Abstract
Understanding the properties and functions of complex biological systems depends upon knowing the proteins present and the interactions between them. Recent advances in mass spectrometry have given us greater insights into the participating proteomes, however, monoclonal antibodies remain key to understanding the structures, functions, locations and macromolecular interactions of the involved proteins. The traditional single immunogen method to produce monoclonal antibodies using hybridoma technology are time, resource and cost intensive, limiting the number of reagents that are available. Using a high content analysis screening approach, we have developed a method in which a complex mixture of proteins (e.g., subproteome) is used to generate a panel of monoclonal antibodies specific to a subproteome located in a defined subcellular compartment such as the nucleus. The immunofluorescent images in the primary hybridoma screen are analyzed using an automated processing approach and classified using a recursive partitioning forest classification model derived from images obtained from the Human Protein Atlas. Using an ammonium sulfate purified nuclear matrix fraction as an example of reverse proteomics, we identified 866 hybridoma supernatants with a positive immunofluorescent signal. Of those, 402 produced a nuclear signal from which patterns similar to known nuclear matrix associated proteins were identified. Detailed here is our method, the analysis techniques, and a discussion of the application to further in vivo antibody production.
1. Introduction
In recent years, advances in genomic sequencing, mRNA microarray, and mass spectrometry techniques have greatly expanded our understanding of gene and protein expression in a wide variety of cell types and pathological conditions. With efforts to further understand the genomic and proteomic data we now have access to, high affinity monoclonal antibodies have become a key asset to scientist to understand the expression, localization, and therefore potential interaction partners of proteins of interest [1]. This is emphasized by the ongoing academic and commercial efforts to further expand available antibody resources [2]. Unfortunately, these efforts have been stymied by the difficulty to rapidly produce reagents that demonstrate high affinity and specificity for their protein targets.
Historically, methods to produce monoclonal antibodies have focused upon either in vivo or in vitro methods. The in vivo use of the mammalian immune system combined with hybridoma generation to produce monoclonal antibodies was first established in 1975 [3]. Because the maturation of the antibody producing B-lymphocytes isolated to produce the immortal hybridoma cell lines occurs in vivo and includes somatic hypermutation, in vivo approaches tend to produce high affinity antibodies at the expense of production time and cost [3,4]. Alternatively, the in vitro use of phage display monoclonal antibody libraries can dramatically decrease production time; however, due to the lack of the hypermutation maturation process, these antibodies tend to have lower binding affinity and often fail when used by cell and developmental biologists in immunofluorescence and immunohistochemistry protocols requiring multiple sample washings [5]. Due to affinity issues and the complexity of library generation and screening, in vivo methods remain the more common methodological approach.
In vivo monoclonal antibody production, however, is not without its own challenges, including the need for a purified immunogens for immunization, long production times (several months), and high cell culture demands that includes specialized media, generation of monoclonal cultures, culture expansion and subsequent frozen storage. Further, the use of chemically synthesized peptides for immunogens, although cost effective and rapidly produced, often do not adequately replicate the natively folded protein resulting in the production of antibodies that recognize only the denatured form of the protein (i.e. western blots) and are of limited utility in cell based assays [4].
A potentially viable approach to overcome the throughput limitations of in vivo antibody generation is the use of pooled or ‘shotgun’ immunizations where a single mouse is immunized with either a small panel of defined purified proteins, semi-purified protein mixtures (e.g., subproteomes), or whole cell preparations. For example, a majority of the antibody reagents available for zebrafish research were generated using tissue lysates as the immunogen. However, due to the presence of highly immunogenic glycans present in the lysates, these studies generally inefficient at producing high quality antibodies to specific targets using standard ELISA based screening methods [6]. Recent efforts have refined the approach using pools of purified protein fragments for in vivo immunization combined with ELISA based screening and recombinant DNA methods to rapidly clone the heavy and light chains into mammalian expression vectors to produce multiple high quality antibodies from a single mouse [4]. Similarly, in work to define targetable antibodies to membrane proteins associated with triple negative breast cancer, researchers immunized mice with intact breast cancer cells and were able to generate over 4,000 hybridoma colonies that were screened by FACS to identify antibodies that were able to bind MDA-MB-231 triple negative breast cancer cells [7]. In both the zebrafish and breast cancer studies, the ability to immunize with complex protein mixtures significantly increased throughput over the single immunogen approach; however, the selection of positive hybridoma clones to expand and characterize was limited by initial screening approaches that failed to differentiate antibody specificity, resulting in the selection of multiple clones that target the same protein [6,7].
High content analysis (HCA) has the potential to remove this limitation in the in vivo generation of monoclonal antibodies. The automated collection and analysis of immunofluorescent images allows the direct examination of antibody binding to target epitopes within the cellular context using protocols that mimic the expected end assay. The current generation of automated imagers can readily capture the required number of auto-focused images from thousands of hybridomas in a library in a single day. When paired with either instrument supplied or stand-alone image analysis software, the intensity and localization of an fluorescent signal at the single cell level can be rapidly determined [8–10]. Using the spatial information inherent to an image based approach, supervised machine learning methods have been developed, where classifiers are trained to recognize different protein labeling patterns [11]. Demonstrating the power of these methods, these classifiers were used to classify the subcelluar patterns observed across the proteome found in the Human Protein Atlas (HPA, www.proteinatlas.org) [12,13].
An important focus to the work done in our lab is understanding the regulation of nuclear receptors involved in human disease. Recent advances in global genome technologies has renewed interest in the manner that nuclear architecture dynamically regulates a diverse set of metabolic functions and integrated transcriptional programs. This dynamic nuclear architecture has been shown to regulate the intranuclear movement of these nuclear receptors in an ATP dependent manner [14]. Further, coregulatory proteins such as SRC-3, SMRT, class II histone deacetylases, and RIF-1 have been shown to have functional and structurally important interactions with the surrounding nuclear architecture [15–17]. Therefore, it stands to reason that understanding the protein, RNA, and DNA components that define the nuclear architecture will be key to understanding the alterations in nuclear receptor biology observed in human disease.
