To the editor:
Tyrosine kinase inhibitors (TKIs) have dramatically changed the natural history of chronic myeloid leukemia (CML), with 10-year overall survival surpassing 80% for patients treated in chronic phase. Unfortunately, most of the world’s CML patients reside in low-resource areas, where treatment, diagnostic, and monitoring options are limited by the cost of TKI treatment and access to capable laboratory facilities. Shipping blood to specialized centers for testing is very costly, and test performance can be hampered by sample degradation during transit.
We developed a procedure to spot peripheral blood of CML patients onto a paper template (dried blood spots [DBSs]), with subsequent shipping, RNA and/or DNA extractions for BCR-ABL1 quantitative testing, and/or ABL1 mutation testing. The storage of blood on filter paper is not a recent discovery, and blood on filter paper is frequently used for genomic testing (ie, DNA testing from Guthrie cards has proven the prenatal origin of some types of leukemia1-6).
Despite the high lability of RNA, its extraction from filter paper was shown in 1992,7,8 and it has been occasionally reported as the method used for the study of infectious diseases such as HIV.9-12 The qualitative detection (present or absent) of BCR-ABL1 rearrangements in RNA from DBS was reported in 1998.13 However, current treatments demand quantification of the BCR-ABL1 transcript for monitoring and mutation studies.
In short, we find that the DBS technique compares favorably to the testing of fresh blood for BCR-ABL1 transcript level quantitation, using the Cepheid GeneXpert, in specimens mailed across the globe and that mutation screening is possible, albeit with sensitivity limitations inherent to the technique.
First, we compared the detection of BCR-ABL1 on 7 fresh CML specimens to the same specimens spotted on filter paper (Whatman 903 Protein Saver Cards [Sigma-Aldrich] with 200 µL blood per card, 50 µL per 4 spots) and “aged” for 41 days on the laboratory bench. RNA was extracted both from fresh blood and from 41-day-old DBSs and run on the GeneXpert BCR-ABL1 monitor test (Cepheid, Sunnyvale, CA). Correlation of BCR-ABL1 transcripts between both groups was r = 0.86 (P = .02) (supplemental Figure 1 and supplemental Table 1, available on the Blood Web site). Ten specimens collected from patients without CML and spotted on filter paper were negative for BCR-ABL1 by the same instrumentation limits of detection.
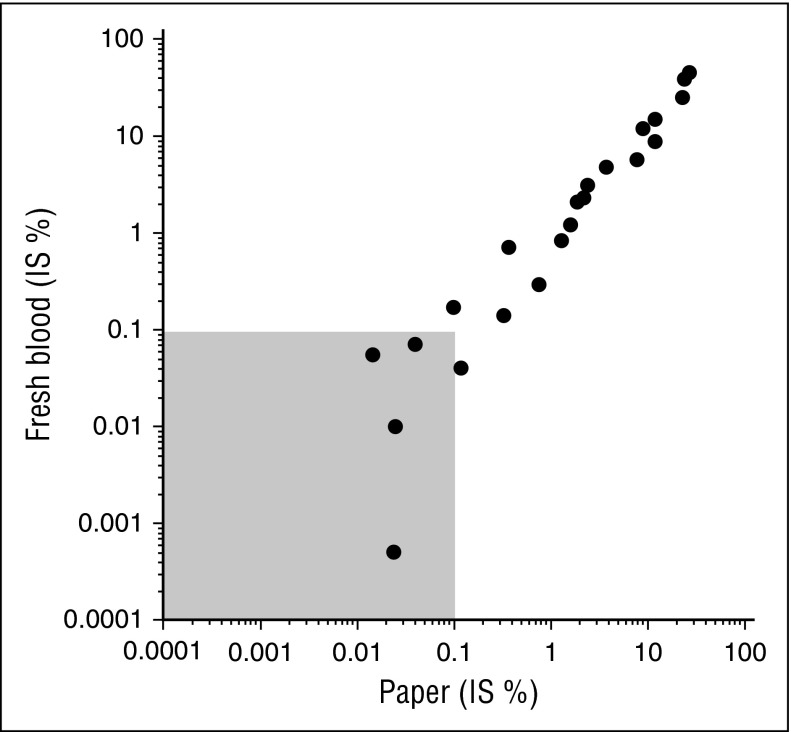
Figure 1.
