Abstract
Fimbrial subunit synthesis, secretion and assembly on the surface of the periodontal pathogen Aggregatibacter actinomycetemcomitans are essential for biofilm formation. A recent quantitative proteomics study employing an afimbriated strain and a developed mutant isogenic for the inner-membrane protein morphogenesis protein C (MorC) revealed that the abundance of the proteins of the fimbrial secretion apparatus in the membrane is dependent on MorC. To investigate further the relationship between MorC and fimbriation, we identified and complemented the defect in fimbriae production in the afimbriated laboratory strain. The transformed strain expressing a plasmid containing genes encoding the WT fimbrial subunit and the prepilin peptidase displayed all of the hallmarks of a fimbriated bacterium including the distinct star-like colony morphology, robust biofilm formation, biofilm architecture composed of discrete microcolonies and the presence of fimbriae. When the identical plasmid was transformed into a morC mutant strain, the bacterium did not display any of the phenotypes of fimbriated strains. Extension of these studies to a naturally fimbriated clinical strain showed that the resulting morC mutant maintained the characteristic colony morphology of fimbriated strains. There was, however, a reduction in the secretion of fimbrial subunits, and fewer fimbriae were observed on the surface of the mutant strain. Furthermore, the morC mutant of the fimbriated strain displayed a significantly altered biofilm microcolony architecture, while maintaining a similar biofilm mass to the parent strain. These results suggest that MorC influences fimbrial secretion and microcolony formation in A. actinomycetemcomitans.
Introduction
Aggregatibacter actinomycetemcomitans, a facultative anaerobic bacterium that colonizes the oral cavity of humans, is typically associated with adult and juvenile periodontal diseases (Zambon et al., 1983). Accumulation of A. actinomycetemcomitans results in tissue inflammation, expression of extracellular matrix remodelling enzymes and bone reduction leading to tooth loss (Bodet et al., 2007; Haubek et al., 2004; Slots et al., 1980). This bacterium is also a member of the HACEK family of pathogens (Haemophilus spp., A. actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, Kingella kingae), which cause culture-negative infective endocarditis (Das et al., 1997). As a member of the family Pasteurellaceae, which includes Haemophilus and Pasteurella spp., A. actinomycetemcomitans displays a rugose outer-membrane morphology in contrast to the flat-membrane morphology displayed by other bacteria (Azari et al., 2013). Loss of morphogenesis protein C (MorC) in the membrane results in conversion of the membrane morphology from rugose to flat, decreasing the surface area of the outer membrane by up to 20 % (Azari et al., 2013; Gallant et al., 2008). In addition to this dramatic morphological change, the absence of MorC results in multiple membrane-related effects including a 30 % decrease in cell size, a 70 % increase in autoaggregation, an 86 % impaired secretion of leukotoxin and a fourfold increase in sensitivity to membrane-destabilizing agents (Smith et al., 2015a).
MorC is a large inner-membrane protein of 1292 aa (141 kDa), first identified in A. actinomycetemcomitans (Gallant et al., 2008). MorC belongs to the COG2911 family of uncharacterized proteins, and homologues are found throughout the Gammaproteobacteria (Selkrig et al., 2012; Smith, 2015). The C terminus of the protein (aa 1053–1292) is homologous to the DUF490 family of unknown proteins and contains the greatest percentage of identical and conserved amino acids within the whole protein (Gallant et al., 2008; Smith et al., 2015c). The morC gene is consistently found in an operon with omp67 (tamA), which encodes an outer-membrane protein paralogous to BamA, a component of the β-barrel assembly module responsible for the export of outer-membrane proteins (Rossiter et al., 2011).
In the Enterobacteriaceae, the MorC homologue, TamB (previously termed YtfN), interacts with TamA (YtfM) to form a protein secretion system (the translocation and assembly module or TAM) (Selkrig et al., 2012). The interaction between TamA and TamB is mediated by the DUF490 domain, which is required for MorC function in A. actinomycetemcomitans and Escherichia coli (Selkrig et al., 2012; Smith et al., 2015c). Disruption of this secretion system results in a reduction of the virulence potential of a number of bacteria including Klebsiella pneumoniae, Proteus mirabilis, Citrobacter rodentium and Salmonella enterica (Burall et al., 2004; Selkrig et al., 2012; Struve et al., 2003). The TamA/B complex is required for the efficient secretion of specific autotransporter proteins to the outer membrane in Enterobacteriaceae (Selkrig et al., 2012), in contrast to what is observed in morC mutants of A. actinomycetemcomitans, which express WT levels of known autotransporters (Smith et al., 2015a), which can easily be visualized by electron microscopy (Azari et al., 2013; Gallant et al., 2008).
A recent quantitative membrane proteomics study revealed that the absence of MorC only affects specific proteins of the membrane proteome of A. actinomycetemcomitans. These effects include a reduction in the abundance of fimbriae-associated proteins (Smith et al., 2015a). Fimbriae of A. actinomycetemcomitans are associated with ‘non-specific’ adherence of the organism to both biotic and abiotic surfaces and favour the formation of a tenacious biofilm (Fine et al., 1999). The structural fimbrial subunit and the secretion machinery are encoded by the conserved 14-gene tight adherence (tad) locus (Kachlany et al., 2000). The fimbriae structural protein (Flp1) is expressed as a pre-protein that is cleaved by the TadV peptidase to generate the mature form of the protein (Tomich et al., 2006). The processed Flp1 subunits are secreted via an 11-protein complex (RcpCAB, TadZ and TadA-G) analogous to a canonical type II secretion system (Kachlany et al., 2000). Under appropriate conditions, fimbriated strains have been demonstrated to generate afimbriated forms. This conversion typically arises from point mutations in the promoter region of the tad locus, which includes the fimbrial transport machinery (Wang et al., 2005). Loss of fimbriae is accompanied by an increase in colony size and loss of the star-shaped colony morphology characteristic of fimbriated strains of A. actinomycetemcomitans (Inouye et al., 1990). Afimbriated strains also display reduced surface adherence and do not colonize host tissues as effectively as the fimbriated form (Schreiner et al., 2003).
In the aforementioned proteomics study (Smith et al., 2015a), the reduction in fimbriae-associated proteins in the morC mutant strain indicated a link between MorC and fimbriae formation. In the present study, we first identified the defect for fimbriae production in a laboratory, afimbriated strain of A. actinomycetemcomitans and complemented fimbriation in this strain by introducing a plasmid expressing the fimbrial structural subunit (Flp1) and the prepilin peptidase (TadV). The effect of MorC on fimbriae production was investigated by introducing an identical plasmid into the morC mutant strain. However, fimbriae were not observed on the surface of these bacteria. Inactivation of morC in a clinical, naturally fimbriated isolate reduced the abundance of fimbriae, which resulted in significant changes in the size and appearance of the biofilm microcolonies. The data suggest that MorC is necessary for optimal fimbriae formation and basic biofilm architecture in A. actinomycetemcomitans. This represents the first report, to our knowledge, of a protein independent of the canonical fimbrial secretion apparatus modulating fimbrial secretion in A. actinomycetemcomitans.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are shown in Table 1. A. actinomycetemcomitans strains were routinely cultured in TSBYE medium (3 % tryptic soy broth, 0.6 % yeast extract, with or without 1.5 % agar; Beckton Dickinson). In all experiments, frozen A. actinomycetemcomitans strains were plated initially on TSBYE agar and grown at 37 °C in a humidified 10 % CO2 atmosphere. Afimbriated cells were inoculated and grown in TSBYE liquid medium. To avoid the potential loss of fimbriation that results from repeated growth in liquid medium (Wang et al., 2005), fimbriated strains were grown and collected from TSBYE solid medium prior to use in the experimental protocol. Where required, chloramphenicol was incorporated into the medium at a concentration of 1 μg ml− 1 and spectinomycin at 50 μg ml− 1. E. coli was propagated on LB medium (1 % tryptone, 0.5 % yeast extract, 0.5 % NaCl; Beckton Dickinson) at 37 °C in ambient air with agitation. For plasmid selection and maintenance in E. coli, ampicillin was used at 100 μg ml− 1 and chloramphenicol at 20 μg ml− 1.