The nucleus, examined by electron microscopy, had two overlapping structural networks: the DNA-containing chromatin and an RNA-containing fibrogranular network. The pioneering electron microscopist Don Fawcett suggested “nuclear matrix” (NM) as a label for the RNA-containing structures [18]. This concept evolved into the understanding that nuclear RNA, far in excess of protein-coding RNA species, is central to NM structure and to nuclear architecture [19,20]. Alternative term for the non-chromatin structures of the nucleus have propagated, initially linked to different protocols to separate the structure from chromatin for ultrastructural or biochemical analysis. The most persistent of these alternatives have probably been “Nuclear Scaffold” and “Nucleoskeleton” [21,22]. Despite the development of alternative isolation protocols, the NM is a structure in the nucleus that can be seen without any isolation protocol by electron microscopy. Decades of evidence supports the idea that the most fundamental processes of nuclear metabolism are associated with the NM, including RNA transcription, RNA processing, and DNA replication [23]. However, the protein and RNA composition of the internal NM remains incompletely understood and there is a need to correct this deficiency with methods that not only identify proteins and RNAs, but also localize them in the structure at high resolution, especially in new super resolution methods and also electron microscopy.
Recently, a SILAC mass spectrometry approach identified a 2800 member nuclear proteome, of which, 272 proteins were determined to belong to a NM central proteome [25]. Interrogation of the HPA revealed that this subset of proteins were associated with nuclear body and nuclear speckle localization patterns, however, suitable antibody reagents were only available for a fraction of the identified proteins. Another recent proteomic study identified DNA repair proteins recruited to the NM following UV-induced DNA damage [26]. This is a more focused approach to NM characterization but, by being hypothesis-driven, it will miss most NM structure-function relationships. A global, non-hypothesis-driven method for identifying NM proteins, determining their localization in the structure of the nucleus, and providing tools for measuring their NM partitioning under different cell manipulations is essential for further progress. Early studies of the NM developed antibodies for use as probes of nuclear organization [27–31]. However, these were made by labor-intensive methods, often from shotgun immunizations, and they repeatedly favored immunodominant antigens due to the screening methodology used. The production of libraries of monoclonal antibodies covering the entire nuclear proteome would be a profoundly important advance.
In this chapter, we describe a reverse proteomics shotgun method coupled with HCA and supervised machine learning to generate a panel of monoclonal antibodies to a variety of targets associated with the NM. Using a single mouse immunized with a purified NM fraction, a hybridoma library was generated and screened using an image-based approach analyzed using a highly customized, web-based software platform. Positive clones were selected using a combination of a rules-based-approach and a learned classifier. We describe the training of this recursive partitioning classifier using a restricted set of metrics defined by screening data and images obtained from the HPA. And finally, we demonstrate how application of the HPA trained classifier can be applied to the hybridoma image set to define distinct antibody binding patterns and define hits of interest to be further evaluated.
2. Methods
2.1. Cell Culture
HeLa cells (cervical cancer, ATCC) were cultured in DMEM media supplemented with 5% fetal bovine serum (FBS, Gemini Bio-Products, West Sacramento, CA) at 37°C in a 95% air/5% CO2 atmosphere.
2.2. NM Preparation
With minor modifications, NM isolation was performed according to Nickerson et al. and performed at 4°C unless otherwise specified. [19]. HeLa cells were seeded into 10 cm dishes (PureCoat Carboxyl, Corning, Corning, NY) at a density ranging from 1.5 to 5 million cells per dish and allowed to adhere overnight under normal growth conditions. Cells were first washed with cold PBS (with Ca++/Mg++) and soluble proteins removed by incubation with cytoskeletal buffer (CSK: 10 mM Pipes, pH 6.8/300 mM Sucrose/100 mM NaCl/3 mM MgCl2/1 mM EGTA/20 mM vanadyl riboside complex (NEB)/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 tablet/10 mls Complete Mini EDTA-Free Protease Inhibitor Cocktail tablet) supplemented with 0.5% Triton X-100 for 5 minutes. CSK buffer was gently aspirated and DNA and its associated proteins digested by incubating cells with 400 units/ml DNase I (RNase-free) in digestion buffer (DIG: 10 mM Pipes, pH 6.8/300 mM sucrose/50 mM NaCl/3 mM MgCl2/1 mM EGTA/20 mM vanadyl riboside complex(NEB)/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 tablet/10 mls Complete Mini EDTA-Free Protease Inhibitor Cocktail tablet) for 1 hour at 32°C. To further extract the digested DNA, the DNase/DIG solution was diluted 1:4 with DIG buffer and 1 M ammonium sulfate in DIG buffer was added drop-wise over a period of minutes with constant agitation to a final concentration of 0.25 M, and then incubated for an additional 5 minutes. A modified second extraction was performed by adding 4 M NaCl in DIG buffer drop-wise to a final concentration of 2 M and incubated for another 5 minutes. All buffers were then carefully aspirated from cells, and then washed once with DIG buffer. Collectively, these steps remove > 95% of cellular proteins and > 95% of DNA, leaving behind the “NM-Intermediate Filament complex.” DIG buffer was replaced with fresh buffer and remaining cellular material scraped and collected into 1.5 ml microcentrifuge tubes. The preparation was then pelleted, washed by suspending in PBS supplemented with protease inhibitors and then re-pelleted. The majority of the PBS buffer was removed prior to freezing at −80°C.
To monitor the quality of DNA removal, poly-D-lysine coated coverslips were included in each 10 cm dish and removed prior to cell scraping. Coverslips were fixed by incubating in a 4% paraformaldehyde solution prepared in PEM buffer (PEM: 80 mM Pipes, pH 6.8/5 mM EGTA, pH 7.0/2 mM MgCl2) for 20 minutes. Coverslips were washed twice with PEM buffer and then DNA/RNA stained by incubating in a 1 μg/ml propridium iodide solution (PI, in PEM) for 5 minutes followed by DNA staining with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, in PEM) for 5 minutes. Coverslips were washed once in PEM and then mounted to glass slides in antifade (SlowFadeGold™, Molecular Probes) and examined by fluorescent microscopy. Nuclei were visualized by PI labeling of RNA present and removal of DNA confirmed by absence of DAPI signal. Samples with a detectable DAPI signal were discarded.
2.3. In Vivo Antibody Generation
A total of 70 NM pellets were thawed and pooled into 2 ml of cold PBS. The entire sample was placed into a glass-glass Dounce homogenizer at 4°C and homogenized into an evenly dispersed suspension. Aliquots of the suspension were analyzed by quantitative Coomassie Blue SDS gels to determine the size distribution of NM proteins and protein concentration was estimated by comparison with BSA standards on SDS gels. By SDS PAGE the NM contains a broad distribution of proteins between 20–200 kDa. A separate Bradford assay was also performed to determine total protein concentration of the NM suspension. The protein concentration was estimated to be 3.8 mg/ml (total protein in 2 ml was 7.62 mg).
