Abstract
Staphylococcus aureus produces several virulence factors that allow it to cause a variety of infections. One of the major virulence factors is the capsule, which contributes to the survival of the pathogen within the host as a way to escape phagocytosis. The production of the capsular polysaccharide is encoded in a 16 gene operon, which is regulated in response to several environmental stimuli including nutrient availability. For instance, the capsule is produced in the late- and post-exponential growth phases, but not in the early- or mid-exponential growth phase. Several regulators are involved in capsule production, but the regulation of the cap operon is still poorly understood. In this study, we show that MsaB activates the cap operon by binding directly to a 10 bp repeat in the promoter region. We show that despite the fact that MsaB is expressed throughout four growth phases, it only activates capsule production in the late- and post-exponential growth phases. Furthermore, we find that MsaB does not bind to its target site in the early and mid-exponential growth phases. This correlates with decreased nutrient availability and capsule production. These data suggest either that MsaB binding ability changes in response to nutrients or that other cap operon regulators interfere with the binding of MsaB to its target site. This study increases our understanding of the regulation of capsule production and the mechanism of action of MsaB.
Introduction
Staphylococcus aureus is an opportunistic human pathogen that causes a wide variety of infections, ranging from relatively minor superficial skin infections to life-threatening diseases, such as osteomyelitis, endocarditis, and bacteraemia or septicaemia (Lowy, 1998, 2011). The ability of this pathogen to cause such infections relies on its many virulence factors and their coordinated regulation (David & Daum, 2010; Lowy, 1998; Cheung et al., 2004; Bronner et al., 2004; Somerville & Proctor, 2009; Novick & Geisinger, 2008). One of the major virulence factors is the capsular polysaccharide (CP) (O'Riordan & Lee, 2004). The production of the capsule has been shown to play a major role in bacterial virulence during infection by facilitating the pathogen's survival inside the host as a way to escape phagocytosis (Karakawa et al., 1988; Thakker et al., 1998; Nanra et al., 2013). This is particularly important during the acute phase of certain types of infection, including bacteraemia (Luong & Lee, 2002; Thakker et al., 1998; O'Riordan & Lee, 2004; Watts et al., 2005; Nilsson et al., 1997; Kampen et al., 2005). Several attempts have been made to produce an effective vaccine to protect individuals at risk of S. aureus infection. Normal human serum contains low levels of antibodies specific for the S. aureus CP5 and CP8 antigens (Fattom et al., 1996), and several animal models of infection have shown that the addition of antibodies specific for capsule serotypes confers some level of protection against S. aureus (Fattom et al., 1996; Lee et al., 1997). However, additional studies that used different models have found that no protection was conferred by the addition of CP-specific antibodies (Nemeth & Lee, 1995). These findings suggest that the efficiency of antibodies specific for capsule in eliminating S. aureus depends on the type of infection model used. Specifically, in the study by Lee et al. (1997) a modified method of the route of inoculum was used in a rat infective endocarditis model. The modified method included a challenge using the intraperitoneal injection route to achieve slow infusion of S. aureus into the blood, compared to the intravenous challenge used by other studies (Nemeth & Lee, 1995). It is becoming more evident that capsule is only produced under certain experimental conditions and under specific conditions associated with the infection process.
The capsule is proposed to act primarily as an antiphagocytic factor that allows the pathogen to persist and escape phagocytic uptake during its movement from one localized site to another in the course of infection or during its circulation through the bloodstream (Thakker et al., 1998). Other studies have suggested that the capsule does not prevent phagocytosis but instead, through an undetermined mechanism, improves the ability of the bacterium to survive within the polymorphonuclear neutrophils (PMNs) after it has been ingested (Voyich et al., 2005). Another hypothesis is that the fine-tuning of capsule production is essential when the infection is progressing to a more chronic long-term infection (Tuchscherr et al., 2010). The initial adherence of the bacterium to surfaces is facilitated by the downregulation in capsule production. When no capsule is present, cell adhesins are exposed on the surfaces of cells. This allows the bacterium to adhere to a surface or a host cell. After adherence to the host cell is achieved, the bacterium can then proceed to internalize within it, where it can readily escape elimination from the components of the host immune system (Tuchscherr et al., 2010). Regardless of the mechanism, it is clear that the production of the capsule is involved in bacterial pathogenesis by enhancing the ability of the bacterium to survive during the acute phase of infection and in the transition to the chronic stage of infection by either resisting phagocytosis or surviving after ingestion (O'Riordan & Lee, 2004; Thakker et al., 1998; Luong & Lee, 2002; Watts et al., 2005; Nilsson et al., 1997; Voyich et al., 2005; Tuchscherr et al., 2010; Kampen et al., 2005).
