Abstract
Vaginal lactobacilli can inhibit colonization by and growth of other bacteria, thereby preventing development of bacterial vaginosis (BV). Amongst the lactobacilli, Lactobacillus crispatus appears to be particularly effective at inhibiting growth of BV-associated bacteria. Nonetheless, some women who are colonized with this species can still develop clinical BV. Therefore, we sought to determine whether strains of L. crispatus that colonize women with lactobacilli-dominated vaginal microbiomes are distinct from strains that colonize women who develop BV. The genomes of L. crispatus isolates from four women with lactobacilli-dominated vaginal microbiomes ( < 1 % 16S rRNA reads above threshold from genera other than Lactobacillus) and four women with microbiomes containing BV-associated bacteria (>12 % 16S rRNA reads from bacterial taxa associated with BV) were sequenced and compared. Lactic acid production by the different strains was quantified. Phage induction in the strains was also analysed. There was considerable genetic diversity between strains, and several genes were exclusive to either the strains from Lactobacillus-dominated microbiomes or those containing BV-associated bacteria. Overall, strains from microbiomes dominated by lactobacilli did not differ from strains from microbiomes containing BV-associated bacteria with respect to lactic acid production. All of the strains contained multiple phage, but there was no clear distinction between the presence or absence of BV-associated bacteria with respect to phage-induced lysis. Genes found to be exclusive to the Lactobacillus-dominated versus BV-associated bacteria-containing microbiomes could play a role in the maintenance of vaginal health and the development of BV, respectively.
Introduction
Lactobacilli are usually the predominant bacteria in the healthy vaginal microbiome. There are at least five Lactobacillus species that can colonize the vagina: Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus jensenii, Lactobacillus vaginalis and Lactobacillus iners (Antonio et al., 1999; Fettweis et al., 2014; Madhivanan et al., 2015; Wilks et al., 2004). Bacterial vaginosis (BV) is a common clinical condition characterized by a dramatic increase in both bacterial species diversity and overall bacterial burden. Bacteria that predominate the vaginal microbiota during BV include Gram-positive and Gram-negative anaerobes such as Gardnerella vaginalis, Prevotella, Bacteroides and Porphyromonas species, as well as Mycoplasma hominis, Mobiluncus species, BV-associated bacteria BVAB1–3 and Atopobium vaginae (Fredricks et al., 2005; Hill, 1993; Hillier et al., 1993; Srinivasan et al., 2012; Thorsen et al., 1998; Zhou et al., 2004). Women with BV are at increased risk for preterm birth, acquisition of sexually transmitted infections, including HIV, and pelvic inflammatory disease (Cherpes et al., 2003; Hillier et al., 1995; Martin et al., 1999; Ness et al., 2004; Sobel, 2000; Wiesenfeld et al., 2003). Despite the clinical significance and high prevalence of BV, the basis for the reduction in the prevalence of lactobacilli and the increase in the prevalence of such anaerobes remain obscure. Species of Lactobacillus vary in their stability and capacity to protect the vagina from colonization by BV-associated anaerobes (Tamrakar et al., 2007; Verstraelen et al., 2009). Stability ensures that the lactobacilli will not be easily displaced by environmental impacts such as hormonal changes, altered nutrient sources, sexual activity and exposure to semen, transient fluctuations in pH, and exposure to non-resident bacterial species. The protective ability reflects the capacity of the species to prevent other bacteria from colonizing the vagina. L. crispatus appears to be one of the most stable and protective species, and women colonized with L. crispatus have been shown to have a fivefold decreased risk for developing BV (Gajer et al., 2012; Verstraelen et al., 2009). Furthermore, a recent study comparing the genomes of 10 L. crispatus isolates found that the core genome of this species includes genes that may play a role in reducing the ability of G. vaginalis to adhere to epithelial cells and possibly to help prevent BV (Ojala et al., 2014).
The mechanisms by which the lactobacilli prevent colonization by other species and maintain a healthy vaginal environment are not completely understood. It is widely believed that lactic acid, H2O2 and bacteriocins produced by the lactobacilli kill other bacterial species as they enter the vagina. However, there is little correlation between the levels of H2O2 produced and the stability or protection conferred by a given strain of lactobacillus insofar as L. vaginalis and L. jensenii produce the highest levels of H2O2, whereas L. crispatus appears to be the most stable and protective species (Tamrakar et al., 2007). In fact, even H2O2-non-producing strains of L. crispatus appear to play a protective role (Verstraelen et al., 2009). Recent studies suggest that lactic acid is the main defence mechanism against BV-associated bacteria and that H2O2 levels are insignificant in the vaginal environment (Kaewsrichan et al., 2006; O'Hanlon et al., 2013; Wilks et al., 2004). In another recent study, the ratios of d- and l-lactic acid (isomers of lactic acid) varied in women with different vaginal communities, suggesting that different lactic acid-producing bacteria produce the two isomers in different ratios (Witkin et al., 2013).
The basis for stability or instability in the vaginal population of Lactobacillus species is also not completely understood. Recent reports document the high prevalence of prophage in vaginal lactobacilli (Baugher et al., 2014; Damelin et al., 2011). Prophage can excise from the chromosome, replicate and lyse bacteria when they enter the lytic cycle. Excision can be spontaneous or induced by DNA-damaging agents that induce the global SOS stress response. One study indicates that CRISPR-related gene expression is elevated in L. iners during BV, suggesting that CRISPR activity may be a response to a phage-loaded environment (Macklaim et al., 2013). Cigarette smoking, which is associated with BV, may expose the vaginal lactobacilli to chemicals capable of inducing phage (Pavlova & Tao, 2000). Thus, lytic temperate bacteriophage could be responsible for the rapid decline in vaginal lactobacilli often associated with BV.
Important strain-to-strain differences that can influence the role of vaginal bacterial species in health and disease can occur (Allen-Daniels et al., 2015). We therefore hypothesized that different strains of L. crispatus vary in their stability and/or protective capacity, possibly explaining the observation that some women colonized by L. crispatus still develop BV (Teixeira et al., 2012). To test this possibility, we isolated and sequenced the genomes of 17 isolates of L. crispatus from four women with lactobacillus-dominated vaginal microbiomes ( < 1 % BV-associated bacteria) and four women with vaginal microbiomes containing >12 % BV-associated bacteria. Genomic sequencing revealed that multiple isolates from the same subject were indistinguishable and therefore the same strain. We therefore restricted our analysis to one isolate per subject, and compared the genomes, quantified lactic acid production and analysed phage induction and phage-induced bacterial lysis.