Table 1. Bacterial strains and plasmids.
| Bacterial strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli | ||
| DH10B | Laboratory strain for general cloning, lac− | Invitrogen |
| A. actinomycetemcomitans | ||
| HK1651 | Sequenced serotype b fimbriated strain | ATCC 700685 |
| VT1257 | Fimbriated clinical isolate, IDHaas10a, serotype b | Maria Saarela, Finland |
| KM700 | Isogenic morC mutant of VT1257, Specr | This study |
| VT1169 | Afimbriated strain derived from SUNY465, serotype b | Mintz & Fives-Taylor (1994) |
| VT1650 | Afimbriated morCmutant of VT1169, Specr | Gallant et al. (2008) |
| KM278 | morC mutant containing pKM2 | Gallant et al. (2008) |
| KM409 | morC mutant containing pKM2/morC | Gallant et al. (2008) |
| KM611 | VT1169 containing pKM586 | This study |
| KM612 | VT1650 containing pKM586 | This study |
| KM609 | VT1257 containing pKM687 | This study |
| KM699 | KM700 containing pKM687 | This study |
| Plasmid | ||
| pGEM | TA cloning vector. Replicates in E. coli, Ampr | Promega |
| pKM2 | E. coli and A. actinomycetemcomitans shuttle vector, Cmr | Gallant et al. (2008) |
| pVT1642 | pKM2 containing JP2 leukotoxin promoter | Tang & Mintz (2010) |
| pKM586 | pVT1642 containing the flp1-tadV region of A. actinomycetemcomitansstrain HK1651 | This study |
| pKM687 | pVT1642 containing T7-flp1-tadV region of A. actinomycetemcomitansstrain HK1651 | This study |
Amp, Ampicillin; Cm, chloramphenicol; Spec, spectinomycin.
flp1-tadV plasmid construction
Primers were designed to generate an amplicon encompassing the flp1-tadV region of the tad locus using the annotation of the A. actinomycetemcomitans strain HK1651 genome in the Human Oral Microbiome Database (http://www.homd.org). The region of interest was amplified using the primers listed in Table 2 (annealing temperature 58 °C, extension time 45 s), ligated into a pGEM TA cloning vector (Promega) and transformed into E. coli DH10B (Invitrogen). The insert was released from the plasmid, purified using a commercial kit (Qiagen), ligated into the pKM2 shuttle vector and transformed into DH10B cells (Gallant et al., 2008). The resulting plasmid was used to transform A. actinomycetemcomitans WT or morC mutant cells (Gallant et al., 2008). PCR-positive transformants were selected, and the plasmid was purified and sequenced. The fidelity of the cloning process was verified by DNA sequencing at the Advanced Genome Technology Core at the University of Vermont, VT, USA.
Table 2. Oligonucleotide primers used in this study.
| Primer name | Sequence (5′ → 3′) | Description |
|---|---|---|
| Flp1Xho1F | CCGCTCGAGTTGAATTTTATTTTCTATAAAC | 5′ Primer, flp1 |
| TadVXho1R | AATCTCGAGATTTGATAGAGCCATGTTTATCATAAAGCC | 3′ Primer, tadV |
| Flp1For | CCGCTCGAGAACAACAAAGGAGCATTAAGACA | 5′ Primer, flp1T7 |
| Flp1Rev | TCTAGATTAACCCATTTGCTGTCCACCAGTCATGCTAGCCATTTTCTTACCGACATCTGC | 3′ Primer, flp1T7 |
| Omp67F | TCTGGACGTATTGCTTTATCCGC | 5′ Primer morC upstream |
| Omp67R | CTTCCTCGAGCTTATTATCCGTTCTTGTTGA | 3′ Primer morC upstream |
| PpxF | CTTCTCTAGATTATGAATAACGAAAATTTA | 5′ Primer morC downstream |
| PpxR | TCAACGTGCCGACAGGCTTA | 3′ Primer morC downstream |
| SpecF | TAAGCTCGAGTGACTAAATAGTGAGG | 5′ Primer pSL60 cassette |
| SpecR | CTTCTCTAGACATGTGATTTTCCTCC | 3′ Primer pSL60 cassette |
Restriction sites are underlined.
Generation of epitope-tagged fimbrial subunits
The Flp1 protein was modified at the C terminus of the protein with a T7 tag according to the procedure of Kachlany et al. (2001). The flp1 gene was amplified by PCR with the primers Flp1For and Flp1Rev (Table 2) using chromosomal DNA isolated from the fimbriated clinical strain VT1257 as a template. The amplicon was ligated into a pGEM TA cloning vector and transformed into E. coli DH10B. The insert was released from the plasmid by restriction with the appropriate enzymes, purified using a commercial kit (Qiagen), ligated into a pKM2 shuttle vector and transformed into DH10B cells. The resulting plasmid was used to transform A. actinomycetemcomitans WT or morC mutant cells (Gallant et al., 2008).
Generation of a morC deletion mutant strain
Inactivation of the morC gene in the clinical isolate VT1257 was performed according to the method of Gallant et al. (2008) with modifications. DNA fragments upstream and downstream of the morC gene were fused to a spectinomycin resistance cassette designed to allow translational readthrough to mitigate potential polar effects (Lukomski et al., 2000) using the primers shown in Table 2. The construct was introduced into the conjugative plasmid pVT1460. A strain of E. coli auxotrophic for diaminopimelic acid was used as the donor strain to transfer the plasmid to A. actinomycetemcomitans (Babic et al., 2008). Conjugation mixtures were plated on medium containing spectinomycin to select for transconjugants. Selected transconjugants were screened genotypically and phenotypically to verify that the morC gene was inactivated.
Crystal violet biofilm assay
Biofilm assays were based on the method of Merritt et al. (2005). A. actinomycetemcomitans strains were grown on solid medium. Cells were collected by scraping with a sterile glass slide and suspended in sterile TSBYE. Cell numbers were standardized based on protein concentration to account for the autoaggregation of the fimbriated strains. Equal cell equivalents were inoculated into sterile 96-well microtitre plates (Nunc) and grown to stationary phase. Following growth, the supernatants were aspirated and the remaining non-adherent cells were removed by three consecutive washes with PBS (pH 7.4; Sigma Aldrich). Biofilms were stained with 0.1 % crystal violet for 20 min and washed three times with PBS. Bound dye was solubilized using a 2 : 1 solution of water : glacial acetic acid. Biomass was quantified by absorbance at 630 nm (A 630) on an ELx800 plate reader (Biotek). Each experiment was performed three times with three replicates in each experiment. ANOVA with Dunnett's post-test was used to identify significant differences (P < 0.05) between the biomass of each strain.