Capsule production is encoded by a single operon composed of 16 genes (Ouyang et al., 1999; Sau et al., 1997b). S. aureus produces four main serotypes of capsule, including the heavily encapsulated serotypes CP1 and CP2, and the microcapsulated serotypes CP5 and CP8. However, in the clinical context, serotypes CP5 and CP8 are considered most significant and account for 70–80 % of clinical isolates (Arbeit et al., 1984; Sompolinsky et al., 1985; Karakawa & Vann, 1982). The DNA that encodes the CP5 and CP8 serotypes is very similar, including serotype-specific genes that are flanked by common, nearly identical genes (Sau et al., 1997a). Given the similarity in the gene sequences of these two serotypes, it is inferred that the regulation of capsule production in the two serotypes is also similar (Sau et al., 1997a). The promoters of the cap operons in these two serotypes are nearly identical, and are located directly upstream from the capA ORF (Ouyang et al., 1999). The regulation of capsule production is complex and depends on the bacterial growth phase. Capsule production is suppressed in the early and mid-exponential growth phases, but is activated in the late- and post-exponential growth phases (Poutrel et al., 1995; Cunnion et al., 2001; Dassy & Fournier, 1996).
Despite the various environmental conditions that regulate capsule production, very little is known yet about the specific regulators that control the expression of the cap operon. Several global regulators have been shown to control the production of the capsule, including agr, sarA, arl and mgrA, as well as others (Luong et al., 2002, 2003, 2011; Luong & Lee, 2006; Chen et al., 2007; Dassy et al., 1993; Gupta et al., 2013). A detailed analysis of the cap operon promoter has revealed that it is highly regulated (Ouyang et al., 1999). A 10 bp inverted repeat that is located directly upstream from the promoter has been predicted to be a binding site for a transcription regulator (activator or repressor) (Ouyang et al., 1999). Additionally, several regulatory proteins have been shown to bind to the promoter region of the cap operon. These include the two-component systems AirSR, KdpDE and SpoVG, as well as nutrient-sensing regulatory proteins CcpE and CodY (Zhao et al., 2010; Majerczyk et al., 2010; Sun et al., 2012; Ding et al., 2014; Jutras et al., 2013). CodY is the best-studied cap regulatory protein and has been shown to sense the availability of nutrients, such as branched-chain amino acids and GTP, during the different phases of bacterial growth. The binding affinity of CodY is highest in an excess of amino acids and GTP, and as the intercellular pools decrease, the binding affinity of CodY decreases, resulting in a derepression effect (Ratnayake-Lecamwasam et al., 2001; Shivers & Sonenshein, 2004; Pohl et al., 2009). Recently, a new DNA-binding regulator, RbsR, has been shown to bind to the cap promoter region and to activate transcription of the cap operon. RbsR is controlled directly by the alternative sigma factor SigB (Lei & Lee, 2015).