A total of three six-week old Balb/c mice were immunized subcutaneously with 400 μg of the NM suspension mixed with complete Fruend’s adjuvant. At three weeks following the initial injection, mice were immunized a second time using an intraperitoneal injection of 400 μg of the NM suspension combined with incomplete Freund’s adjuvant. A final immunization occurred at 6 weeks consisting of a subcutaneous injection of 400 μg of the NM suspension combined with incomplete Freund’s adjuvant. The immunized mice were tested for an antibody response by collecting serum and examining immunofluorescent labeling in HeLa cells as described below. The mice were given a final immunization without adjuvant 4 days before the spleen was removed. Splenocytes (108) were fused to SP2/0 myeloma cells (107) in 50% PEG 1500 (Roche, Hertfordshire, UK) using standard procedures. The hybridoma mixture was plated over twelve 96-well plates and supernatants were harvested after 10–15 days for screening. All animal work performed by the Monoclonal Antibody/Recombinant Protein Expression Shared Resource at Baylor College of Medicine and approved by the Institutional Animal Care and Use Committee ((IACUC number: 15-0008-S1A0).
2.4. Immunofluorescence (IF) Sample Processing
For direct visualization of antibody binding, mouse bleeds or subsequent hybridoma supernatants were evaluated by IF using paraformaldehyde-fixed Hela cells. Unless otherwise stated, all steps were performed at room temperature and two PBS (with Ca++/Mg++) washes were performed between each step. HeLa cells were seeded into 384-well microtiter plates at a plating density of 2,000 cells/well in 25 ul of growth media supplemented with regular FBS. After cells had adhered and grown overnight, assay plates were washed with PBS and fixed using a 4% solution of electron-microscopy-grade paraformaldehyde (PFA) in PEM buffer solution for 25 minutes at room temperature. Auto-fluorescence was quenched using a 0.1 M NH4Cl (in PEM) solution for 10 minutes. Cell membranes were disrupted using a 0.5% Triton-X100 detergent (in PEM) solution for 30 minutes. Non-specific binding was blocked using a 5% powdered milk solution (Blotto, prepared in TBS-T buffer) for 30 minutes. For initial characterization of bleeds collected from the immunized mice, blocking solution was removed and replaced with Blotto containing serum dilutions ranging from 1:200 to 1:400,000. For screen of hybridoma supernatants, blocking solution was removed and replaced with hybridoma supernatants (approximately a 1:1.5 dilution in blotto). Samples were incubated at 37°C for 1 hour. After washing three times using TBS-T buffer, a secondary antibody solution containing a 1:2000 dilution of anti-Ms IgG A647 (Molecular Probes) was added to wells and incubated for 1 h at room temperature. Samples were washed 4 times with TBS-T and post-fixed by incubating with a 4% PFA (in PEM) solution for 10 minutes. Nuclei were labeled using a 1 μg/ml DAPI (in TBS-T) solution for 10 minutes, washed, and plates sealed to minimize the evaporation of the remaining PBS buffer. All plate processing was performed using a Biomek NX liquid handling platform equipped with an AP96 96-channel pipettor.
2.5. Image Collection
IF samples were imaged using an epifluorescent IC100 image cytometer (Beckman Coulter) equipped with a 40X/0.9 NA S-fluor objective. A total of 16 fields were imaged per well with DAPI and A647 signals captured. Due the high range of signal intensities observed in the A647 channel, a short and long exposure image were captured and later merged in the image analysis software. All images were stored as an uncompressed 8-bit grayscale bitmap images with data from each channel saved as separate files.
Reference confocal images where retrieved from the HPA (www.proteinatlas.org) using an algorithm designed within Biovia Pipeline Pilot (PLP) software package. Using the list of gene names associated with the 2381 proteins identified as members of the nuclear proteome [25], the algorithm accesses the Ensembl database to retrieve the Ensembl gene ID for each protein. The Ensembl gene ID is then used to retrieve the HPA XML data file associated with that protein containing the URL for each IF image file describing the subcellular localization of the protein. Images are subsequently downloaded as 24-bit RGB JPEG images, split into individual channels, and stored as uncompressed 8-bit grayscale TIF images with individual files for each data channel. Of the 2381 proteins identified, 808 images were retrieved from the HPA for comparison with hybridoma screening data.
2.6. Image Analysis
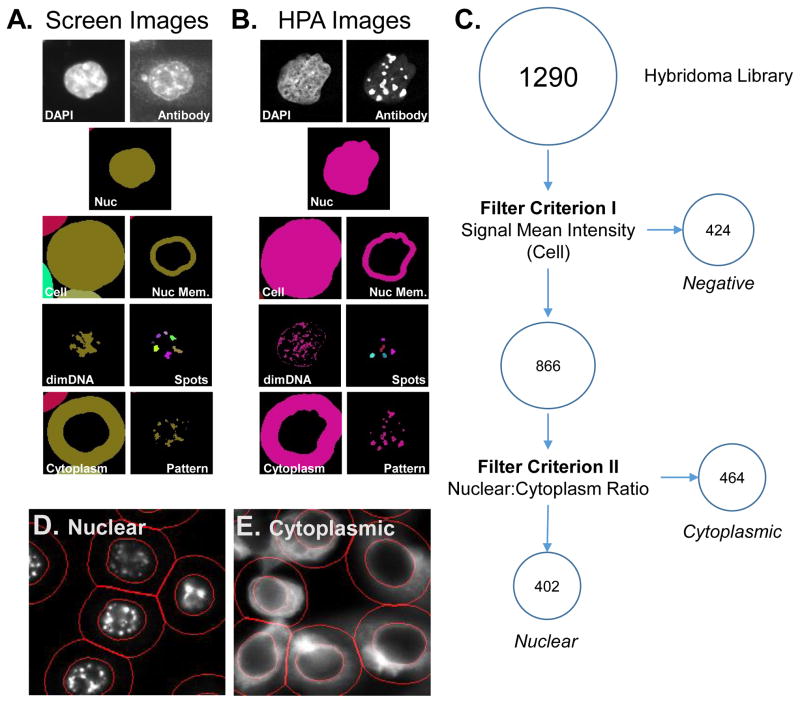
All images were analyzed using the myImageAnalysis web application which is based upon the Biovia PLP software package with a total of 7 regions defined for each cell. All images were background corrected using a rolling ball algorithm with a neighborhood radius corresponding to ¼ of the image size. For hybridoma screening data, short and long exposure A647 images were merged into a single integer image with an expanded pixel value range. DAPI Images were used to define the nuclear (1, Nuc) region of each cell. To account for differences in magnification used during imaging, HPA images were resized so that the median nuclear area equaled the median nuclear area observed in the hybridoma screening data. A cell (2, Cell) region was defined by tessellation, with an expansion radius equal to the median nuclear diameter. The cytoplasmic (3, Cytoplasm) region was defined as the difference between the cell and nuclear regions for each object. A nuclear membrane (4, NucMem) region was defined by dilating and eroding the nuclear region a distance equal to 1/10th of the nuclear diameter. The DAPI staining images were also used to determine the DNA poor (5, dimDNA) regions in each nucleus by defining those pixels with a signal intensity 0.25 median absolute deviations (MAD) from the median DAPI signal measured per nucleus. Nuclear spots (6, Spots) in the A647/antibody channel were detected using a K-means segmentation algorithm on pixel intensities located within the nuclear region. Finally, a general pattern (7, Pattern) mask was determined corresponding to the brightest 5% of pixels in the A647/antibody image within the nuclear region. Examples of the identified regions are shown in Figure 2A–B.