Confocal microscopy
Cells were grown in glass-bottomed dishes (MatTek). Following growth, the supernatants were aspirated, and non-adherent cells were removed by three washes with TBS (pH 7.4; Sigma). Biofilms were stained for 30 min with 5 μM SYTO 9 (Invitrogen) in TBS. The staining solution was decanted, and unbound stain was removed by four washes with TBS. Images were recorded at the University of Vermont Microscopy Imaging Center using a Zeiss LSM 510 META confocal microscope with a plan-apochromatic × 63 objective and an excitation wavelength of 488 nm. Random fields were selected, Z-slices were acquired at increments of 0.37 μm and Z-stacks were generated. Stack sizes depended on biofilm depth and ranged from 5.2 to 40 μm. Surface coverage of the biofilm was determined using the particle analysis function of the ImageJ program (Schneider et al., 2012). The shape, volume and surface area of individual microcolonies were quantified using the Volocity software package (PerkinElmer). All experiments were performed in triplicate. For comparisons between strains, ANOVA with Dunnett's post-test was used with significance defined as P < 0.05.
Transmission electron microscopy
A. actinomycetemcomitans cells were visualized by transmission electron microscopy based on the method described by Azari et al. (2013). Briefly, appropriate strains were streaked for isolation on solid medium. Colonies were transferred directly to carbon-coated grids and stained using Nano-W (Nanoprobes). Images were collected on a 2048 × 2048 pixel charge-coupled camera with a pixel size of 14 μm (TVIPS) at 52 000 nominal magnification using a Tecnai 12 electron microscope operating at 100 000 V (FEI).
Quantification of surface fimbrial subunits
The relative number of fimbriae present on the surface of the bacterial strains was determined using an ELISA and antibodies specific for the T7 protein tag associated with the Flp1 fimbrial subunit. Overnight bacterial colonies containing the plasmid expressing T7-flp1 were collected from a confluent agar plate and suspended in 2 ml medium. Suspensions were allowed to settle for 10 min, diluted (1 : 5) and a 100 μl aliquot was added to the wells of sterile 96-well plates. Following overnight incubation, the medium was removed and the wells were blocked with a 1 % solution of BSA (Sigma) in PBS. Following three washes with PBS, 100 μl anti-T7 mAb (1 μg ml− 1; EMD Millipore) in 1 % BSA was added. Immune complexes were detected by addition of 100 μl horseradish peroxidase-labelled goat anti-mouse antibody (1 μg ml− 1; Jackson Laboratory). The reaction was initiated by addition of hydrogen peroxide and stopped by the addition of an excess of 4 M sulfuric acid (Mintz, 2004). The mean protein concentration of the biofilm formed was determined using a bicinchoninic acid assay (Thermo Scientific) and used to normalize the absorbance values. Experiments were repeated three times with a minimum of three wells per assay. Data are expressed as the percentage of the WT strain. Student's t-test was used to compare the WT and morC mutant strains, with significance defined as P < 0.05.
Results
Identification of the defect in fimbriae production
A recent quantitative membrane proteomic study revealed a consistent decrease in the abundance of the proteins of the fimbrial secretion apparatus in the morC mutant strain when compared with the parent strain (Smith et al., 2015a). To fully comprehend the significance of these results, it was necessary first to determine the molecular basis of the defect that caused the afimbriation of the parent strain and secondly to complement and restore fimbrial production.
In the proteomics study of Smith et al. (2015a), TadZ, a protein encoded in the tad locus but not associated with the fimbriae secretion apparatus (Perez-Cheeks et al., 2012), was found at equal abundance in the morC mutant and the parent strain. This finding eliminated point mutations in the promoter region of the tad locus as a potential mechanism causing afimbriation. Sequencing of the fimbrial structural subunit gene (flp1) of the parent strain (VT1169) revealed a base deletion at nt 105. This deletion resulted in a frame shift, which introduced four stop codons at nt 121–123, 142–144, 165–167 and 231–233 (based on the HK1651 sequence). This shift resulted in a truncated form of the protein (Fig. 1). No premature stop codons were identified in the sequences of the non-functional Flp2 subunit and the TadV peptidase. However, based on the proteomic data, TadV was not identified in the membrane of either the WT (VT1169) or morC mutant (VT1650) strains (Smith et al., 2015a, b). Therefore, both strains were transformed with a replicating plasmid expressing the flp1-tadV region of the tad locus, under the control of an endogenous A. actinomycetemcomitans promoter, and investigated for fimbriae production.
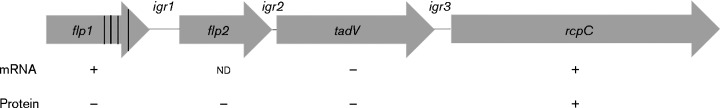
Fig. 1.
Partial organization of the tad operon. Protein-coding genes are represented by arrows. Horizontal lines represent intergenic regions. Vertical lines in flp1 represent premature stop codons. Symbols below each gene indicate the detection (+) or absence ( − ) of mRNA or protein in A. actinomycetemcomitans VT1169. igr, Intergenic region; nd, not determined.
Colony morphology of transformed parent and morC mutant strains
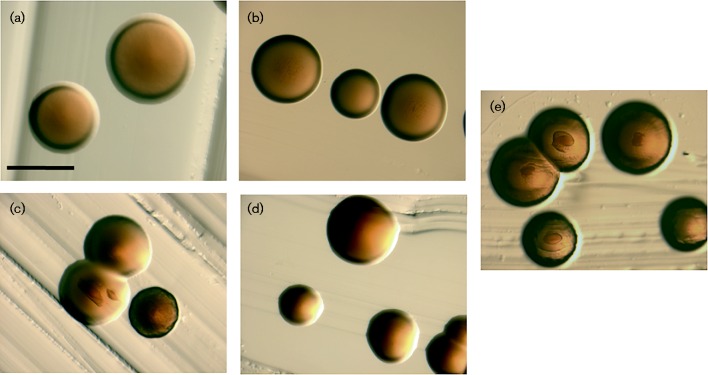
Colonies of afimbriated strains are large (>1 mm) and translucent. Afimbriated colonies have a regular edge and lack internal structures as represented by the laboratory strain VT1169 (Fig. 2a). Transformation of the WT strain (VT1169) with the flp1-tadV plasmid in trans (KM611, Fig. 2c) resulted in small ( < 1 mm), opaque colonies with irregular edges. Some colonies additionally displayed internal structures, all characteristic of the colony morphology typical of fimbriated clinical isolates (VT1257, Fig. 2e). Replating of individual colonies with or without observable internal structures resulted in a majority of the colonies (83 %) with no discrete structures, whereas 17 % of the colonies displayed the morphology associated with the clinical isolate.
Fig. 2.