We previously identified the msaABCR operon, which is involved in the regulation of virulence, biofilm development and antibiotic resistance in S. aureus (Sambanthamoorthy et al., 2006, 2008; Samanta & Elasri, 2014; Sahukhal & Elasri, 2014; Sahukhal et al., 2015). The deletion of the msaABCR operon alters the expression of several global regulators, including sarA, agr and sigB, all of which have been shown to be indirectly involved in capsule production (Bischoff et al., 2004; Pané-Farré et al., 2006; Meier et al., 2007; Dassy et al., 1993; Luong et al., 2002; Sahukhal & Elasri, 2014). The regulatory mechanism for msaABCR has not yet been defined. In this study, we show that msaABCR is essential for capsule production, and that MsaB is a transcriptional activator of the cap operon, binding specifically to the 10 bp inverted repeat located upstream from the cap operon. We also show that the regulation of capsule production is analogous in both clinical serotypes CP5 and CP8.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are described in Table 1. We used a representative strain for each of the two clinically significant S. aureus capsule serotypes, CP5 and CP8: Mu50, a known vancomycin-intermediate S. aureus (VISA) isolate that produces capsule serotype 5; and UAMS-1, a vancomycin-sensitive S. aureus (VSSA) osteomyelitis isolate that produces capsule serotype 8. A restriction-deficient laboratory strain of S. aureus, RN4220, and the DH5α strain of Escherichia coli were used to move plasmid constructs into the strains of choice through transformation and phage transduction, as described elsewhere (Bae & Schneewind, 2006; Sahukhal & Elasri, 2014; Samanta & Elasri, 2014). S. aureus strains were grown on tryptic soy agar (TSA) or in tryptic soy broth (TSB). When required, either erythromycin (10 μg ml− 1) or chloramphenicol (10 μg ml− 1) was added to media for selection. The E. coli strain DH5α was grown in Luria–Bertani (LB) medium, with ampicillin (100 μg ml− 1) used for selection when required.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Relevant feature | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | F− ϕ80lacZΔM15 recA1 | Life Technologies |
| S. aureus | ||
| Mu50 | CP5-producing strain | NARSA |
| Mu50ΔmsaABCR | msaABCR operon knock-out | Samanta & Elasri (2014) |
| Mu50ΔmsaABCR comp. | Mu50ΔmsaABCR (pCN34-msaABCR operon) | Samanta & Elasri (2014) |
| Mu50ΔmsaB | msaB knock-out | This study |
| Mu50ΔmsaB comp. | Mu50ΔmsaB (pCN34-msaABCR operon) | This study |
| Mu50 | Mu50 (pCN58-cap promoter) | This study |
| Mu50ΔmsaABCR | Mu50ΔmsaABCR (pCN58-cap promoter) | This study |
| Mu50ΔmsaB | Mu50ΔmsaB (pCN58-cap promoter) | This study |
| UAMS-1 | CP8-producing strain | |
| UAMS-1ΔmsaABCR | msaABCR operon knock-out | This study |
| UAMS-1ΔmsaABCR comp. | UAMS-1ΔmsaABCR (pCN34-msaABCR operon) | This study |
| UAMS-1ΔmsaB | msaB knock-out | This study |
| UAMS-1ΔmsaB comp. | UAMS-1ΔmsaB (pCN34-msaABCR operon) | This study |
| UAMS-1 | UAMS-1 (pCN58-cap promoter) | This study |
| UAMS-1ΔmsaABCR | UAMS-1ΔmsaABCR (pCN58-cap promoter) | This study |
| UAMS-1ΔmsaB | UAMS-1ΔmsaB (pCN58-cap promoter) | This study |
| USA300 JE2 | capA : : Tn transposon mutant (CP negative) | NARSA |
| UAMS-1ΔcapA | capA : : Tn transposon mutant (CP negative) | This study |
| Plasmids | ||
| pKOR1 | Ampr Cmr; shuttle vector, temperature-sensitive | Bae & Schneewind (2006) |
| pKOR1-ΔmsaABCR | Upstream and downstream fragment of msaABCR operon cloned into pKOR1 for mutagenesis | Samanta & Elasri (2014) |
| pCN34 | Ampr Ermr; shuttle vector, low copy number | Charpentier et al. (2004); NARSA |
| pCN34(Cmr)-msaABCR operon | 1.7 kb PCR fragment containing msaABCR operon cloned into pCN34(Cmr) | Samanta & Elasri (2014) |
| pCN58-vector control | Promoterless LuxAB as a reporter gene for promoter–gene fusion studies | NARSA |
| pCN58-cap promoter | Promoterless LuxAB as a reporter gene for promoter–gene fusion studies of cap | This study |
Generation of deletion mutants and complementation
The allelic replacement method was used to generate the msaABCR and msaB deletion mutants in strains Mu50 and UAMS-1 (Bae & Schneewind, 2006). For trans-complementation, the msaABCR region was cloned into the pCN34 low-copy vector with the modification of changing the kanamycin selectable marker to a chloramphenicol-resistance marker as described elsewhere (Samanta & Elasri, 2014; Charpentier et al., 2004). The capsule mutant was generated by insertion of a transposon in the capA ORF. Briefly, strain NE302 (SAUSA300 0152) was obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) collection (bei Resources). This strain contains the bursa aurealis mariner-based erythromycin-resistance expression transposon within the encoding capA region. The mutation was moved by generalized transduction using bacteriophage Φ11 (Sambanthamoorthy et al., 2006; Samanta & Elasri, 2014). Introduction of the capA : : Tn mutation into the recipient strain was verified by PCR, sequencing and capsule production assay. The primers used in this study are listed in Table 2.