Figure 2. Workflow for HCA analysis of image data sets associated with nuclear matrix hybridoma screening.
The hybridoma screening (A) or HPA reference set (B) image sets were analyzed using the myImageAnalysis application to identify nuclear (Nuc), cellular (cell), nuclear membrane (Nuc Mem), cytoplasmic (Cytoplasm) regions and DNA poor regions (dimDNA) from the DAPI images for each cell. In addition, the antibody/hybridoma signal was used to define bright spots (spots) and a general pattern (pattern) masks. (C) A schematic overview of a criterion-based strategy used to initially characterize the labeling observed with hybridoma supernatants. The mean pixel intensity per cell (Criterion I)was used to define positive and negative clones. The ratio between nuclear mean pixel intensity to cytoplasmic mean pixel intensity (Criterion II) was used to define nuclear and cytoplasmic clones. Example images of pattern classified as nuclear (D) or cytoplasmic (E) using the above criterion. Red lines indicated identified nuclear and cell regions.
2.7. Feature Extraction and Selection
Various features from the segmented cell regions were collected from each cell. Basic intensity metrics were used to determine antibody binding intensity in the A647/antibody channel and key ratios including the nuclear: cytoplasmic ratio and the nuclear membrane: nuclear ratio of signal intensity. Shape statistics were collected for the pattern region and included solidity, Euler number, and extent. Intensity statistics were collected from the Nuc, dimDNA, and Pattern regions using an intensity normalized A647/antibody image and included features such as pixel kurtosis, entropy, and Hu moments. Finally, Haralick texture features of the nuclear A647/antibody signal were calculated at multiple scales as these features have been useful in subcellular pattern recognition in the past [11].
2.8. Generation of Classification Model
For developing the classification model, relative standard deviation (RSD) of each feature observed in the hybridoma screening data set and the HPA reference image set were evaluated and only those features with similar RSD values were used for model training.
Based upon methods published previously [11], a random partitioning/forest (RF) classifier framework was implemented. The RF approach is less sensitive to individual training samples, inherently performs feature selection, and has been shown to have greater accuracy when used to classify HPA images. We used 200 trees with each tree using 11 features at each decision node. The output of the RF model is a classifier, a series of discriminative features, and the probabilities that each sample belongs to each class. Classification was implemented using version 4.5–30 of the randomForest package found in the R GNU project and accessed through PLP.
2.9. Western Blot Assay
Protein samples from HeLa cells grown in 10-cm dishes were solubilized in NP-40 extraction buffer, separated on a 4–12% Bis-TRIS gradient gel, using MOPS running buffer (Life Technologies, Corning, NY), and transferred to PVDF membrane using a Trans-Blot SD electrophoretic transfer cell (Biorad, Hercules, CA) using standard protocols. Hybridoma supernatants were used in concentrations ranging between undiluted and 1:5. Secondary HRP-coupled antibodies (Biorad, Hercules, CA) were used for detection with a commercial ECL solution (Western Lightning, Perkin Elmer, Waltham, MA).
3. Results
3.1. In vivo Generation of NM Antibodies
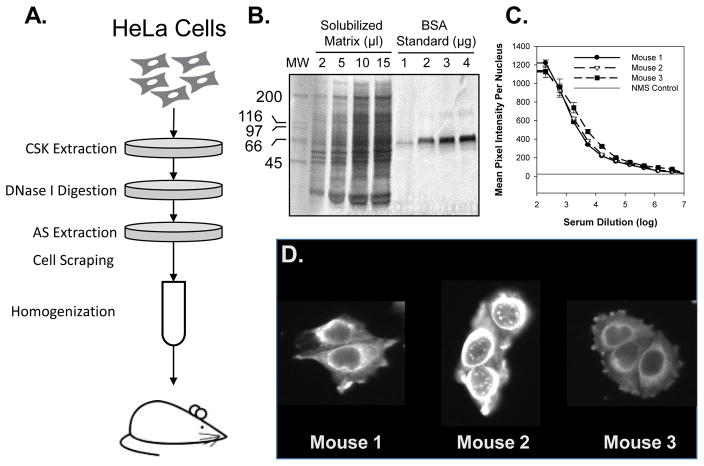
NM fractions were prepared using a modified version of the protocol according to Nickerson et al. (Figure 1A) It was necessary to adapt the method to use dishes as opposed to test tubes due to the occasional tendency of the HeLa extract material to clump prior to/during DNase I digestion. Following homogenization of the NM fraction, Coomassie Blue SDS gel analysis demonstrates a complex protein fraction containing proteins ranging in size from 20 – 200 kDa. (Figure 1B)
Figure 1. In vivo strategy for producing nuclear matrix antibody panel.
(A) A schematic outline of nuclear matrix preparation prior to mouse immunization. Soluble proteins in HeLa cells plated onto 10 cm dishes were extracted by use of CSK extraction buffer. The extracted cells were subsequently digested with Dnase I followed by extraction with 250 mM ammonium sulfate (AS). Nuclear matrix fraction was collected by scraping and homogenized prior to mouse injection. (B) Quantitative Coomassie Blue SDS gel of homogenized nuclear matrix demonstrates a complex protein fraction with proteins ranging between 20-200kDa and an estimated protein concentration of 3.8mg/ml. (C) HeLa cells were processed for immunofluorescence analysis using a titration of mouse serum at dilutions ranging between 1:200 to 1:400,000. Samples were imaged and intensity of labeling quantified relative to control mouse serum. (D) Typical labeling pattern observed with each mouse serum tested.