A. actinomycetemcomitans colony morphology. Bacteria were grown on solid TSBYE medium for 3 days at 37 °C in a humidified 10 % CO2 atmosphere. Images were taken using a Leica MZ16F stereomicroscope. (a) WT (VT1169); (b) morC mutant (VT1650); (c) WT/pKM2/flp1-tadV (KM611); (d) morC mutant/pKM2/flp1-tadV (KM612); (e) fimbriated clinical isolate (VT1257). Bar, 1 mm.
Colonies of the morC mutant (VT1650, Fig. 2b) were identical to the WT afimbriated strain (VT1169). In contrast, the morC mutant strain transformed with the flp1-tadV plasmid (KM612, Fig. 2d) formed colonies indistinguishable from the parent strain (VT1169, Fig. 2a). Nucleotide sequences of the plasmids isolated from the two transformed strains (KM611 and KM612) were found to be identical, suggesting that the lack of fimbriation in strain KM612 was not due to mutations in the amplicon introduced during the cloning process.
Fimbriae expression in transformed parent and morC mutant strains
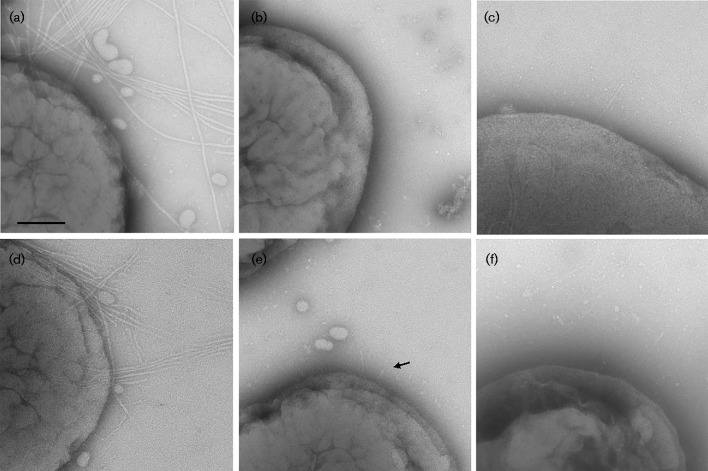
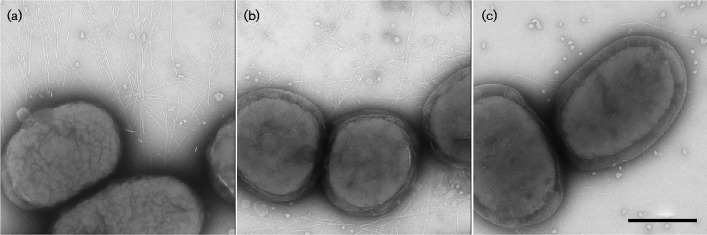
The internal colony structure associated with the transformed WT (KM611) and clinical (VT1257) strains is related to the expression of fimbriae (Inouye et al., 1990). Visualization of individual cells by transmission electron microscopy revealed fimbriae associated with the clinical strain (VT1257, Fig. 3a). Fimbriae were not detected in preparations of the afimbriated WT (VT1169) or the morC mutant (VT1650) strains (Fig. 3b, c, respectively). In contrast, fimbriae were observed in the WT afimbriated strain transformed with the flp1-tadV plasmid (KM611, Fig. 3d). However, there were noticeable differences in the characteristics of the fimbriae between the clinical (VT1257) and the complemented strain (KM611) strains. The fimbriae associated with VT1257 appeared overall to be longer and more abundant than the fimbriae of the complemented strain. In addition, a large heterogeneity in fimbrial length and abundance was observed among cells of strain KM611 (Fig. 4). Some of the cells (20 % of the inspected bacteria) produced fimbriae with the characteristics of the clinical strain VT1257 fimbriae. The remaining bacteria were observed to contain very short stub-like protrusions (∼100 nm, Fig. 3e) associated with the cell surface. In contrast, the morC afimbriated mutant strain transformed with the identical flp1-tadV plasmid (KM612) did not express fimbriae, as determined by the absence of fimbriation in all mutant cells imaged by transmission electron microscopy (Fig. 3f). Visualization of acellular regions of the grids did not reveal the presence of fimbrial fragments or fimbrial bundles detached from the cell envelope, which are commonly observed in preparations of the clinical strain VT1257, further confirming the lack of fimbriae of the KM612 cells.
Fig. 3.
Transmission electron microscopy of A. actinomycetemcomitans strains. Whole-mount negatively stained preparations of A. actinomycetemcomitans strains grown on solid medium were analysed by transmission electron microscopy. (a) Fimbriated clinical isolate (VT1257); (b) WT (VT1169); (c) morC mutant (VT1650); (d, e) WT (VT1169)/pKM2/flp1-tadV (KM611); (f) morC mutant/pKM2/flp1-tadV (KM612). The arrow indicates a fimbrial stub. Bar, 100 nm.
Fig. 4.
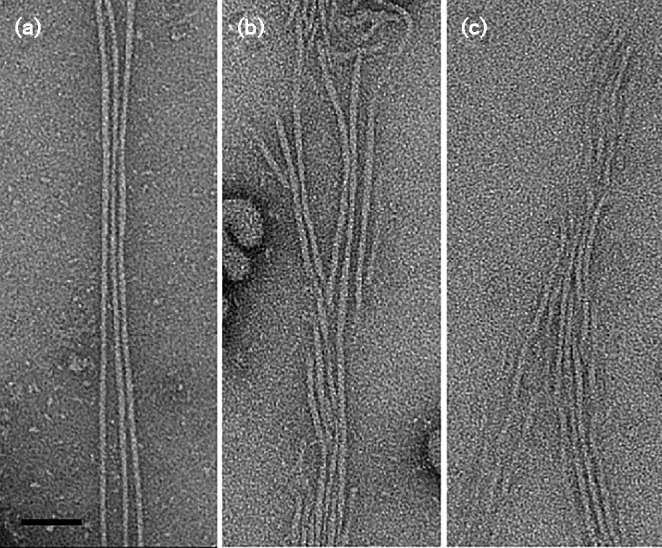
Transmission electron microscopy of fimbriae. Whole-mount negatively stained preparations of A. actinomycetemcomitans strains grown on solid medium were analysed by transmission electron microscopy and inter-bacterial regions containing fimbriae were extracted. (a) Fimbriated clinical isolate (VT1257); (b, c) WT (VT1169)/pKM2/flp1-tadV (KM611). Bar, 50 nm.
