 110–118. [DOI] [PubMed] [Google Scholar]
- Kim H. Y., Prasenjit S., Macarena F., Li B. S., Sadka A., Eduardo B. (2015). RNA-seq analysis of spatiotemporal gene expression patterns during fruit development revealed reference genes for transcript normalization in plums. Plant Mol. Biol. Rep. 33, 1634–1649. 10.1007/s11105-015-0860-3 [DOI] [Google Scholar]
- Kong Q. S., Yuan J. X., Gao L. Y., Zhao S., Jiang W., Huang Y., et al. (2014). Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 9:e90612. 10.1371/journal.pone.0090612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. X., Chen C. L., Chen Z. G. (2001). Progress in biotechnology research in longan. Acta Hort. 558, 137–141. 10.17660/ActaHortic.2001.558.18 [DOI] [Google Scholar]
- Lee J. M., Roche J. R., Donaghy D. J., Thrush A., Sathish P. (2010). Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol. Biol. 11:8. 10.1186/1471-2199-11-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. D., Sladek R., Greenwood C. M., Hudson T. J. (2002). Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 12, 292–297. 10.1101/gr.217802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Q., Wang Y., Huang X. M., Li J., Wang H. C., Li J. G. (2015b). An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 6:439. 10.3389/fpls.2015.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. G., Yuan R. C. (2008a). Effect of sprayable 1-MCP, AVG, and NAA on ethylene biosynthesis, preharvest fruit drop, fruit maturity, and quality of ‘Delicious’ apples. HortScience 43, 1454–1460. [Google Scholar]
- Li J. G., Yuan R. C. (2008b). NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. J. Plant Growth Regul. 27, 283–295. 10.1007/s00344-008-9055-6 [DOI] [Google Scholar]
- Li M. Y., Wang F., Jiang Q., Wang G. L., Tian C., Xiong A. S. (2016). Validation and comparison of reference genes for qPCR normalization of celery (apiumgraveolens) at different development stages. Front. Plant Sci. 7:313. 10.3389/fpls.2016.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. S., Zhang D. Y., Li H. Y., Gao B., Yang H. L., Zhang Y. M., et al. (2015a). Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 6:38. 10.3389/fpls.2015.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Thibivilliers S., Bilgin D. D., Radwan O., Benitez M., Clough S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome 1, 44–54. 10.3835/plantgenome2008.02.0091 [DOI] [Google Scholar]
- Lin Y. L., Lai Z. X. (2010). Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 178, 359–365. 10.1016/j.plantsci.2010.02.005 [DOI] [Google Scholar]
- Lin Y. L., Lai Z. X. (2013). Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol. Biochem. 66, 20–25. 10.1016/j.plaphy.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Liu Q., Wei C., Zhang M. F., Jia G. X. (2016). Evaluation of putative reference genes for quantitative real-time PCR normalization in Lilium regale during development and under stress. Peer J. 4:e1837. 10.7717/peerj.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pardo R, Ruiz de Galarreta, J. I., Ritter E. (2012). Selection of housekeeping genes for qRT-PCR analysis in potato tubers under cold stress. Mol. Breed. 31, 39–45. 10.1007/s11032-012-9766-z [DOI] [Google Scholar]
- Ma R., Xu S., Zhao Y. C., Xia B., Wang R. (2016). Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 7:536. 10.3389/fpls.2016.00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafra V., Kubo K. S., Alves-Ferreira M., Ribeiro-Alves M., Stuart R. M., Boava L. P., et al. (2012). Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE 7:e31263. 10.1371/journal.pone.0031263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10:4. 10.1186/1471-2229-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino E. R., Borges A. A., Perez A. B., Perez J. A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8:131. 