Following three cycles of immunizations, the response of each mouse was determined using IF analysis of collected mouse bleeds. We observed a strong response in all mice with detected signal above background at dilutions greater than 1:200,000. The intensities of observed binding were similar between immunized mice and the calculated EC50 titer ranged between 1:1,500 and 1:3,000. (Figure 1C) Direct visual inspection of the labeling pattern demonstrated differences between the mice responses with mouse 2 generating a distinct nuclear pattern, demonstrating the advantages of an image based approach even at this early point in a project designed to generate imaging compatible antibodies. (Figure 1D) These results indicate that immunization with a complex protein mixture results in strong immune response from each mouse that can be differentiated by direct visual inspection of the IF pattern observed when target cells are labeled with collected serum.
3.2. Criterion-Based Analysis of Immunofluorescent Signal
We initially classified the labeling patterns observed with hybridoma supernatants from a library generated from mouse 2 using a rules/criterion-based method. Using the myImageAnalysis web application, images were segmented into three key regions, a nuclear region, a cell region, and a cytoplasmic region (Figure 2A). In parallel, we applied the same image analysis algorithm to the HPA image set corresponding to nuclear and NM proteins (Figure 2B). Positive labeling by the supernatant was determined by a threshold of a cellular mean pixel intensity 3-fold above the level observed with naïve control normal mouse serum samples (Criterion I). This threshold was set by manual inspection of the images and resulted in the identification of 866 (61.7%) supernatants with positive labeling. A second threshold, in which a nuclear to cytoplasmic ratio of mean pixel intensity of at least 3 was used to define supernatants with a predominant nuclear labeling pattern, was used to further divide the positive supernatants. This resulted in 402 (46.4%) of the positive supernatants being classified as nuclear in pattern. (Figure 2C) As expected, manual inspection of the images following classification confirmed that a rules/criterion based classification method was able to accurately define basic populations of responses. (Figure 2D–E)
3.3. RF Classification Model for Nuclear Patterns
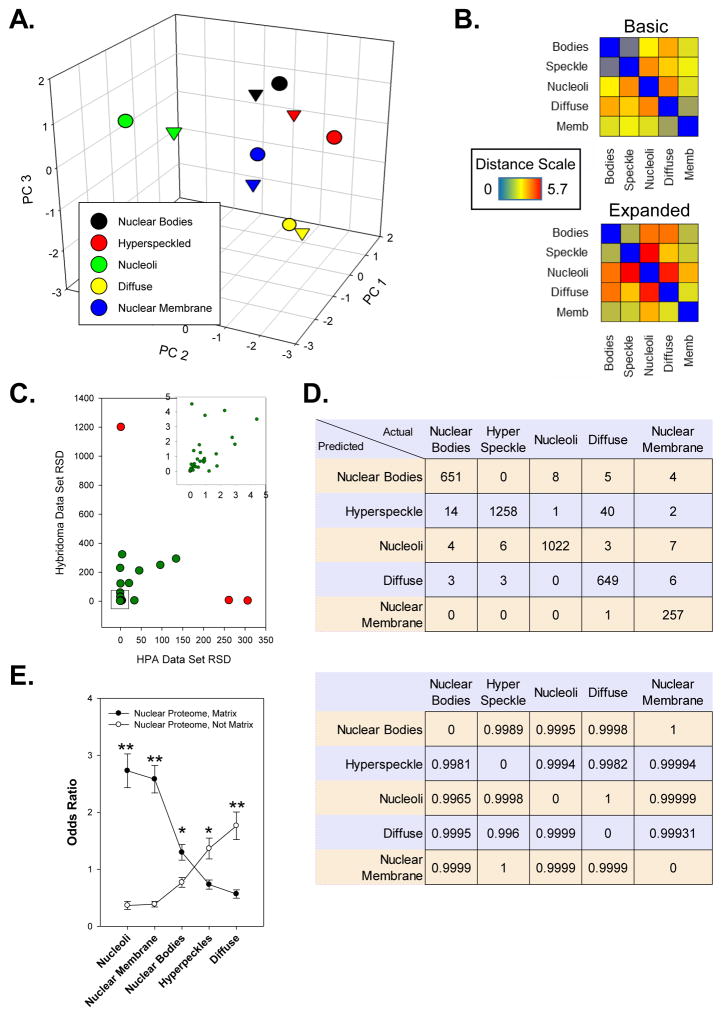
Next, we next developed a RF model to further classify nuclear patterns observed with hybridoma supernatant labeling. Initially, we used HPA reference images to generate a training set of approximately 4000 individual cells manually classified by 3 independent scorers into 5 classes: nucleoli, nuclear bodies (including Cajal bodies, PML bodies, and splicing speckles), nuclear membrane, nuclear hyperspeckles, and nuclear diffuse patterns. For inclusion into the training set, two or more scorers must agree on the pattern observed. Principle component analysis (PCA) was used to reduce the 25 features (Supplement Table 1) extracted from the nuclear region of each cell into 3 principle components (Supplement Table 3). When summarized for each class and visualized, each nuclear pattern class occupied a distinct neighborhood (Figure 3A, “▽” symbols, Supplement Figure 1). We next modified the image analysis to include the identification of 4 additional regions/masks in the HPA and hybridoma screening images: dimDNA, nuclear membrane, nuclear spots, and nuclear pattern masks (Figure 2A–B). Inclusion of these masks expanded our panel of features to 60 (Supplement Table 2) and subsequent PCA analysis demonstrated variation in the data set as percentage of variance explained by three principle components decreased from 75% to 58%. When visualized, further separation between pattern classes is seen (Figure 3A, “○” symbols, Supplement Figure 1). When a distance matrix is calculated, we observed increased distance (p < 0.05) between classes with the expanded panel of features with the exception of the nuclear membrane: nuclear body parings (p = 0.23, Figure 3B), suggesting that the additional metrics derived from the added regions added meaningful variance to the data set.
Figure 3. Development of a random forest machine learning model to classify nuclear labeling patterns.
(A) A 3D scatter plot shows the principle component analysis of each nuclear pattern class identified in the HPA traning set of images.. (▽ = 3 cellular regions used; ○ = 7 cellular regions used) (B) Distance matrix showing the distance between each class after principle component analysis using either 3 (basic) or 7 (expanded) regions. (C) A scatter plot of the relative standard deviations (RSD) observed with collected features in the HPA and hybridoma image sets. Red indicates features excluded and not used for model generation. Inset represents expanded view of data contained in the indicated box. (D) Confusion matrix (top) and pair-wise ROC matrix of generated random forest model. (E) Odd ratios of patterns observed when random forest model is used to classify patterns observed in HPA nuclear proteome image data set. Error bars represent standard error with “**” indicating a p-value < 0.01 and “*” indicating a p-value < 0.05.