Biofilm production and architecture of transformed parent and morC mutant strains
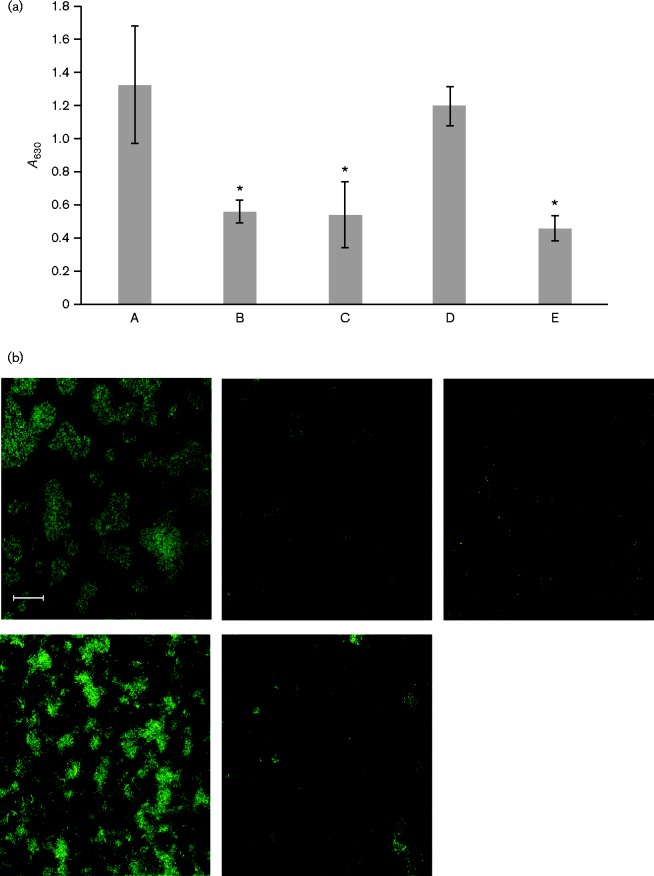
Biofilm formation is dependent on the fimbriation state of A. actinomycetemcomitans. The naturally fimbriated clinical strain (VT1257) formed a relatively robust biofilm with a greater biomass relative to the afimbriated WT (VT1169) and morC mutant (VT1650) strains, as determined by a standard static biofilm assay based on crystal violet retention (Fig. 5a). The biofilm formed by the WT strain transformed with the flp1-tadV plasmid (KM611) was equivalent to the one formed by the clinical strain (VT1257). However, the biomass of the biofilm formed by the morC mutant expressing the same construct (KM612) was unchanged when compared with the one formed by the parent strain (VT1169).
Fig. 5.
Quantification and confocal microscopy of biofilm formation of strains of A. actinomycetemcomitans. (a) Quantification of biofilm mass using a crystal violet assay. A. actinomycetemcomitans strains were grown as biofilms in glass-bottomed dishes and stained with SYTO 9. A series of Z-stack images was generated with a Zeiss LSM 510 META confocal microscope, and biofilm area was quantified using ImageJ. Asterisks indicate significant difference from the WT fimbriated strain (ANOVA with Dunnett's post-test, P < 0.05). A, Fimbriated clinical isolate (VT1257); B, WT (VT1169); C, morC mutant (VT1650); D, WT/pKM2/flp1-tadV (KM611); E, morC mutant/pKM2/flp1-tadV (KM612). Results are shown as means ± sd. (b) Representative fields of confocal microscopy of biofilms formed by the strains in (a). Upper left: Fimbriated clinical isolate (VT1257); middle: WT (VT1169; upper right: morC mutant (VT1650); lower left: WT/pKM2/flp1-tadV (KM611); lower right: morC mutant/pKM2/flp1-tadV (KM612). Bar, 20 μm.
Crystal violet biofilm assays quantify the total amount of organic material composing a biofilm, which includes cells, proteins and exopolysaccharides. To visualize the structure of the cellular components of the biofilm, we utilized confocal microscopy (Fig. 5b). The WT afimbriated strain (VT1169) lacked obvious microcolony formation. Numerous individual cells were observed, and coverage of the glass surface was minimal. These biofilm attributes were unaffected by inactivation of morC (VT1650). After transformation of the WT afimbriated strain with the flp1-tadV plasmid (KM611), the biofilm morphology displayed different characteristics from the original afimbriated strain (VT1169). Microcolonies with irregular edges were observed in addition to numerous individual cells in the KM611 strain, while the morC mutant transformed with the identical plasmid (KM612) displayed biofilm features similar to VT1169.
The architecture of the biofilm formed by the WT afimbriated strain expressing the flp1-tadV construct (KM611) differed from the biofilm produced by the clinical, fimbriated strain (VT1257) (Fig. 5b). Strain VT1257 formed large, discrete microcolonies, with few free cells in between. In contrast, the KM611 strain formed diffuse microcolonies with individual cells in between the microcolonies. The total biofilm surface coverage of both strains was equivalent and represented 8042 μm2 per field for strain KM611 and 8962 μm2 per field for strain VT1257. However, the mean volume of the biofilm per field was significantly lower (ANOVA, P < 0.05) for the afimbriated strain transformed with flp1-tadV (KM611, 32 715 μm3) than for the naturally fimbriated strain (VT1257, 114 134 μm3).
MorC is required for maximal fimbriae production and biofilm microcolony formation in A. actinomycetemcomitans
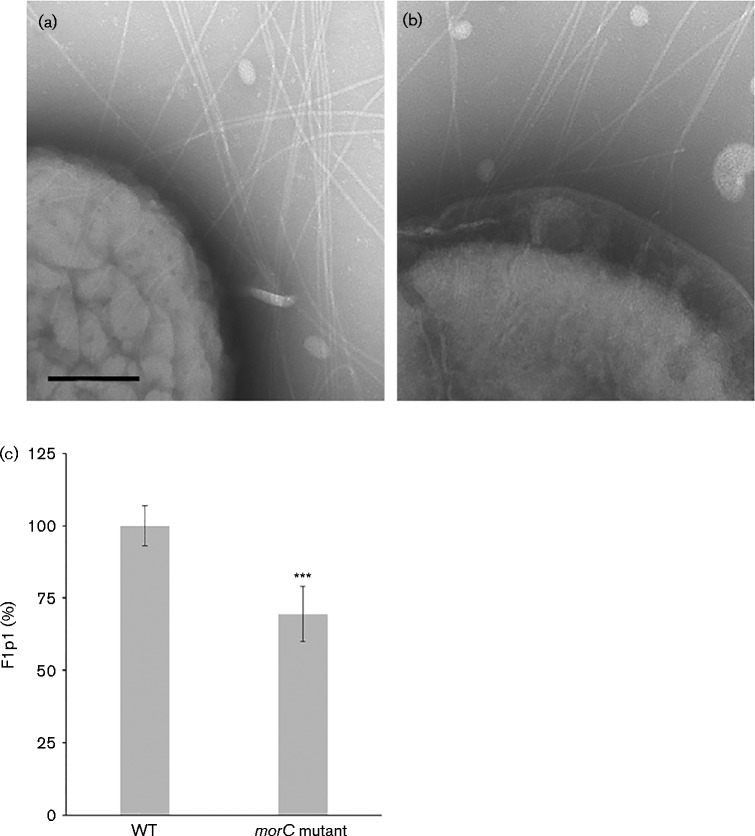
The data so far suggested that MorC is required for the surface expression of fimbriae and biofilm microcolony formation in a laboratory strain of A. actinomycetemcomitans. To investigate the role of morC in the fimbriae synthesis and biofilm formation of a clinical, naturally fimbriated strain, a morC deletion mutant was constructed in a clinical strain (KM700). This strain demonstrated the same measurable phenotypes that are associated with the afimbriated morC mutant strain (VT1650), including: decrease in toxin secretion, sensitivity to bile salt, altered membrane morphology and absence of MorC as determined by immunoblotting. Examination of the KM700 strain by transmission electron microscopy suggested a lower abundance of fimbriae associated with this strain compared with the parent VT1257 strain (Fig. 6 and Fig. 7a, b). To quantify the abundance of fimbriae associated with the clinical parent and mutant cells, a T7-tagged version of the fimbrial subunit, Flp1, was expressed in these strains. In the strains expressing the T7-tagged Flp1 protein, a statistically significant 30 % decrease (t-test, P < 0.001) in the abundance of fimbriae on the surface of the morC mutant strain (KM700) was observed when compared with the parent strain (VT1257) (Fig. 7c). Strains containing empty plasmid or no plasmid generated little detectable signal in this assay format.