10.1186/1471-2229-8-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marum L., Miguel A., Ricardo C. P., Miguel C. (2012). Reference gene selection for quantitative real-time PCR normalization in quercus suber. PLoS ONE 7:e35113. 10.1371/journal.pone.0035113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio G. A., Miguel G. R., Alex D. G., José C., Alejandro M., Ariel O., et al. (2013). Identification of two putative reference genes from grapevine suitable for gene expression analysis in berry and related tissues derived from RNA-Seq data. BMC Genomics 14:878. 10.1186/1471-2164-14-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Ivanova A., Whelan J. (2010). Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 10:56. 10.1186/1471-2229-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N., Hausman J. F., Hoffmann L., Evers D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. 10.1093/jxb/eri285 [DOI] [PubMed] [Google Scholar]
- Obrero A., Die J. V., Roman B., Gomez P., Nadal S. (2011). Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J. Agr. Food Chem. 59, 5402–5411. 10.1021/jf200689r [DOI] [PubMed] [Google Scholar]
- Reid K. E., Olsson N., Schlosser J., Peng F., Lund S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27 10.1186/1471-2229-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha G. C., Vandemark G. J. (2012). Evaluation of expression stability of candidate references genes among green and yellow pea cultivars (Pisum sativum L.) subjected to abiotic and biotic stress. Am. J. Plant Sci. 3, 235–242. 10.4236/ajps.2012.32028 [DOI] [Google Scholar]
- Schmidt G. W., Delaney S. K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics 283, 233–241. 10.1007/s00438-010-0511-1 [DOI] [PubMed] [Google Scholar]
- Shi S. Y., Li W. C., Zhang H. N., Liu L. Q., Shu B., Liang Q. Z., et al. (2015). Application of extended biologische bundesantalt, bundessortenamt and chemische industrie scale for phenological studies in longan (Dimocarpus longan). Ann. Appl. Biol. 167, 127–134. 10.1111/aab.12214 [DOI] [Google Scholar]
- Shuai L., Li J., Niu J. J., Qian P. H., Liu W. H., Xue X. Q., et al. (2016). Sucrose-metabolizing enzymes and their genes in the arils of two Dimocarpus longan cultivars. Biol. Plant. 20, 1–8. 10.1007/s10535-016-0602-x [DOI] [Google Scholar]
- Shuai L., Xue X. Q., Niu J. J., Gu Y. J., Han D. M., Wu Z. X. (2015). Analyses of the fructokinase activity and its gene expression during the development of longan fruits. J. S. China Agr. Univ. 36, 99–104. 10.7671/j.issn.1001-411X.2015.05.017 [DOI] [Google Scholar]
- Tatiane T. S., Camila P., Taciane F., Vera Q., Cesar V. R., César L. G. (2015). Identification of a novel reference gene for apple transcriptional profiling under postharvest conditions. PLoS ONE 10:e0120599. 10.1371/journal.pone.0120599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Jiang Q., Wang F., Wang G. L., Xu Z., Xiong A. S. (2015). Selection of suitable reference genes for qPCR normalization nunder abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 10:e0117569. 10.1371/journal.pone.0117569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjasa R., Natasa S., Dunja B., Branka J., Jernej J. (2013). Validation of candidate reference genes in RT-qPCR studies of developing olive fruit and expression analysis of four genes involved in fatty acids metabolism. Mol. Breed. 32, 211–222. 10.1007/s11032-013-9863-7 [DOI] [Google Scholar]
- Tong Z., Gao Z., Wang F., Zhou J., Zhang Z. (2009). Selection of reliable reference genes for gene expression studies in peach using realtime PCR. BMC Mol. Biol. 10:71 10.1186/1471-2199-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Zhang X., Liu D., Jin S., Cao J., Zhu L., et al. (2007). Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin. Sci. Bull. 52, 3110–3117. 10.1007/s11434-007-0461-0 [DOI] [Google Scholar]