We next determined the relative standard deviation (RSD) for the panel of 60 features in cells from both the HPA (N = 68,732) and hybridoma screening (N = 97,698) image sets (Figure 3C). This was done to exclude metrics with a high degree of variation in only one of the two data sets, thereby skewing the classification model inappropriately. This was important due to the different types of instruments used to collect the HPA reference images and the hybridoma screening images. Of the 60 features, only 3 were found to have a RSD difference greater than 250 and thereby excluded from the feature list prior to model generation. The excluded metrics were:
INTENSITY_Pattern_mask_ch01_norm_HuMoment_5,
INTENSITY_Pattern_mask_ch01_norm_HuMoment_6, and
INTENSITY_dimDNA_mask_ch01_norm_Kurtosis
The first two metrics are based on the Hu set of pixel intensity moments that are invariant under translation and changes in object scale and rotation. The third metric is a measure of pixel intensity distribution. It is likely that these metrics remain sensitive to the differences in object resolution and contrast differences between HPA images and screening images despite attempts to scale HPA resolutions to match that seen in the screening image set.
Using the recursive partitioning forest (RF) classification model components within PLP, we generated a classification model based on the HPA derived training set. The overall accuracy using the restricted panel of 57 features was 97.3% with class accuracies ranging from 95.7–99.7% (Figure 3D, top). Pairwise analysis of receiver operating characteristic (ROC) scores demonstrate high model performance with scores exceeding 0.99 (Figure 3D, bottom).
We next used the RF model to classify the 68,732 cells within the HPA image set. For statistical analysis, the cells were randomly split into 5 groups. Interestingly, when the odds ratio of each nuclear pattern was calculated, a clear bias was observed (Figure 3E). For proteins identified as part of the NM proteome, the odds of either a nucleoli (p < 0.01), nuclear membrane (p < 0.01), and nuclear bodies (p < 0.05) pattern were increased relative to the other pattern classes. In contrast, for proteins associated with the nuclear protein proteome but not the NM, the nuclear hyperspeckles (p < 0.05) and nuclear diffuse (p < 0.01) patterns were more common. This is consistent to what others have observed by manual analysis and suggest that identifying hybridoma supernatants with nucleoli or nuclear membrane patterns would be more suggestive of a target associated with the NM [25].
3.4. RF Classification of Hybridoma Supernatant Labeling Patterns
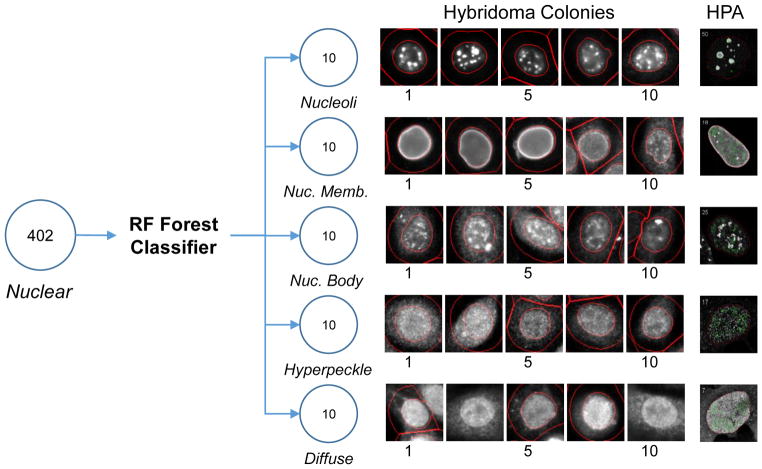
We next used the RF model to classify cells from the 402 hybridoma supernatant samples there were classified as nuclear using a criterion-based method. Due to the inherent differences in images from different types instruments used to collect the HPA images used to train the model, and those obtained from the hybridoma screening, the primary use of the RF model is to define the hybridoma supernatants most likely to belong to each labeling pattern. This is consistent with the primary goal of screening the hybridoma library, rapidly defining a diverse subset of antibodies that function in a standard IF labeling protocol, then followed by culture expansion and further characterization of only a subset of the initial library.
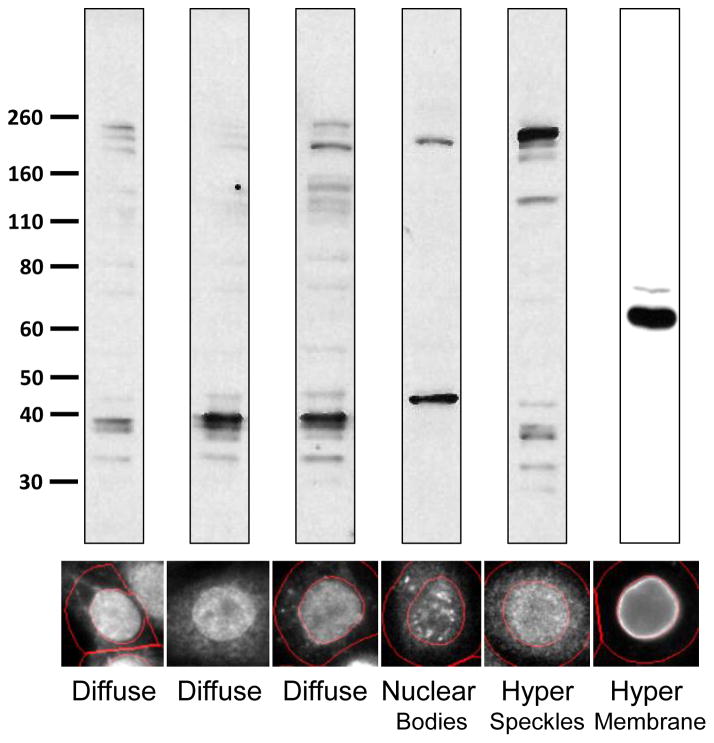
Using this approach, the probability of each cell belonging to each pattern class is determined and averaged for all cells belonging to a single hybridoma sample. The samples were then ranked ordered for each pattern class and the top 10 were classified as members of that pattern class (Figure 4). Inspection of the images by independent scorers confirmed that the pattern classification of these cells demonstrated 100% accuracy indicating a larger number of samples could be retained with minimal loss in accuracy if desired. Due to the bias in labeling patterns observed with NM proteins in the HPA image set, hybridoma cultures with supernatants that produced a labeling pattern belonging to the nucleoli, nuclear membrane, and nuclear body pattern classes were favored for further expansion and characterization. Consistent with the idea that differing patterns represents different target specificity, western blot analysis of hybridoma supernatants with a similar labeling pattern by IF tend to share predominant bands whereas supernatants with different IF labeling patterns produce bands of differing sizes (Figure 5).