Fig. 6.
Transmission electron micrographs of A. actinomycetemcomitans strains. Whole-mount negatively stained preparations of A. actinomycetemcomitans strains grown on solid medium were analysed by transmission electron microscopy and showed whole bacteria and inter-bacterial regions of WT (VT1257) (a) and the morC mutant (KM700) (b, c). Bar, 0.5 μm.
Fig. 7.
Transmission electron micrographs and quantification of fimbriae production of clinical and isogenic morC mutant strains of A. actinomycetemcomitans. (a, b) Whole-mount negatively stained preparations of A. actinomycetemcomitans strains grown on solid medium were analysed by transmission electron microscopy for WT (VT1257) (a) and the morC mutant (KM700) (b) Bar, 100 nm. (c) Quantification of surface-associated fimbriae assessed by ELISA. Asterisks indicate a significant difference from the WT strain (t-test, ***P < 0.001).
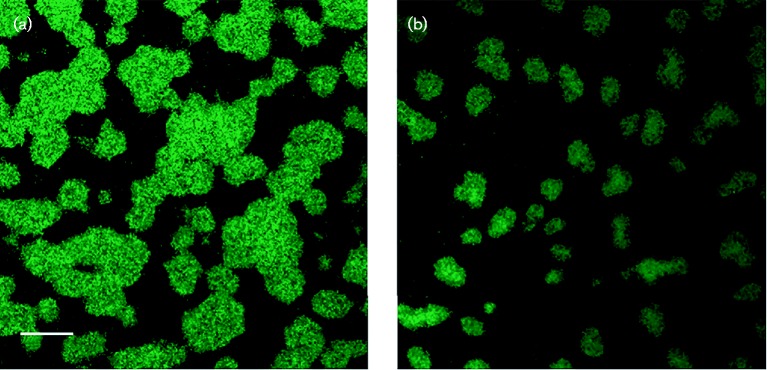
The decrease in the abundance of fimbriae did not affect the mass of the biofilm formed by the mutant (KM700) compared with the parent strain as determined by the crystal violet assay (data not shown). However, there were significant differences in the microcolony architecture of VT1257 and KM700 (Fig. 8a, b, respectively). Large, amorphous microcolonies were commonly observed in the biofilm formed by strain VT1257, while smaller and rounder microcolonies were observed in the mutant strain. The mean volume of a microcolony formed by VT1257 was 2219 μm3 and that by the morC mutant strain (KM700) was 1309 μm3, which represented an approximate twofold reduction in microcolony size.
Fig. 8.
Confocal microscopy of biofilms generated by clinical strains of A. actinomycetemcomitans. A. actinomycetemcomitans strains were grown as biofilms in glass-bottomed dishes and stained with SYTO 9. A series of Z-stack images was generated with a Zeiss LSM 510 META confocal microscope. (a) Fimbriated clinical isolate (VT1257); (b) morC mutant (KM700). Images are representative of three separate experiments. Bar, 20 μm.
Discussion
The protein composition of the A. actinomycetemcomitans membrane envelope has evolved to fit the specific environmental niche that the bacterium colonizes in the oral cavity. A membrane proteome study revealed the presence of specific secretion systems responsible for secretion and incorporation of proteins into the inner and outer membranes (Smith et al., 2015a, b). Although the macromolecular composition of secretion complexes is well characterized in other bacterial systems, the accessory proteins that modulate secretion are understudied (Silhavy et al., 2010). In A. actinomycetemcomitans, the absence of MorC impairs leukotoxin secretion (Gallant et al., 2008). However, the reduction in toxin secretion in the mutant strain is not associated with changes in the abundance of the proteins forming the secretory apparatus (Smith et al., 2015a). Interestingly, the abundances of known autotransporter proteins (e.g. EmaA, Aae and ApiA) are also unchanged in the morC mutant strain (Smith et al., 2015a). This is in contrast to what was found in Enterobacteriaceae for a mutant strain of a morC homologue (tamB) where the Flu autotransporter proteins were affected (Selkrig et al., 2012). Moreover, transformation of the A. actinomycetemcomitans mutant (VT1650) with homologues of morC from other Gammaproteobacteria complemented some but not all of the mutant phenotypes (Smith et al., 2015c). This implies that the role of MorC in membrane biogenesis may vary among Gammaproteobacteria and thus can display a wide variety of phenotypes in different bacteria.
In contrast to the observed equivalent abundance of autotransporter and leukotoxin secretion apparatus proteins in the morC mutant and parent strains, the amount of fimbriae-associated proteins was reduced in the morC mutant strain (Smith et al., 2015a). Proteomic and DNA sequence analysis of the genetic locus associated with fimbrial production in this study provided a basis for the complementation of fimbriae expression in the afimbriated strains under study. The effect on fimbriation appears to occur post-transcriptionally, as both the WT (VT1169) and morC mutant (VT1650) afimbriated strains show equal amounts of TadZ (Smith et al., 2015a), the product of the seventh gene of the 14-gene tad operon (Perez-Cheeks et al., 2012). In trans expression of a prototypic flp1-tadV sequence promoted the production of fimbriae in the afimbriated WT strain. However, microscopic examination of this strain (KM611) revealed the presence of two different colony morphologies. The majority of colonies (>95 %) were small, rough in appearance and adherent to agar. However, the internal star-shaped structure characteristic of clinically isolated fimbriated strains was lacking. Colonies displaying these phenotypes have been observed previously and are described as an intermediate form between fimbriated and afimbriated strains (Inouye et al., 1990). The remaining colonies displayed a star-shaped internal structure characteristic of fimbriated strains and were indistinguishable from a low-passage-number clinical strain (VT1257) isolated directly from the oral cavity. Heterogeneity in colony morphology of strain KM611 was reproduced upon replating individual colonies, implying that the fimbriation phenotype in this laboratory strain exhibited incomplete penetrance or stochasticity in gene expression, phenomena that are well described in bacteria (Elowitz et al., 2002).
Fimbriae are associated with biofilm formation in A. actinomycetemcomitans (Kachlany et al., 2000). Differences in biofilm mass are apparent between fimbriated and afimbriated strains. The total biomass of the biofilm formed by the afimbriated strain transformed with the flp1-tadV plasmid (KM611) was enhanced compared with the parent strain (VT1169) and was equivalent to the naturally fimbriated strain (VT1257). Interestingly, only 20 % of the KM611-transformed cells expressed fimbriae that resembled the fimbriae associated with VT1257. The majority of cells expressed short fimbriae or long fimbriae in lesser abundance. This observation suggests maximal fimbriae length and/or abundance per cell is not required to maintain optimal biofilm mass (as measured by a crystal violet assay) under the laboratory conditions used in this study.