- Vandesompele J., Pattyn F., Poppe B., Van R. N. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H. J., Zhao Z. G., Qian C. T., Sui Y. H., Malik A. A., Chen J. F. (2010). Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261. 10.1016/j.ab.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Y., Zhou P. (2013). Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiol. Biochem. 70, 304–310. 10.1016/j.plaphy.2013.05.038 [DOI] [PubMed] [Google Scholar]
- Wu J. Y., Peng G., Li C. Q., Lu W. J., Wang Z. H., Li J. G. (2011). A new rapid and effective method for RNA isolation from litchi tissues of fruitlet and abscission zone. Acta Hortic. Sin. 38, 1191–1196. 10.16420/j.issn.0513-353x.2011.06.025 [DOI] [Google Scholar]
- Wu T., Zhang R. P., Gu C., Wu J. Y., Wan H. J., Zhang S. J., et al. (2012). Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr. J. Agr. Res. 7, 3701–3704. 10.5897/AJAR11.1842 [DOI] [Google Scholar]
- Xu Y. Y., Li H., Li X. G., Lin J., Wang Z. H., Yang Q. S., et al. (2015). Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol. Plant 37, 1738–1754. 10.1007/s11738-015-1784-0 [DOI] [Google Scholar]
- Yang Z. M., Chen Y., Hu B. Y., Tan Z. Q., Huang B. T. (2015). Identification and validation of reference genes for quantification of target gene expression with quantitative real-time PCR for tall fescue under four abiotic stresses. PLoS ONE 10:e0119569. 10.1371/journal.pone.0119569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X. R., Zhang Y. Y., Li L., Li Z. C., Li M. J., Li C. B., et al. (2014). Cloning and molecular characterization of phospholipase D (PLD) delta gene from longan (Dimocarpus longan Lour.). Mol. Biol. Rep. 41, 4351–4360. 10.1007/s11033-014-3306-3 [DOI] [PubMed] [Google Scholar]
- Yperman J., De V. G., Holvoet P., Flameng W. (2004). Beta-actin cannot be used as a control for gene expression in ovine interstitial cells derived from heart valves. J. Heart Valve Dis. 13, 848–853. [PubMed] [Google Scholar]
- Zhang H. N., Shi S. Y., Li W. C., Shu B., Liu L. Q., Wei Y. Z., et al. (2016). Transcriptome analysis of ‘Sijihua’ longan (Dimocarpus longan L.) based on next-generation sequencing technology. J. Hortic. Sci. Biotechnol. 91, 180–188. 10.1080/14620316.2015.1133539 [DOI] [Google Scholar]
- Zhao M. L., Kuang J. F., Lu W. J., Chen J. Y. (2011). Expression analysis of pectin methylesterase genesin postharvest longan fruit during aril breakdown. J. Trop. Subtrop. Bot. 19, 135–141. 10.3969/j.issn.1005-3395.2011.02.006 [DOI] [Google Scholar]
- Zhong H. Y., Chen J. W., Li C. Q., Chen L., Wu J. Y., Chen J. Y., et al. (2011). Selection of reliable reference genes for expression studies by reverse transcription quantitative realtime PCR in litchi under different experimental conditions. Plant Cell Rep. 30, 641–653. 10.1007/s00299-010-0992-8 [DOI] [PubMed] [Google Scholar]
- Zhu X., Li X., Chen W., Chen J., Lu W., Chen L., et al. (2012). Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE 7:e44405. 10.1371/journal.pone.0044405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H. H., Fu Y. P., He W., Wang L., Wei Y. H. (2015). Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 6:475. 10.3389/fpls.2015.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single-peaked melt curves for all reference genes from three technical replicates of one set of cDNA pools with non-template control.
Cq values of all samples in longan.
Data statistics of Cq values of candidate reference genes in longan. Mean, median, minimum, and maximun of Cq values were determined by statistical analysis.
Candidate genes ranked according to their expression stability values (M) estimated using the geNorm algorithm.
Ranking of the candidate reference genes in different samples according to their stability value calculated by NormFinder analysis.
Comparison of the ranking of candidate reference genes according to their stability value calculated by geNorm and NormFinder analysis.