Methods
Bacterial strains and growth conditions
Participants were recruited from outpatient clinics at the Virginia Commonwealth University Medical Center, the Virginia Department of Health and the Mid-Atlantic Twin Registry following written, informed consent from 2009 to 2013. Inclusion criteria included women 18–50 years old who were able to provide informed consent and who were willing or already scheduled to undergo a vaginal examination using a speculum. The Institutional Review Boards for Human Subjects Research at Virginia Commonwealth University (Panel B) and the Virginia Department of Health reviewed and approved this study. Participants filled out a detailed questionnaire that included questions about ethnicity, education, employment, health habits, dietary habits and sexual history.
Clinicians used CultureSwab EZ polyurethane foam swabs (BD) to obtain specimens from the mid-vaginal wall during a speculum examination. DNA was extracted from the swabs within 4 h of collection using a Powersoil kit (MoBio). Surveys of the 16S rRNA genes present in the samples were generated as part of the Vaginal Human Microbiome Project (Fettweis et al., 2011). Sequences were classified using a local installation of rdp Classifier (0.8 cut-off) and the STIRRUPS analysis platform (Fettweis et al., 2012). Samples containing L. crispatus and variable amounts of non-lactobacilli were cultured on Lactobacilli MRS Agar. Single-colony isolates were identified by 16S rRNA gene sequencing and 17 selected L. crispatus isolates were grown in Lactobacilli MRS Broth. DNA was isolated using a Genomic-tip 500/G (Qiagen) according to the manufacturer's instructions. The DNA samples were prepared for multiplexed sequencing using standard Illumina protocols and Illumina paired-end adapters. Libraries containing mean insert sizes of 300 bp were sequenced on an Illumina GAIIx with a paired-end cluster generation kit version 4 and TruSeq SBS version 5 sequencing kits. Sequencing was performed following a 2 × 75 bp cycle recipe. De novo assembly using high-quality reads was performed using CLC Bio software version 4.1.
Comparative genomics pipeline
Using the gene predictions from the National Center for Biotechnology Information (NCBI) and blast+ (Camacho et al., 1990), a Python pipeline was generated to compare the genes from each strain. Annotated coding sequences for each strain were aligned in pairs with every other strain using blastp. blastp and tblastn output was generated in tab-delimited format (outfmt 6) and Python was used to parse through the data. Alignments with >85 % amino acid identity and >90 % coverage of the length of the annotated gene were clustered as orthologues. For genes below this threshold, tblastn was used to query the entire contig sequences for an alignment match with >85 % nucleotide identity and >90 % of the length of the annotated gene. Genes found in all strains were categorized as core genes. Genes found in a single strain were categorized as a singletons.
Phylogenetic analysis
The web-based phylogenetic analysis tool realphy (preference alignment-based phylogeny builder) was used to compare the L. crispatus genomes (Bertels et al., 2014). realphy generates phylogenetic trees by merging alignments obtained by mapping to multiple reference genomes. The contigs from the eight strains of L. crispatus sequenced in this work and the 10 strains available from the NCBI at the time of the study were submitted to the server using default settings, and aligned and merged to generate a core sequence that was then used to reconstruct a rooted maximum-likelihood tree.
Bacteriocin gene and insertion sequence identification
The web-based tool bagel (version 2) was used to predict putative bacteriocins in the L. crispatus strains (van Heel et al., 2013). bagel uses a combination of ORF prediction tools, databases of known bacteriocins and motif databases to identify potential bacteriocin genes. The contig sequences were submitted to the online tool and analysed using the default setting. The web-based tool ISsaga (issaga.biotoul.fr/ISsaga/issaga_index.php) was used to predict putative insertion sequences (Varani et al., 2011). Both the nucleic acid contigs and the amino acid fasta file were uploaded and analysed using the default settings. Results were obtained from the annotation report of predicted insertion sequences, as distributed by family section.
l-/d-Lactic acid quantification
To approximate vaginal conditions, a chemically defined medium (CDM) modified from that described by Geshnizgani & Onderdonk (1992) was used. The CDM contained 0.1 % glycogen, 0.025 % mucin, 0.02 % Tween, 0.05 % urea, 2 % albumin, 0.03 % MgSO4, 4 % human serum and 1 % 100 × HyClone solution. Starter cultures of each strain grown overnight in CDM were diluted in fresh CDM to OD600 0.01 and cultured for an additional 24 h under anaerobic conditions. The OD600 values of the cultures were determined, bacteria were collected by centrifugation and spent media samples were diluted with double-distilled H2O based on the OD600 of the culture. Diluted media samples were filter-sterilized, and l- and d-lactic acid in the supernatants were measured using EnzyChrom l-lactate and EnzyChrom d-lactate kits (BioAssay Systems) according to the manufacturer's instructions.
Phage induction and detection
Overnight cultures were diluted 1 : 10 in fresh medium, incubated for 3 h anaerobically and induced with 2 μg mitomycin C ml–1 for an additional 7 h. The OD600 was determined for each sample and the bacteria were removed by centrifugation. In initial experiments, equal portions of the spent medium were unfiltered or filtered to remove intact bacteria remaining in the media following centrifugation, but the results from filtered and unfiltered medium were indistinguishable, so the step was omitted for the remaining assays. The amount of spent medium used for DNA isolation was calculated based on OD600 and samples were diluted with MRS Broth to equilibrate the volumes. DNA was purified using a QIAmp kit (Qiagen). To detect phage, quantitative PCR (qPCR) was used. The lysin gene was highly conserved relative to other phage genes, and primer sets were designed to detect all of the predicted lysin genes in each of the L. crispatus isolates (Table S1, available in the online Supplementary Material). Reactions were in a total volume of 25 μl and consisted of 1 μl DNA, 12.5 μl iTaq Universal SYBR Green Supermix (Bio-Rad) and 1 μM each primer. The reactions were denatured for 3 min at 95 °C and cycled 40 times for 15 s at 95 °C, 10 s at 55 °C and 30 s at 60 °C using an iQ5 real-time thermal cycler (Bio-Rad). Relative values representative of the concentrations of 16S rRNA and lysin genes present in the spent medium were calculated from the cycle threshold values (C t) as: 2–Ct. The resulting values for samples with mitomycin C (induced) were divided by the samples without mitomycin C (uninduced), and these ratios are represented in the Results. Pearson's method was used to determine the correlation between the ratio of induced/uninduced soluble lysin gene (a marker of phage release) and the ratio of induced/uninduced soluble 16S rRNA gene (a marker of bacterial lysis).