Clinical strains of A. actinomycetemcomitans form biofilms composed of well-defined, discrete microcolonies with few free cells in between the microcolonies (Haase et al., 2006) (Figs 5a and 8a). When compared with the clinical strain (VT1257) used in this study, the WT transformed strain (KM611) displayed a different biofilm phenotype with less-well-defined microcolonies and a greater abundance of free cells. In addition to altered biofilm architecture, as mentioned above, this strain displayed reduced abundance of fimbriae that appeared more fragile when compared with those present on the naturally fimbriated strain (VT1257). These observations suggest that, although maximum fimbriae production and integrity is not critical for total biofilm mass, biofilm architecture formed by the A. actinomycetemcomitans strains used in this study is dependent on fimbriae abundance and length.
In similar fimbrial secretion systems, subunits are secreted via a piston-like mechanism mediated by inner-membrane proteins of the complex, which ensure the proper spatial organization of the polymerizing structure (Craig et al., 2006). The abnormal fimbriae observed in the transformed strain (KM611) may be due to defects in the export mechanism of the fimbrial subunit. The laboratory-adapted strain used in this study (VT1169) may have accumulated mutations in one or more of the fimbrial secretion proteins, resulting in a defect in subunit export. A reduction in fimbriae export may explain the absence of fimbriae production in the morC mutant strain (VT1650). As the VT1169 cells are impaired in fimbrial secretion, the further reduction of the abundance of the export apparatus proteins in the mutant (Smith et al., 2015a) would result in a rate of subunit export not compatible with fimbrial formation.
The data associated with the strain KM612 suggested that inactivation of morC would lead to complete loss of fimbriae in a naturally fimbriated strain (VT1257). However, a partial loss of surface-exposed fimbriae in the fimbriated mutant (KM700) was observed, and may be attributed to the extensive genomic variation between strains, as reported for A. actinomycetemcomitans (Kittichotirat et al., 2011). However, a reduced abundance and altered microcolony architecture was evident in the isogenic morC mutant of the clinical strain (KM700). The overall appearance of the fimbriae of the parent and mutant strains was comparable. Therefore, the change in the microcolony phenotype may result from the decrease in the abundance of the secretion apparatus in the membrane of the mutant strain leading to fewer functional fimbriae expressed on the surface of the bacteria.
The specific 3D structure of MorC is unknown, and the molecular details of the interaction with other cell-envelope proteins have yet to be determined. The secondary protein structure is predicted to comprise 9 % helices, 48 % β-strands and 41 % coils. Structural prediction programs, based on homology and de novo modelling, have returned minimal information. Only one program, RaptorX (Källberg et al., 2012), returned a low-confidence 3D model prediction which depicts the molecule as having a boomerang-shaped periplasmic region, composed of two domains, that spans 15 nm and is anchored to the inner membrane by a non-interrupted α-helix containing residues 21–42. This structure is not significantly reliable enough to propose a detailed molecular model of MorC and its interacting partners in the cell envelope.
Two more comprehensive models can be envisioned to construe the role of MorC in membrane protein secretion: a direct and an indirect model. The direct model relies on the high sequence homology that the C terminus of MorC shares with AsmA, an E. coli inner-membrane protein involved in the insertion of proteins into the outer membrane (Deng & Misra, 1996). TamB, the Enterobacteriaceae orthologue of MorC, is proposed to interact with Tam A (Omp67), which functions as a lever arm to drive proteins into bacterial outer membranes (Selkrig et al., 2015). If the Tad structural proteins of A. actinomycetemcomitans are substrates for this secretion system, the reduced abundance of outer-membrane Tad proteins (e.g. RcpA and -B and TadD), which has been observed in our quantitative proteomic study (Smith et al., 2015a), would result in fewer fimbriae observed on the surface of the morC mutant strain. The moderate effect on fimbriae surface expression of the mutant may be attributed to a redundant but lower-affinity membrane secretion system, e.g. Bam, which does not fully compensate for the absence of MorC. The indirect model is based on the reduction of both the height of the outer-membrane convolutions and the number of connections between the inner and outer membrane associated with the morC mutant strain (Azari et al., 2013). The fourfold reduction in the number of membrane connections and the twofold decrease in the height of the membrane convolutions lead to a more moderate curvature of the outer membrane and a twofold decrease in the volume/area ratio of the periplasmic space. Thus, these fundamental effects would change the environment of the periplasmic compartment, compromising the equilibrium between the formation and degradation of assembling proteins. Concomitantly, these changes decrease the surface area of the regions of the cell envelope with the optimal inter-membrane distance for supporting interactions between the components of bacterial secretion systems. The reported morphological and physiological alterations would result in a change in the number of Tad-associated proteins and be transmuted into a decrease in the abundance of fimbriae observed on the surface of the mutant strain. It is conceivable that the actual mechanism incorporates aspects of both the direct and the indirect models, with the relative contribution of each being species dependent. In the case of the members of the families Pasteurellaceae and Moraxellaceae, characterized by the high rugosity of the bacterial surface, the indirect method might be preponderant.
To date, genes outside the tad locus are not known to have an effect on fimbriation of A. actinomycetemcomitans. In this work, we have demonstrated that the novel inner-membrane protein MorC is associated with fimbrial secretion and microcolony formation in A. actinomycetemcomitans. This finding builds on our previous observations that MorC plays an important role in the membrane physiology of this pathogen. As MorC homologues are present in Gammaproteobacteria, these proteins may contribute to the fimbriae secretion and biofilm architecture of these bacteria.
Acknowledgements
We would like to thank Thomas Freeman and Yan Xing for their contributions to this study. This study was supported by NIH grant RO1-DE018889 (K. P. M.) and has benefited from developments supported by NIH grant RO1-DE017474 (T. R.). We would like to thank Nicole Bouffard for her assistance in confocal imaging and 3D analysis of biofilms. Confocal microscopy experiments were performed in the Microscopy Imaging Center at the University of Vermont, College of Medicine. National Center for Research Resources award number 1S10RR019246 (Douglas Taatjes, University of Vermont) allowed the purchase of the Zeiss 510 META confocal scanning laser microscope.