Figure 4. Classification of hybridoma supernatants with nuclear labeling using RF model.
After criterion classification of a nuclear distribution, hybridoma supernatant labeling pattern was further sub-divided using a RF forest medel trained using images from the Human Protein Atlas (HPA). The top 10 supernatants for each pattern were identified and cell image shown. Number indicates assigned rank. For reference, a cell image from the HPA used for training is shown at the far right.
Figure 5. Western Blot Analysis of Selected Hybridoma Supernatants.
After single cell cloning and expansion of hybridoma cultures, hybridoma supernatant labeling was observed for selected clones using cell extracts prepared from HeLa cells. A representative image of the observed IF labeling from the initial screen is shown below.
4. Conclusions
In summary, these results demonstrate in vivo antibody generation using enriched protein mixtures, single mouse injection, and a HCA screening approach coupled with a machine learning algorithm was highly successful in identifying multiple hybridomas producing imaging compatible antibodies that are specific for distinct nuclear structures including nucleoli, nuclear membranes, nuclear bodies, nuclear hyperspeckles and diffuse nucleoplasm. (Figure 4) While it remains unknown exactly how many distinct antibodies to unique antigens were generated, based upon IF staining pattern we suspect there are many (i.e. dozens). With the panel of antibodies in hand, it will now be possible to capture and identify antigens by mass spectrometry sequencing and to better define the protein composition of the NM and other nuclear structures. Since the antibodies are all qualified for cell imaging based assays, they provide a potential wealth of new reagents to detect biomarkers of the nuclear matrix under different experimental conditions.
In addition, adoption of a HCA approach to hybridoma screening is highly complementary to existing efforts to maximize the efficiency of in vivo antibody generation. HCA allows for the miniaturization of the process using multiwell assay plates and the high sensitivity of the image based approach allows for earlier hybridoma screening which reduces time of production and reagent use. The ability to multiplex monoclonal antibody generation from a single animal and cell fusion is well suited to be combined with reported methods that allow for the rapid recombinant cloning of the antibody variable regions from hybridoma cultures and microprinting of protein arrays that allow for the rapid identification of antibody binding targets [4,32]. The ability of HCA to define distinct hybridomas in primary and secondary characterization would only further increase the efficiency of these methods.
The ability to use the HPA to retrieve reference images to train a machine learning algorithm has several important advantages. Most importantly, HPA images resolve the conflict between the time needed to generate a high quality annotated image set to train the machine learner and the need to rapidly obtain results to direct further hybridoma culture work. For this project, scorers required 5–8 hours to classify the 4,000 cells used in the training set and it was observed that the chance of discrepancy between scorers increased with viewing/scoring time (data not shown), suggesting fatigue may alter the results. By using HPA images as opposed to control images obtained during the screening process, manual image annotation can occur prior to screening activity and in multiple short sessions. Second, a robust classification model can score an entire hybridoma library at the single cell level within minutes where manual classification would take days, if not weeks, and would not be compatible with the hybridoma process. Further, when combined with proteomic data, the HPA image set can define distinct labeling pattern that will facilitate the identification of antibodies recognizing unknown members of complex biological interaction networks. Finally, the diversity of images in the HPA, including multiple cell types for each protein, results in a more robust classification model.
As our ability to survey the genomic and proteomic landscape continues to increase, the need for high quality monoclonal antibodies will continue to arise. This is particularly true in the context of using monoclonal antibodies under challenging conditions, including fluorescent in situ hybridization for nuclei acids (DNA, mRNA, lncRNA, miRNA, etc), and also in highly cross-linked samples for the best ultrastructure in super resolution light microscopy and immunogold electron microscopy applications. While it is impossible that the traditional monoclonal antibody approach of “one antigen-one animal” for in vivo antibody generation will be able to meet this demand, the combining of pooled animal immunizations with HCA and machine learners to classify observed patterns may remove obstacles to increasing throughput via the generation of multiple high quality antibodies through parallel selection.
Supplementary Material
Supplement Figure 1. Principle component analysis of nuclear pattern classes using basic and expanded region metrics. Three 2D scatter plots for each principle component pairs showing the values of each nuclear pattern class identified in the HPA traning set of images.. (▽ = 3 cellular regions used; ○ = 7 cellular regions used). Error bars indicate standard deviation about class mean value.
Acknowledgments
The authors would like to thank Kurt Christensen, Karen Moberg and Celetta Callaway for their contributions to the hybridoma work required for the experiments presented in this chapter. This project was supported by the Monoclonal Antibody/Recombinant Protein Expression Shared Resource at Baylor College of Medicine with funding from NIH Cancer Center Support Grant P30 CA125123 as well by the Integrated Microscopy Core at Baylor College of Medicine with funding from the NIH (HD007495, DK56338, and CA125123), the Dan L. Duncan Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics. This work was also supported by the Diana Helis Henry Medical Research Foundation (MAM) through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine. Work was also supported by NIEHS grants NIEHS R01 (1R01ES023206-01; Cheryl L. Walker, Bert W. O’Malley, M.A.M. and Mark T. Bedford), and NIEHS P30 (ES023512-01; Center of Excellence in Environmental Health, C.L.W.). ATS is a K12 Scholar supported by NIH grant K12DK0083014, the multidisciplinary K12 Urologic Research (KURe) Career Development Program awarded to Dr. Dolores J. Lamb.