Results
Microbiome profiles
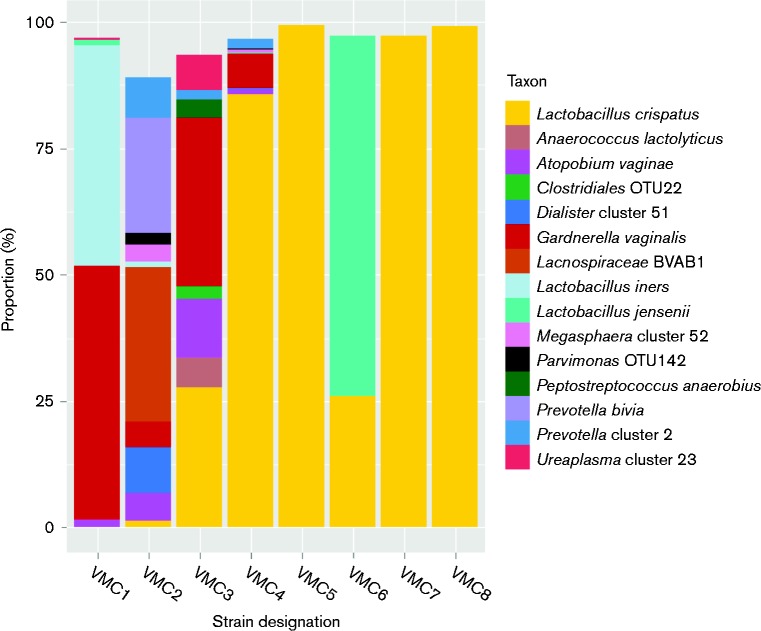
Our goal was to isolate L. crispatus strains from subjects with different vaginal microbiome profiles. Thus, 16S rRNA gene survey data from the Vaginal Microbiome Project (Fettweis et al., 2011) were analysed prior to sample selection (Fig. 1). Metadata from the selected subjects, collected through health history questionnaires, are listed in Table S2. Three microbiomes containing < 50 % lactobacilli and >50 % BV-associated taxa, one microbiome with ∼86 % L. crispatus and ∼12 % BV-associated taxa from a subject with a history of BV, three microbiomes with >90 % L. crispatus and ≤ 10 % BV-associated bacterial taxa, and one dominated by L. jensenii (another Lactobacillus species associated with vaginal health) and L. crispatus (four lactobacillus-dominated profiles) from women with no reported history of BV were selected. L. crispatus abundance in the chosen samples ranged from 0.043 to 99.66 % (Fig. 1). Single colonies were isolated and identified as L. crispatus by 16S sequence analysis.
Fig. 1.
Microbiome of samples used for isolation of L. crispatus. Microbial profiles for each of the eight subjects chosen for isolation of L. crispatus are shown. The y-axis represents the prevalence of different bacterial taxa as a function of percentage of total reads of the 16S rRNA gene. Only bacterial taxa representing ≥ 2 % of the reads from 16S rRNA gene surveys are shown. The x-axis labels are the strain designations given to the L. crispatus isolates chosen from each vaginal swab sample for study.
Whole-genome sequence analysis
DNA from each L. crispatus isolate was purified and sequenced to >25-fold coverage and analysed as described in Methods. General characteristics of the genomes are listed in Table 1. Genomes ranged in size from 2.073 to 2.25 Mbp and were predicted to encode 2146–2488 genes. Of these, 1307 genes that encoded proteins consisting of >75 amino acids were common to all strains and the remaining genes were present on only a subset of the genomes analysed. The phylogenetic relationship amongst these isolates and the 10 L. crispatus strains listed in NCBI was determined by comparing the whole genomes with realphy (Bertels et al., 2014). First, the genomes of all 18 strains were merged into a core genome that was used as a reference for the analysis. A phylogenetic tree was then reconstructed using single-nucleotide polymorphisms to determine relatedness (Fig. 2). The results suggested that the strains isolated from lactobacilli-dominated microbiomes were no more closely related to one another than they were to the strains isolated from microbiomes containing BV-associated bacteria.
Table 1. General characteristics of the L. crispatus genomes.
| Strain | Whole Genome Shotgun project accession no. | Genome size (Mb) | Contigs | Contigs >500 bp | Predicted coding sequences | Proteins >75 aa | tRNA genes | Secreted proteins | GC content | Mean coverage | BV* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VMC1 | LJCZ00000000 | 2.224 | 272 | 221 | 2344 | 2077 | 60 | 47 | 36.8 | 128 | Hx |
| VMC2 | LJDA00000000 | 2.073 | 283 | 241 | 2146 | 1900 | 51 | 38 | 36.8 | 28 | Dx |
| VMC3 | LJGP00000000 | 2.207 | 92 | 91 | 2261 | 2012 | 59 | 42 | 36.58 | 90 | Dx |
| VMC4 | LJGQ00000000 | 2.321 | 245 | 215 | 2488 | 2168 | 64 | 50 | 36.81 | 207 | Hx |
| VMC5 | LJOK00000000 | 2.250 | 261 | 231 | 2428 | 2059 | 61 | 44 | 36.93 | 236 | No |
| VMC6 | LJOL00000000 | 2.350 | 254 | 227 | 2455 | 2130 | 67 | 48 | 38.81 | 215 | No |
| VMC7 | LJOM00000000 | 2.108 | 248 | 228 | 2130 | 1898 | 61 | 39 | 36.66 | 232 | No |
| VMC8 | LJON00000000 | 2.334 | 256 | 240 | 2453 | 2126 | 63 | 41 | 36.85 | 92 | No |
Hx, Self-reported history of BV; Dx, clinical diagnosis of BV based on Amsel criteria (Amsel et al., 1983); No, subjects with no diagnosis or known history of BV.