References
- Azari F., Nyland L., Yu C., Radermacher M., Mintz K. P., Ruiz T. (2013). Ultrastructural analysis of the rugose cell envelope of a member of the Pasteurellaceae family J Bacteriol 195 1680–1688 10.1128/JB.02149-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic A., Guérout A. M., Mazel D. (2008). Construction of an improved RP4 (RK2)-based conjugative system Res Microbiol 159 545–549 10.1016/j.resmic.2008.06.004 . [DOI] [PubMed] [Google Scholar]
- Bodet C., Andrian E., Tanabe S., Grenier D. (2007). Actinobacillus actinomycetemcomitans lipopolysaccharide regulates matrix metalloproteinase, tissue inhibitors of matrix metalloproteinase, and plasminogen activator production by human gingival fibroblasts: a potential role in connective tissue destruction J Cell Physiol 212 189–194 10.1002/jcp.21018 . [DOI] [PubMed] [Google Scholar]
- Burall L. S., Harro J. M., Li X., Lockatell C. V., Himpsl S. D., Hebel J. R., Johnson D. E., Mobley H. L. (2004). Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold Infect Immun 72 2922–2938 10.1128/IAI.72.5.2922-2938.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. (2006). Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions Mol Cell 23 651–662 10.1016/j.molcel.2006.07.004 . [DOI] [PubMed] [Google Scholar]
- Das M., Badley A. D., Cockerill F. R., Steckelberg J. M., Wilson W. R. (1997). Infective endocarditis caused by HACEK microorganisms Annu Rev Med 48 25–33 10.1146/annurev.med.48.1.25 . [DOI] [PubMed] [Google Scholar]
- Deng M., Misra R. (1996). Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins Mol Microbiol 21 605–612. [DOI] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002). Stochastic gene expression in a single cell Science 297 1183–1186 10.1126/science.1070919 . [DOI] [PubMed] [Google Scholar]
- Fine D. H., Furgang D., Kaplan J., Charlesworth J., Figurski D. H. (1999). Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite Arch Oral Biol 44 1063–1076 10.1016/S0003-9969(99)00089-8 . [DOI] [PubMed] [Google Scholar]
- Gallant C. V., Sedic M., Chicoine E. A., Ruiz T., Mintz K. P. (2008). Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans J Bacteriol 190 5972–5980 10.1128/JB.00548-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase E. M., Bonstein T., Palmer R.J.,, Jr, Scannapieco F. A. (2006). Environmental influences on Actinobacillus actinomycetemcomitans biofilm formation Arch Oral Biol 51 299–314 10.1016/j.archoralbio.2005.09.002 . [DOI] [PubMed] [Google Scholar]
- Haubek D., Ennibi O. K., Poulsen K., Benzarti N., Baelum V. (2004). The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss J Dent Res 83 767–770 10.1177/154405910408301006 . [DOI] [PubMed] [Google Scholar]
- Inouye T., Ohta H., Kokeguchi S., Fukui K., Kato K. (1990). Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans FEMS Microbiol Lett 57 13–17 10.1111/j.1574-6968.1990.tb04167.x . [DOI] [PubMed] [Google Scholar]
- Kachlany S. C., Planet P. J., Bhattacharjee M. K., Kollia E., DeSalle R., Fine D. H., Figurski D. H. (2000). Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and Archaea J Bacteriol 182 6169–6176 10.1128/JB.182.21.6169-6176.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany S. C., Planet P. J., Desalle R., Fine D. H., Figurski D. H., Kaplan J. B. (2001). flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans Mol Microbiol 40 542–554 10.1046/j.1365-2958.2001.02422.x . [DOI] [PubMed] [Google Scholar]
- Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. (2012). Template-based protein structure modeling using the RaptorX web server Nat Protoc 7 1511–1522 10.1038/nprot.2012.085 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittichotirat W., Bumgarner R. E., Asikainen S., Chen C. (2011). Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis PLoS One 6 e22420 10.1371/journal.pone.0022420 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S., Hoe N. P., Abdi I., Rurangirwa J., Kordari P., Liu M., Dou S. J., Adams G. G., Musser J. M. (2000). Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization Infect Immun 68 535–542 10.1128/IAI.68.2.535-542.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H., Kadouri D. E., O'Toole G. A. (2005). Growing and analyzing static biofilms 10.1002/9780471729259.mc01b01s00 Curr Protoc Microbiol, 22:B:1B.1:1B.1.1–1B.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz K. P. (2004). Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen Microbiology 150 2677–2688 10.1099/mic.0.27110-0 . [DOI] [PubMed] [Google Scholar]
- Mintz K. P., Fives-Taylor P. M. (1994). Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line Infect Immun 62 3672–3678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cheeks B. A., Planet P. J., Sarkar I. N., Clock S. A., Xu Q., Figurski D. H. (2012). The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans Mol Microbiol 83 694–711 10.1111/j.1365-2958.2011.07955.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter A. E., Leyton D. L., Tveen-Jensen K., Browning D. F., Sevastsyanovich Y., Knowles T. J., Nichols K. B., Cunningham A. F., Overduin M., other authors (2011). The essential β-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis J Bacteriol 193 4250–4253 10.1128/JB.00192-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis Nat Methods 9 671–675 10.1038/nmeth.2089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner H. C., Sinatra K., Kaplan J. B., Furgang D., Kachlany S. C., Planet P. J., Perez B. A., Figurski D. H., Fine D. H. (2003). Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model Proc Natl Acad Sci U S A 100 7295–7300 10.1073/pnas.1237223100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J., Mosbahi K., Webb C. T., Belousoff M. J., Perry A. J., Wells T. J., Morris F., Leyton D. L., Totsika M., other authors (2012). Discovery of an archetypal protein transport system in bacterial outer membranes Nat Struct Mol Biol 19 506–510, S1 [DOI] [PubMed] [Google Scholar]
- Selkrig J., Belousoff M. J., Headey S. J., Heinz E., Shiota T., Shen H. H., Beckham S. A., Bamert R. S., Phan M. D., other authors (2015). Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module Sci Rep 5 12905 10.1038/srep12905 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D., Walker S. (2010). The bacterial cell envelope Cold Spring Harb Perspect Biol 2 a000414 10.1101/cshperspect.a000414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J., Reynolds H. S., Genco R. J. (1980). Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation Infect Immun 29 1013–1020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. P. (2015). A conserved inner membrane protein of Aggregatibacter actinomycetemcomitans is integral for membrane function Thesis, University of Vermont, VT, USA. . [Google Scholar]
- Smith K. P., Fields J. G., Voogt R. D., Deng B., Lam Y. W., Mintz K. P. (2015a). Alteration in abundance of specific membrane proteins of Aggregatibacter actinomycetemcomitans is attributed to deletion of the inner membrane protein MorC Proteomics 15 1859–1867 10.1002/pmic.201400505 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. P., Fields J. G., Voogt R. D., Deng B., Lam Y. W., Mintz K. P. (2015b). The cell envelope proteome of Aggregatibacter actinomycetemcomitans Mol Oral Microbiol 30 97–110 10.1111/omi.12074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. P., Voogt R. D., Ruiz T., Mintz K. P. (2015c). The conserved carboxyl domain of MorC, an inner membrane protein of Aggregatibacter actinomycetemcomitans, is essential for membrane function Mol Oral Microbiol 31 43–58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Forestier C., Krogfelt K. A. (2003). Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection Microbiology 149 167–176 10.1099/mic.0.25833-0 . [DOI] [PubMed] [Google Scholar]
- Tang G., Mintz K. P. (2010). Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway J Bacteriol 192 1395–1404 10.1128/JB.01453-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich M., Fine D. H., Figurski D. H. (2006). The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins J Bacteriol 188 6899–6914 10.1128/JB.00690-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu A., Chen C. (2005). Genetic basis for conversion of rough-to-smooth colony morphology in Actinobacillus actinomycetemcomitans Infect Immun 73 3749–3753 10.1128/IAI.73.6.3749-3753.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon J. J., Slots J., Genco R. J. (1983). Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease Infect Immun 41 19–27 . [DOI] [PMC free article] [PubMed] [Google Scholar]