ABBREVIATIONS
- HCA
High Content Analysis
- HPA
Human Protein Atlas
- IF
Immunofluorescence
- NM
Nuclear Matrix
- PCA
Principle Component Analysis
- PLP
Pipeline Pilot
- RF
Random Partitioning/Forest
- RSD
Relative Standard Deviation
References
- 1.Goldman RD. Antibodies: indispensable tools for biomedical research. [accessed July 12, 2015];Trends Biochem Sci. 2000 25:593–5. doi: 10.1016/s0968-0004(00)01725-4. http://www.ncbi.nlm.nih.gov/pubmed/11116184. [DOI] [PubMed] [Google Scholar]
- 2.Taussig MJ, Stoevesandt O, Borrebaeck CAK, Bradbury AR, Cahill D, Cambillau C, et al. ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods. 2007;4:13–7. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 3.KÖHLER G, MILSTEIN C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 4.Crosnier C, Staudt N, Wright GJ. A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biol. 2010;8:76. doi: 10.1186/1741-7007-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple SD, Clarke KF, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- 7.Rust S, Guillard S, Sachsenmeier K, Hay C, Davidson M, Karlsson A, et al. Combining phenotypic and proteomic approaches to identify membrane targets in a “triple negative” breast cancer cell type. Mol Cancer. 2013;12:11. doi: 10.1186/1476-4598-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcelli M, Stenoien DL, Szafran AT, Simeoni S, Agoulnik IU, Weigel NL, et al. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem. 2006;98:770–88. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- 9.Ashcroft FJ, Newberg JY, Jones ED, Mikic I, Mancini Ma. High content imaging-based assay to classify estrogen receptor-α ligands based on defined mechanistic outcomes. Gene. 2011;477:42–52. doi: 10.1016/j.gene.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udono M, Kadooka K, Yamashita S, Katakura Y. Quantitative analysis of cellular senescence phenotypes using an imaging cytometer. Methods. 2012;56:383–388. doi: 10.1016/j.ymeth.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Newberg JY, Li J, Rao A, Pontén F, Uhlén M, Lundberg E, et al. Automated analysis of human protein atlas immunofluorescence images. Proc - 2009 IEEE Int Symp Biomed Imaging From Nano to Macro ISBI. 2009;2009:1023–1026. doi: 10.1109/ISBI.2009.5193229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlén M, Björling E, Agaton C, Szigyarto CAK, Amini B, Andersen E, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Xiong L, Schneider J, Murphy RF. Protein subcellular location pattern classification in cellular images using latent discriminative models. Bioinformatics. 2012;28:32–39. doi: 10.1093/bioinformatics/bts230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuda K, Nishi M, Takaya H, Kaku N, Kawata M. Intranuclear mobility of estrogen receptor alpha and progesterone receptors in association with nuclear matrix dynamics. J Cell Biochem. 2008;103:136–48. doi: 10.1002/jcb.21393. [DOI] [PubMed] [Google Scholar]
- 15.Amazit L, Pasini L, Szafran AT, Berno V, Wu RC, Mielke M, et al. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol. 2007;27:6913–32. doi: 10.1128/MCB.01695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino H, Nishino TG, Tashiro S, Miyazaki M, Ohmiya Y, Igarashi K, et al. Co-repressor SMRT and class II histone deacetylases promote Bach2 nuclear retention and formation of nuclear foci that are responsible for local transcriptional repression. J Biochem. 2007;141:719–27. doi: 10.1093/jb/mvm073. [DOI] [PubMed] [Google Scholar]
- 17.Li HJ, Haque ZK, Chen A, Mendelsohn M. RIF-1, a novel nuclear receptor corepressor that associates with the nuclear matrix. J Cell Biochem. 2007;102:1021–35. doi: 10.1002/jcb.21340. [DOI] [PubMed] [Google Scholar]
- 18.Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–45. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- 19.Nickerson Ja, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci U S A. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson JA, Krochmalnic G, Wan KM, Penman S. Chromatin architecture and nuclear RNA. [accessed August 20, 2015];Proc Natl Acad Sci U S A. 1989 86:177–81. doi: 10.1073/pnas.86.1.177. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=286427&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. [accessed June 10, 2015];Cell. 1984 39:223–32. doi: 10.1016/0092-8674(84)90208-3. http://www.ncbi.nlm.nih.gov/pubmed/6091913. [DOI] [PubMed] [Google Scholar]
- 22.Jackson DA, Cook PR. Transcription occurs at a nucleoskeleton. EMBO J. 1985;4:919–25. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson DA, Cook PR. Transcription occurs at a nucleoskeleton. [accessed August 20, 2015];EMBO J. 1985 4:919–25. doi: 10.1002/j.1460-2075.1985.tb03719.x. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=554280&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickerson J. Experimental observations of a nuclear matrix. [accessed August 20, 2015];J Cell Sci. 2001 114:463–74. doi: 10.1242/jcs.114.3.463. http://www.ncbi.nlm.nih.gov/pubmed/11171316. [DOI] [PubMed] [Google Scholar]
- 24.Berezney R, Coffey DS. Identification of a nuclear protein matrix. [accessed June 29, 2015];Biochem Biophys Res Commun. 1974 60:1410–7. doi: 10.1016/0006-291x(74)90355-6. http://www.ncbi.nlm.nih.gov/pubmed/4214419. [DOI] [PubMed] [Google Scholar]
- 25.Engelke R, Riede J, Hegermann J, Wuerch A, Eimer S, Dengjel J, et al. The Quantitative Nuclear Matrix Proteome as a Biochemical Snapshot of Nuclear Organisation. J Proteome Res. 2014 doi: 10.1021/pr500218f. [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Quaresma aJC, Nickerson Ja, Green KM, Shaffer Sa, Imbalzano aN, et al. Subnuclear domain proteins in cancer cells support the functions of RUNX2 in the DNA damage response. J Cell Sci. 2015;128:728–740. doi: 10.1242/jcs.160051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. [accessed August 20, 2015];Genes Dev. 1998 12:996–1009. doi: 10.1101/gad.12.7.996. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=316672&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. [accessed August 20, 2015];J Cell Biol. 1994 127:593–607. doi: 10.1083/jcb.127.3.593. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2120221&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickerson JA, Krockmalnic G, Wan KM, Turner CD, Penman S. A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles, and midbodies. [accessed August 20, 2015];J Cell Biol. 1992 116:977–87. doi: 10.1083/jcb.116.4.977. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2289346&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan KM, Nickerson JA, Krockmalnic G, Penman S. The B1C8 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. [accessed August 20, 2015];Proc Natl Acad Sci U S A. 1994 91:594–8. doi: 10.1073/pnas.91.2.594. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=42995&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaly N, Sabour MP, Silver JC, Aitchison WA, Little JE, Brown DL. Monoclonal antibodies against nuclear matrix detect nuclear antigens in mammalian, insect and plant cells: an immunofluorescence study. [accessed August 20, 2015];Cell Biol Int Rep. 1986 10:421–8. doi: 10.1016/0309-1651(86)90037-8. http://www.ncbi.nlm.nih.gov/pubmed/3527452. [DOI] [PubMed] [Google Scholar]
- 32.Staudt N, Müller-Sienerth N, Wright GJ. Development of an antigen microarray for high throughput monoclonal antibody selection. Biochem Biophys Res Commun. 2014;445:785–790. doi: 10.1016/j.bbrc.2013.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Figure 1. Principle component analysis of nuclear pattern classes using basic and expanded region metrics. Three 2D scatter plots for each principle component pairs showing the values of each nuclear pattern class identified in the HPA traning set of images.. (▽ = 3 cellular regions used; ○ = 7 cellular regions used). Error bars indicate standard deviation about class mean value.