Fig. 2.
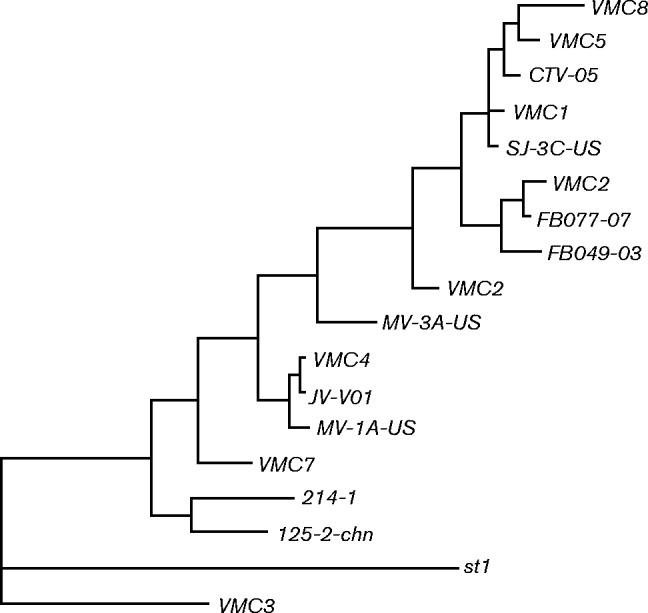
Phylogenetic analysis of L. crispatus isolates. The entire genomes of the 10 L. crispatus strains analysed by Ojala et al. (2014) and the eight strains isolated in this study were compared using realphy (Bertels et al., 2014).
Mobile elements
Insertion sequences in the strains were identified using ISsaga (Varani et al., 2011). Each strain was found to contain multiple insertion sequences from different insertion sequence families (Table S3). None of the insertion sequence families identified correlated with the presence of BV-associated bacteria in the source microbiome. However, there was a trend, although not statistically significant (Student's t-test P = 0.07), towards a greater total number of insertion sequences in the strains from microbiomes containing BV-associated bacteria. When VMC4, which was isolated from a microbiome containing only ∼12 % BV-associated bacteria, was removed from the analysis, the prevalence of mobile elements in the strains from lactobacilli-dominated microbiomes (VMC5–VMC8) was significantly greater than that in the strains from microbiomes with >50 % BV-associated bacteria (VMC1–VMC3) (P = 0.0039).
The strains contained a number of antibiotic and heavy metal resistance genes, although many of these were within the core genome. Phage lysin genes were also common. As most phage contain a lysin gene, we estimated the number of phage within each genome using the number of lysin genes. Strains VMC3 and VMC8 each contained two lysin genes, VMC1, 2, 5 and 7 each contained three lysin genes, and strains VMC4 and 6 each contained four lysin genes. Therefore there was no correlation between the number of lysin genes and the presence of BV-associated bacteria in the source microbiome.
Genes associated with Lactobacillus dominance
We hypothesized that genes present in the strains from lactobacilli-dominated microbiomes, but absent in strains from microbiomes containing BV-associated bacteria, might play a role in strain stability or in the exclusion of BV-associated bacteria. There were five genes present in the strains from lactobacilli-dominated microbiomes, but absent in strains from the microbiomes containing BV-associated bacteria, including three hypothetical proteins and two transposase genes. All of the strains from microbiomes containing BV-associated bacteria contained a number of genes absent in the strains from lactobacilli-dominated microbiomes, including seven genes predicted to be involved in cellobiose transport (two for phosphtransferase system cellobiose transporter subunit IIA, one for phosphotransferase system cellobiose transporter subunit IIB and two for phosphotransferase system cellobiose transporter subunit IIC), a putative glucosidase (6-phosphobeta-glucosidase) and one hypothetical protein (Table S4).
Exclusion of bacteria could be associated with specific bacterial toxins or bacteriocins. An examination of putative bacteriocin-encoding genes present in the different isolates, performed as described previously by Ojala et al. (2014), identified multiple genes encoding putative bacteriocins in each isolate (Table 2). However, none of the bacteriocin genes were found exclusively in strains from lactobacilli-dominated microbiomes. In addition, the total number of bacteriocin genes was not associated with lactobacillus dominance versus presence of BV-associated bacteria.
Table 2. Putative bacteriocin genes.
| Bacteriocin type | VMC1 | VMC2 | VMC3 | VMC4 | VMC5 | VMC6 | VMC7 | VMC8 |
|---|---|---|---|---|---|---|---|---|
| Enterolysin A 1 | 1, 2, 3 | 1, 2 | 1, 2, 3 | 1, 2, 3 | 1, 2 | 1, 1, 2 | 1 | |
| Helveticin-J 1 | 1, 2 | 1, 2, 3 | 2, 4 | 1, 2 | 1, 2 | 1, 2 | 1 | 1, 2, 4 |
| Penocin A 1 | 2 | 2 | 2 | 1, 2 | 1, 2 | |||
| Thermophilin A | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Durancin Q | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| CoagulinA | 1 | |||||||
| Staphylococcin C55β | 1 |
bagel (van Heel et al., 2013) was used to identify bacteriolysin genes. The genes were given random identifiers 1–4 for inter-strain comparison.