Correlation of fresh blood and paper results (BCR-ABL1% IS, log scale). Shaded area represents major molecular response levels.
We then compared BCR-ABL1 in fresh samples analyzed in Adelaide, Australia, to the results in DBSs after shipping to our laboratory from overseas. Both Australia and Seattle tested for BCR-ABL1 using the same GeneXpert platform. This experiment consisted of 22 DBS cards, the majority sent as letters via regular (“snail”) mail. RNA was extracted from DBSs upon arrival.
DBSs were 43 days old on average (8-73 days; median, 34 days) at the time of RNA extraction. The mean RNA concentration measured was 16.1 ng/µL (0-110 ng/µL; median, 6.0 ng/µL) (one sample had undetectable RNA levels). The yield of RNA showed a correlation with the white blood cells (WBCs) of the sample (r = 0.80), although after log-transforming RNA levels, the correlation was reduced (r = 0.54). There was also a correlation, yet weaker, between RNA and the shipping time (r = −0.42), although with RNA log-transformed, the correlation with shipping time is a bit stronger (r = −0.52). The correlation between RNA/WBC and shipping time is r = −0.48.
The average amount of RNA used for the GeneXpert assay was 200 ng. Amplification was successful in all specimens. Supplemental Table 2 shows the results of the 22 international samples, with the results of the test performed in fresh blood in Adelaide and in DBSs after shipping to Seattle. Of note, in 3 cases, the GeneXpert reported an invalid result because of late ABL1 amplification (after cycle 20), but the BCR-ABL1% could be calculated using the cartridge lot efficiency (E) with the formula BCR-ABL1% International Scale (IS) = EΔCt × 100.
There was a strong correlation between the BCR-ABL1% IS assays performed in fresh samples versus RNA from DBSs (r = 0.97), same as when the BCR-ABL1% IS were log-transformed (r = 0.94, Figure 1). Supplemental Figure 2 displays the bias as a function of the average log (% IS) with a Bland-Altman plot, where the bias is simply the difference between the log-transformed values of the fresh and DBS samples.
Finally, we performed mutation analysis in a third set of specimens received as DBSs from a low-resource area. Results performed on fresh specimens in Adelaide were available on these samples for comparison. We extracted DNA from a single DBS (50 µL blood) and amplified and sequenced ABL1 exons 4, 5, and 6 in 3 independent reactions. We obtained concordant results in 28 out of 30 samples (93%). We detected Y253H in 2 patients, E255K and E255V in 1 patient each, T315I in 2 more cases, and F359V in 1 case. Our results were discordant in 2 cases, where we could not detect mutations although fresh samples tested positive for the Y253H and E355G mutations. We attribute the disagreement to a sensitivity limitation, inherent to sequencing ABL1 DNA. In the first case, the GeneXpert BCR-ABL1 determination was “invalid” (no BCR-ABL1 amplification and ABL1 detection after cycle 20), and for the second one, BCR-ABL1 was 11%.
The ability to accurately perform BCR-ABL1 testing of dried spots has cost and sample stability as major advantages. Samples can potentially be batched and sent via regular mail, which eliminates the need of costly “next day” specialized shipping requirements of blood specimens. It would also be reasonable to spot multiple cards for patients with a low WBC count, or if more RNA was needed, as shipping and storage cost increases would be next to nil. Regarding sample stability, we show here that the quantification of BCR-ABL1 transcripts is not significantly affected across different disease levels, and the age at which samples are too old to be tested is not known. Currently, the oldest specimen we have tested is a 5-month-old card we received from a low-resource area. With our protocol, we detected BCR-ABL1 at 2.9%, whereas the result obtained locally on the fresh specimen at the time of collection was 5%. Lastly, DNA and RNA can be recovered from DBSs, thus also potentially allowing the testing of ABL1 resistance mutations in the low-resource setting.
Spotted blood can potentially increase the number of CML patients in resource-poor areas who can be diagnosed and monitored during TKI therapy. The method could potentially also be used to facilitate and cheapen molecular diagnostic for clinical trials in these areas, which could allow for earlier access of new TKI agents for CML. Further studies are in progress to field test in multiple countries, as well as developing assays for other “actionable” mutations in other types of leukemia. We would welcome collaborators to help optimize DBS for a broader range of indications and uses.