d-/l-Lactic acid
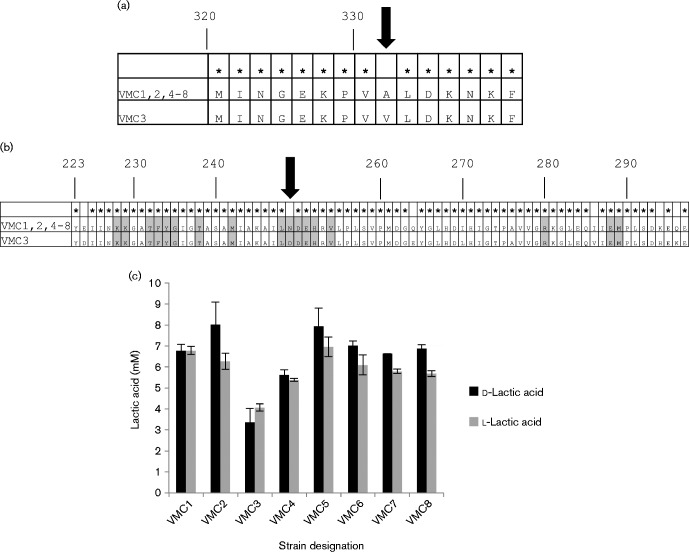
Reportedly, lactic acid is the main inhibitor of BV-associated bacteria produced by lactobacilli (O'Hanlon et al., 2013). L. crispatus encodes two l-lactate dehydrogenase genes and one d-lactate dehydrogenase genes. Lactate dehydrogenase genes in the eight strains were compared. The d- and l-lactate dehydrogenase genes were conserved at the amino acid level in all of the strains except VMC3, which had one missense mutation in the d-lactate dehydrogenase gene and six amino acid substitutions in one of the l-dehydrogenase genes (Fig. 3a, b). The mutation in the d-lactate dehydrogenase gene was predicted to be ‘Probably damaging’, with a score of 0.994 by PolyPhen-2 (Adzhubei et al., 2010). One of the mutations in the l-lactate dehydrogenase gene was a conversion from an asparagine (amide) to an aspartic acid (acidic) located at a residue that is conserved amongst the ldh gene family.
Fig. 3.
Lactic acid production. (a) The amino acid sequences of the d-lactate dehydrogenase genes of strains VMC1, 2 and 4–8 were identical. There was a single amino acid substitution in strain VMC3, at position 331. (b) The amino acid sequences of the l-lactate dehydrogenase genes of strains VMC1, 2 and 4–8 were identical. Amino acids conserved amongst members of the LDH Conserved Protein Domain Family (NCBI) are shaded in grey. There were six amino acid substitutions in strain VMC3, at positions 224, 249, 264, 286, 294 and 296. (c) Lactic acid produced during culture in CDM resembling vaginal secretions.
We quantified the levels of d- and l-lactic acid produced by each isolate in vitro using a CDM designed to replicate vaginal secretions (Geshnizgani & Onderdonk, 1992). There were no consistent or significant differences in lactic acid production when comparing the strains from lactobacilli-dominated microbiomes with those from microbiomes containing BV-associated bacteria. VMC3, the isolate with polymorphisms in its d- and l-lactate dehydrogenase genes, was capable of producing both isomers of lactic acid, although the quantities of each were considerably lower than the other strains (Fig. 3b).
Phage replication
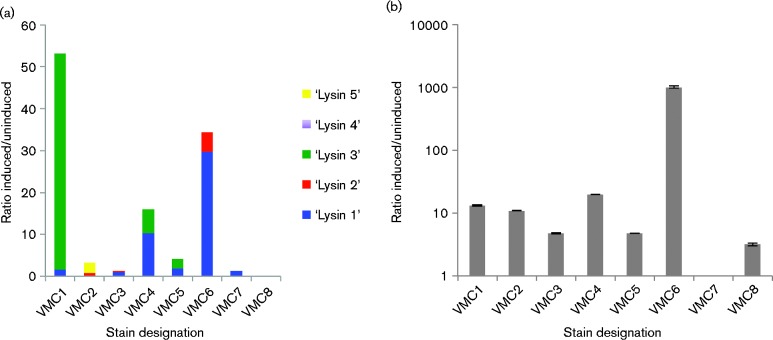
It is not always possible to predict whether a lysogenic phage is capable of replication through in silico analyses. Defective prophage are often retained in the bacterial chromosome, unable to excise, replicate or lyse the bacteria. To determine whether our isolates of L. crispatus contained replicative phage, we assessed spontaneous phage release and phage induction by the DNA damage-inducing antibiotic mitomycin C using qPCR of the relatively conserved lysin gene. Five sets of primers were used to amplify all of the lysin genes of the different phage detected in the eight isolates (Table S1). As indicated in Fig. 4(a), most of the phage present in the eight strains were not induced by mitomycin C, although several were induced up to 50-fold. We also analysed bacterial lysis through the quantification of 16S rRNA-encoding DNA in spent medium from mitomycin C-induced cultures (Fig. 4b). There was not a strong correlation between phage induction and bacterial lysis (Pearson's correlation 0.478). The fold increase in lysis in response to the SOS-inducing stimulus ranged from 0 to >1000. The amount of bacterial lysis did not differ between the group of strains from lactobacilli-dominated microbiomes and the group of strains from microbiomes containing BV-associated bacteria.
Fig. 4.
Phage induction. Mitomycin C was used to induce the strains and DNA was extracted from spent medium. Relative levels of DNA encoding the phage lysin genes were quantified by qPCR as a measure of phage replication. Relative levels of DNA encoding the 16S rRNA gene were quantified as a measure of bacterial lysis. The ratio of bacterial or phage DNA in the spent medium from cultures induced with mitomycin C to uninduced was calculated as (2Ctinduced)/(2Ctuninduced) and is shown on the y-axis.
Discussion
L. crispatus produces lactic acid and other compounds that are potent inhibitors of BV-associated bacterial species. Therefore, women who are vaginally colonized by L. crispatus are less likely to develop BV. However, for reasons that are not yet clear, some women do develop BV despite L. crispatus colonization. One possible explanation is that genetic variations between strains affect the bacteriostatic and bactericidal activity against other bacteria or their stability in the face of fluctuating vaginal conditions. Bacteriostatic and bactericidal activities are largely due to lactic acid, a product of lactate dehydrogenase enzymes, and to bacteriocins. Studies have shown that stability can be influenced by phage induction (Macklaim et al., 2013; Pavlova & Tao, 2000) but it may also be affected by things that confer resistance to harmful environmental stimuli, such as efflux pumps, or by factors that function in adaptation to changes in nutrient availability. We tested this by examining the whole genomes, the bactericidal activity (lactic acid production) and the stability (phage induction) of strains from women who were predominated by L. crispatus and did not have a history of BV, and comparing these parameters with strains from women with more diverse vaginal microbial profiles.
Recent reports implicate lactic acid as the most important product of lactobacilli with regard to preventing growth of BV-associated bacterial species (O'Hanlon et al., 2013). L. crispatus encodes two l-lactate dehydrogenase genes and one d-lactate dehydrogenase gene. L. jensenii, another species that is associated with the healthy vaginal microbiome, has two d-lactate dehydrogenase genes and one l-lactate dehydrogenase gene, whereas L. iners, which is frequently found in women with recurrent BV, only encodes one d-lactate dehydrogenase gene. These genes and their promoters were completely conserved at the amino acid level amongst all of the strains except for VMC3. Furthermore, lactic acid was produced at similar levels by all strains except VMC3. Therefore, the results from this study suggest that strain-to-strain differences in lactic acid production may affect the capacity of some strains to prevent BV, but that other strains with a strong capacity to produce lactic acid can be found in women colonized with BV-associated bacteria. It should be noted, however, that our study measured lactic acid production under a single set of defined conditions that do not precisely replicate the conditions encountered in vivo. Therefore, in vivo, and in the presence of other bacteria, the strains could produce different amounts of lactic acid.
Some vaginal lactobacilli secrete bacteriocins, which also prevent growth of other bacterial species and help to prevent BV (Sabia et al., 2014; Stoyancheva et al., 2014). All of the strains characterized in this study encoded multiple bacteriocins, but the number and type of bacteriocins did not correlate with the level of BV-associated bacteria in the microbiome. However, VMC3 was the only strain that lacked a gene for a bacteriocin, similar to Enterolysin A from Enterococcus faecalis (Nilsen et al., 2003). Thus, VMC3, isolated from a subject with BV at the time of sampling, could have diminished bactericidal capacity due to diminished lactic acid production and the lack of an Enterolysin A family bacteriocin. However, we could not detect any genetic differences in the remaining isolates from women with microbiomes containing >10 % BV-associated bacteria, i.e. VMC1, 2 and 4, that would be predicted to make these strains less bactericidal than the strains from lactobacilli-dominated microbiomes.
All of the strains contained genes predicted to be involved in cellobiose transport, but identities between a number of the genes were so low that they were annotated as distinct genes. Thus, we found five genes predicted to be involved in cellobiose transport that were present only in the BV-associated strains. Ojala et al. (2014) also found that genes involved in cellobiose transport differ across strains and suggested that these genes may have been acquired by horizontal transfer. The significance of this is not yet clear. There were five genes present in the strains from lactobacilli-dominated microbiomes that were absent in all of the strains isolated from microbiomes with >10 % BV-associated bacteria; however, two of these appeared to encode transposase and are therefore unlikely to play a role in maintaining vaginal health, and the other three did not display significant identity to any genes of known function. Therefore, a hypothesis about their role in health cannot be made without further study. In addition to the genes identified, there may be genes that differ at the level of expression rather than by presence or absence. Another possibility is that there are a number of variable genes that are essential for a strain to prevent BV and the strains from microbiomes containing BV-associated bacteria in our set lack different essential genes, which would prevent detection of the genes as exclusive to the strains from lactobacilli-dominated microbiomes. Finally, the genetic diversity within the L. crispatus species suggests that many strains may need to be analysed in order to identify genes that contribute to lactobacilli dominance.
There was a trend towards a greater number of mobile elements in L. crispatus strains from microbiomes that contained BV-associated bacteria. This could be an effect of the increased bacterial diversity in these microbiomes leading to more sharing of mobile elements. Phage genes and CRISPR repeats were detected in all strains, and phage that were inducible by the SOS response were detected in most. Therefore, prior exposure to phage could influence strain stability in the face of an inducing stimulus or a newly introduced phage (Burmistrz & Pyrć, 2015). If this were the case, then it would be a combination of genetic composition of the strain and the environment of the bacterial population that would cause a decline in the numbers of lactobacilli. A longitudinal study that carefully monitored the subjects' behaviour, diet, menstrual cycle and other parameters would be necessary to address this possibility. Our study focused on the genotypic and phenotypic characteristics of the L. crispatus strains, but did not encompass complex interactions between the strains and their environment. Therefore, phage may influence the stability of L. crispatus, but differences in stability are not simply explained by the presence or absence of inducible phage.
A recent study by Ojala et al. (2014) compared the 10 L. crispatus genomes that were publicly available at the time. The objective of that study was not to identify differences in strains from women with and without BV or to compare phenotypic traits, but rather to characterize the core and pan genomes of this species. Similar to our study, they found that the L. crispatus genome is highly variable, with the core genome making up only ∼60 % of the genetic material of a particular strain. They also found that phage are prevalent in this species, although the functionality of the phage was not investigated. Evidence from that study suggested that L. crispatus adhesins, in particular the Lactobacillus epithelium adhesin, may also play a role in exclusion of BV-associated bacteria, through inhibition of adherence rather than direct killing. Our study focused on bactericidal and bacteriostatic qualities of L. crispatus rather than on competitive inhibition of adherence. However, we did find that all of the strains from lactobacilli-dominated microbiomes and strains from microbiomes containing BV-associated bacteria encoded the LEA protein (data not shown).
A potential therapeutic approach to recurrent BV is the use of a probiotic vaginal Lactobacillus strain (Ya et al., 2010). For a probiotic approach to be highly effective, an understanding of what makes vaginal lactobacilli stable and bactericidal against other species is key. L. crispatus is a promising species because it is associated with vaginal health, and negatively associated with BV and preterm birth. We investigated the hypothesis that certain L. crispatus strains may be inferior and less likely to prevent BV. Clear genetic deficiencies, which included mutations in one of the d-lactate dehydrogenase genes and in the l-lactate dehydrogenase gene, and absence of an Enterolysin A gene, were detected in only one out of four strains from women with BV. However, all of the strains from microbiomes containing BV-associated bacteria lack two transposase genes and three genes of unknown function that were detected in all of the strains from Lactobacillus-dominated microbiomes; whilst it is impossible to assign a hypothetical role based on this limited information, these genes could potentially be involved in limiting the stability or protective capacity of these strains. The development of BV could be caused, in part, by genetic differences between the L. crispatus strains and/or by differences in the bacterial environment or in the microbiome. Whilst BV is not generally considered a sexually transmitted disease, the number of sexual partners amongst the group with microbiomes containing BV-associated bacteria was greater, suggesting that exposure to certain bacteria may play a role. Additionally, genetic differences that led to variations in the stability and/or protective capacity of the strains may have been present but were not detected in this study.