Paper or plastic, indeed.
All methods are available as supplemental information on the Blood Web site.
Footnotes
The online version of this article contains a data supplement.
Authorship
Acknowledgments: This work was partially supported by grant CA-18029 from the National Institutes of Health National Cancer Institute (J.P.R.).
Contribution: O.S.T., J.L.S., A.P., and J.P.R. designed the experiments; O.S.T., L.B., L.W., J.G., R.K., and K.G. collected and assembled data; C.C.S.Y. helped procure specimens; O.S.T., L.B., S.B., T.A.G., and J.P.R. analyzed and interpreted the data; and all authors helped write the manuscript and approved the final version.
Conflict-of-interest disclosure: S.B. is a consultant to Cepheid and an advisory board member for Qiagen and Novartis and received honoraria and research funding from Novartis, honoraria from Bristol Myers-Squibb, and research funding from Ariad. J.P.R. is a consultant to Novartis, BMS, and Ariad and received research support from Novartis and Cepheid. The remaining authors declare no competing financial interests.
Correspondence: Olga Sala Torra, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: osala@fhcrc.org.
References
- 1.Fasching K, Panzer S, Haas OA, Marschalek R, Gadner H, Panzer-Grümayer ER. Presence of clone-specific antigen receptor gene rearrangements at birth indicates an in utero origin of diverse types of early childhood acute lymphoblastic leukemia. Blood. 2000;95(8):2722–2724. [PubMed] [Google Scholar]
- 2.Gale KB, Ford AM, Repp R, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA. 1997;94(25):13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maia AT, Ford AM, Jalali GR, et al. Molecular tracking of leukemogenesis in a triplet pregnancy. Blood. 2001;98(2):478–482. doi: 10.1182/blood.v98.2.478. [DOI] [PubMed] [Google Scholar]
- 4.Wiemels JL, Cazzaniga G, Daniotti M, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354(9189):1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 5.Wiemels JL, Xiao Z, Buffler PA, et al. In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood. 2002;99(10):3801–3805. doi: 10.1182/blood.v99.10.3801. [DOI] [PubMed] [Google Scholar]
- 6.Yagi T, Hibi S, Tabata Y, et al. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood. 2000;96(1):264–268. [PubMed] [Google Scholar]
- 7.Matsubara Y, Ikeda H, Endo H, Narisawa K. Dried blood spot on filter paper as a source of mRNA. Nucleic Acids Res. 1992;20(8):1998. doi: 10.1093/nar/20.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YH, McCabe ER. RNA analysis from newborn screening dried blood specimens. Hum Genet. 1992;89(3):311–314. doi: 10.1007/BF00220548. [DOI] [PubMed] [Google Scholar]
- 9.Ou CY, Yang H, Balinandi S, et al. Identification of HIV-1 infected infants and young children using real-time RT PCR and dried blood spots from Uganda and Cameroon. J Virol Methods. 2007;144(1-2):109–114. doi: 10.1016/j.jviromet.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Lofgren SM, Morrissey AB, Chevallier CC, et al. Evaluation of a dried blood spot HIV-1 RNA program for early infant diagnosis and viral load monitoring at rural and remote healthcare facilities. AIDS. 2009;23(18):2459–2466. doi: 10.1097/QAD.0b013e328331f702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilian RR, Bhowan K, Sherman GG. Early diagnosis of human immunodeficiency virus-1 infection in infants with the NucliSens EasyQ assay on dried blood spots. J Clin Virol. 2010;48(1):40–43. doi: 10.1016/j.jcv.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Brambilla D, Jennings C, Aldrovandi G, et al. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J Clin Microbiol. 2003;41(5):1888–1893. doi: 10.1128/JCM.41.5.1888-1893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suttorp M, Ritgen M, von Neuhoff N, Schoch R, Schmitz N. Blood on filter paper as a readily available source of bcr-abl rearranged mRNA. Blood. 1997;90(4):1713–1715. [PubMed] [Google Scholar]