Acknowledgements
This work was supported by National Institutes of Health grants 4UH3AI083263 (The Vaginal Microbiome: Disease, Genetics and the Environment), P60 MD002256 (VCU NIMHD Comprehensive Center of Excellence) and U54 DE023786-01 (A Multi-'omic Analysis of the Vaginal Microbiome during Pregnancy).
Supplementary Data
Supplementary Data
Abbreviations:
- BV
bacterial vaginosis
- CDM
chemically defined medium
- NCBI
National Center for Biotechnology Information
- qPCR
quantitative PCR.
References
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., Sunyaev S. R. (2010). A method and server for predicting damaging missense mutations Nat Methods 7 248–249 10.1038/nmeth0410-248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Daniels M. J., Serrano M. G., Pflugner L. P., Fettweis J. M., Prestosa M. A., Koparde V. N., Brooks J. P., Strauss J. F., III, Romero R., other authors (2015). Identification of a gene in Mycoplasma hominis associated with preterm birth and microbial burden in intraamniotic infection Am J Obstet Gynecol 212 779.1–779.e13 10.1016/j.ajog.2015.01.032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsel R., Totten P. A., Spiegel C. A., Chen K. C., Eschenbach D., Holmes K. K. (1983). Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations Am J Med 74 14–22 10.1016/0002-9343(83)91112-9 . [DOI] [PubMed] [Google Scholar]
- Antonio M. A., Hawes S. E., Hillier S. L. (1999). The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species J Infect Dis 180 1950–1956 10.1086/315109 . [DOI] [PubMed] [Google Scholar]
- Baugher J. L., Durmaz E., Klaenhammer T. R. (2014). Spontaneously induced prophages in Lactobacillus gasseri contribute to horizontal gene transfer Appl Environ Microbiol 80 3508–3517 10.1128/AEM.04092-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertels F., Silander O. K., Pachkov M., Rainey P. B., van Nimwegen E. (2014). Automated reconstruction of whole-genome phylogenies from short-sequence reads Mol Biol Evol 31 1077–1088 10.1093/molbev/msu088 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmistrz M., Pyrć K. (2015). CRISPR–Cas systems in prokaryotes Pol J Microbiol 64 193–202 . [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L. (2009). BLAST+: architecture and applications BMC Bioinformatics 10 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherpes T. L., Meyn L. A., Krohn M. A., Lurie J. G., Hillier S. L. (2003). Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis Clin Infect Dis 37 319–325 10.1086/375819 . [DOI] [PubMed] [Google Scholar]
- Damelin L. H., Paximadis M., Mavri-Damelin D., Birkhead M., Lewis D. A., Tiemessen C. T. (2011). Identification of predominant culturable vaginal Lactobacillus species and associated bacteriophages from women with and without vaginal discharge syndrome in South Africa J Med Microbiol 60 180–183 10.1099/jmm.0.024463-0 . [DOI] [PubMed] [Google Scholar]
- Fettweis J., Alves J., Borzelleca J., Brooks J. P., Friedline C. J., Gao Y., Gao X., Girerd P., Harwich M. D., other authors (2011). The vaginal microbiome: disease, genetics and the environment.http://precedings.nature.com/documents/5150/version/2.
- Fettweis J. M., Serrano M. G., Sheth N. U., Mayer C. M., Glascock A. L., Brooks J. P., Jefferson K. K., Buck G. A., Vaginal Microbiome Consortium (2012). Species-level classification of the vaginal microbiome BMC Genomics 13 (Suppl 8), S17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis J. M., Brooks J. P., Serrano M. G., Sheth N. U., Girerd P. H., Edwards D. J., Strauss J. F., III, Jefferson K. K., Buck G. A., Vaginal Microbiome Consortium (2014). Differences in vaginal microbiome in African American women versus women of European ancestry Microbiology 160 2272–2282 10.1099/mic.0.081034-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks D. N., Fiedler T. L., Marrazzo J. M. (2005). Molecular identification of bacteria associated with bacterial vaginosis N Engl J Med 353 1899–1911 10.1056/NEJMoa043802 . [DOI] [PubMed] [Google Scholar]
- Gajer P., Brotman R. M., Bai G., Sakamoto J., Schütte U. M., Zhong X., Koenig S. S., Fu L., Ma Z. S., other authors (2012). Temporal dynamics of the human vaginal microbiota Sci Transl Med 4 132ra52 10.1126/scitranslmed.3003605 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshnizgani A. M., Onderdonk A. B. (1992). Defined medium simulating genital tract secretions for growth of vaginal microflora J Clin Microbiol 30 1323–1326 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill G. B. (1993). The microbiology of bacterial vaginosis Am J Obstet Gynecol 169 450–454 10.1016/0002-9378(93)90339-K . [DOI] [PubMed] [Google Scholar]
- Hillier S. L., Krohn M. A., Rabe L. K., Klebanoff S. J., Eschenbach D. A. (1993). The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women Clin Infect Dis 16 (Suppl 4), S273–S281 10.1093/clinids/16.Supplement_4.S273 . [DOI] [PubMed] [Google Scholar]
- Hillier S. L., Krohn M. A., Cassen E., Easterling T. R., Rabe L. K., Eschenbach D. A. (1995). The role of bacterial vaginosis and vaginal bacteria in amniotic fluid infection in women in preterm labor with intact fetal membranes Clin Infect Dis 20 (Suppl 2), S276–S278 10.1093/clinids/20.Supplement_2.S276 . [DOI] [PubMed] [Google Scholar]
- Kaewsrichan J., Peeyananjarassri K., Kongprasertkit J. (2006). Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens FEMS Immunol Med Microbiol 48 75–83 10.1111/j.1574-695X.2006.00124.x . [DOI] [PubMed] [Google Scholar]
- Macklaim J. M., Fernandes A. D., Di Bella J. M., Hammond J. A., Reid G., Gloor G. B. (2013). Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis Microbiome 1 12 10.1186/2049-2618-1-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan P., Alleyn H. N., Raphael E., Krupp K., Ravi K., Nebhrajani R., Arun A., Reingold A. L., Riley L. W., Klausner J. D. (2015). Identification of culturable vaginal Lactobacillus species among reproductive age women in Mysore, India J Med Microbiol 64 636–641 10.1099/jmm.0.000070 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H. L., Richardson B. A., Nyange P. M., Lavreys L., Hillier S. L., Chohan B., Mandaliya K., Ndinya-Achola J. O., Bwayo J., Kreiss J. (1999). Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition J Infect Dis 180 1863–1868 10.1086/315127 . [DOI] [PubMed] [Google Scholar]
- Ness R. B., Hillier S. L., Kip K. E., Soper D. E., Stamm C. A., McGregor J. A., Bass D. C., Sweet R. L., Rice P., Richter H. E. (2004). Bacterial vaginosis and risk of pelvic inflammatory disease Obstet Gynecol 104 761–769 10.1097/01.AOG.0000139512.37582.17 . [DOI] [PubMed] [Google Scholar]
- Nilsen T., Nes I. F., Holo H. (2003). Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333 Appl Environ Microbiol 69 2975–2984 10.1128/AEM.69.5.2975-2984.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hanlon D. E., Moench T. R., Cone R. A. (2013). Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota PLoS One 8 e80074 10.1371/journal.pone.0080074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala T., Kankainen M., Castro J., Cerca N., Edelman S., Westerlund-Wikström B., Paulin L., Holm L., Auvinen P. (2014). Comparative genomics of Lactobacillus crispatus suggests novel mechanisms for the competitive exclusion of Gardnerella vaginalis BMC Genomics 15 1070 10.1186/1471-2164-15-1070 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova S. I., Tao L. (2000). Induction of vaginal Lactobacillus phages by the cigarette smoke chemical benzo[a]pyrene diol epoxide Mutat Res 466 57–62 10.1016/S1383-5718(00)00003-6 . [DOI] [PubMed] [Google Scholar]
- Sabia C., Anacarso I., Bergonzini A., Gargiulo R., Sarti M., Condò C., Messi P., de Niederhausern S., Iseppi R., Bondi M. (2014). Detection and partial characterization of a bacteriocin-like substance produced by Lactobacillus fermentum CS57 isolated from human vaginal secretions Anaerobe 26 41–45 10.1016/j.anaerobe.2014.01.004 . [DOI] [PubMed] [Google Scholar]
- Sobel J. D. (2000). Bacterial vaginosis Annu Rev Med 51 349–356 10.1146/annurev.med.51.1.349 . [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Hoffman N. G., Morgan M. T., Matsen F. A., Fiedler T. L., Hall R. W., Ross F. J., McCoy C. O., Bumgarner R., other authors (2012). Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria PLoS One 7 e37818 10.1371/journal.pone.0037818 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyancheva G., Marzotto M., Dellaglio F., Torriani S. (2014). Bacteriocin production and gene sequencing analysis from vaginal Lactobacillus strains Arch Microbiol 196 645–653 10.1007/s00203-014-1003-1 . [DOI] [PubMed] [Google Scholar]
- Tamrakar R., Yamada T., Furuta I., Cho K., Morikawa M., Yamada H., Sakuragi N., Minakami H. (2007). Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women BMC Infect Dis 7 128 10.1186/1471-2334-7-128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira G. S., Carvalho F. P., Arantes R. M., Nunes A. C., Moreira J. L., Mendonça M., Almeida R. B., Farias L. M., Carvalho M. A., Nicoli J. R. (2012). Characteristics of Lactobacillus and Gardnerella vaginalis from women with or without bacterial vaginosis and their relationships in gnotobiotic mice J Med Microbiol 61 1074–1081 10.1099/jmm.0.041962-0 . [DOI] [PubMed] [Google Scholar]
- Thorsen P., Jensen I. P., Jeune B., Ebbesen N., Arpi M., Bremmelgaard A., Møller B. R. (1998). Few microorganisms associated with bacterial vaginosis may constitute the pathologic core: a population-based microbiologic study among 3596 pregnant women Am J Obstet Gynecol 178 580–587 10.1016/S0002-9378(98)70442-9 . [DOI] [PubMed] [Google Scholar]
- van Heel A. J., de Jong A., Montalbán-López M., Kok J., Kuipers O. P. (2013). bagel3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides Nucleic Acids Res 41 (W1), W448–W453 10.1093/nar/gkt391 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani A. M., Siguier P., Gourbeyre E., Charneau V., Chandler M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes Genome Biol 12 R30 10.1186/gb-2011-12-3-r30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraelen H., Verhelst R., Claeys G., De Backer E., Temmerman M., Vaneechoutte M. (2009). Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora BMC Microbiol 9 116 10.1186/1471-2180-9-116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfeld H. C., Hillier S. L., Krohn M. A., Landers D. V., Sweet R. L. (2003). Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection Clin Infect Dis 36 663–668 10.1086/367658 . [DOI] [PubMed] [Google Scholar]
- Wilks M., Wiggins R., Whiley A., Hennessy E., Warwick S., Porter H., Corfield A., Millar M. (2004). Identification and H2O2 production of vaginal lactobacilli from pregnant women at high risk of preterm birth and relation with outcome J Clin Microbiol 42 713–717 10.1128/JCM.42.2.713-717.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin S. S., Mendes-Soares H., Linhares I. M., Jayaram A., Ledger W. J., Forney L. J. (2013). Influence of vaginal bacteria and d- and l-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections MBio 4 e00460–e00413 10.1128/mBio.00460-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ya W., Reifer C., Miller L. E. (2010). Efficacy of vaginal probiotic capsules for recurrent bacterial vaginosis: a double-blind, randomized, placebo-controlled study Am J Obstet Gynecol 203 120e1–120e6 10.1016/j.ajog.2010.05.023 . [DOI] [PubMed] [Google Scholar]
- Zhou X., Bent S. J., Schneider M. G., Davis C. C., Islam M. R., Forney L. J. (2004). Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods Microbiology 150 2565–2573 10.1099/mic.0.26905-0 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data