Table 2. Primers used in this study.
| Primer | Sequence (5′ → 3′ end) | Reference |
|---|---|---|
| Primer for msaABCR deletion and complementation | ||
| Operon del 1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTTTAAATCAGCGATTAATGTTCGTTTG | Samanta & Elasri (2014) |
| Operon del 2 | ATGACTGGATCCTATTAAAGACCCCTTCCATACTTCAAAAAC | Samanta & Elasri (2014) |
| Operon del 3 | ATGACTGGATCCTTTCATGATGCTTGTTTAAAGTGTGGTAT | Samanta & Elasri (2014) |
| Operon del 4 | GGGGACCACTTTGTACAAGAAAGCTGGGTAGTTTGGATTATCAATTCAATATGGCTTAGC | Samanta & Elasri (2014) |
| Comp-F1 | GGGGGATCCTTTTACCACCTCATAATGTTAT | Samanta & Elasri (2014) |
| Comp-R1 | CCCGAATTCAAATAAACAAAGTAATCCCCGA | Samanta & Elasri (2014) |
| Cm-F | GTTTAAGGGCCCACCTAGGTATTATCAAGATAAGAAAGAAAAG | Samanta & Elasri (2014) |
| Cm-R | CTATGACTCGAGGCCGCGGCCTTCTTCAACTAACGGGG | Samanta & Elasri (2014) |
| msaB del 1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTTTAAAT CAGCGATTAATGTTCGTTTG | |
| msaB del 2 | AACGTTGTTAAAGGATCCTTCTTAGATTTGAATCATTGAT | |
| msaB del 3 | CCTTGTTTCAGGATCCGAAACCTCCAAGACTAAAATTCAT | |
| msaB del 4 | GGGGACCACTTTGTACAAGAAAGCTGGGTAGTTTGGATTATCAATTCAATATGGCTTAGC | |
| Primers for real-time PCR | ||
| RT gyrA F | GCCGTCAGTCTTACCTGCTC | |
| RT gyrA R | AATAACGACACGCACACCAG | |
| RT capE F | ACATTGGTGATGTGCGTGAT | |
| RT capE R | TCACATGACGGCACTTGTTT | |
| Primers for cap promoter activity | ||
| cap promoter F | ATGCATGGTACCAGTAAAAAATGCCACATAAACTTTAAGTC | |
| cap promoter R | AGTCTAGGATCCTGTACTTTCCATTATTTACCTCCCTTAAA | |
| Primers for ChIP | ||
| cap-ChIP F | CTACTTTAGAGTATAATTATTTTTAATTTC | |
| cap-ChIP R | CCCTTAAAAATTTTCATTAAAATTG | |
| Primers for recombinant MsaB | ||
| pH6HTN-msaB-F | GAATGTCTAGAATGAAACAAGGTACAGTTAAATGGTTT | |
| pH6HTN-msaB-R | ATTAAGGGCCCTTATAGTTTAACAACGTTTGCAGCTT | |
RNA extraction, reverse transcription and quantitative reverse transcriptase PCR (qRT-PCR)
The expression of the capE gene was measured with qRT-PCR in both wild-type strains and isogenic mutants (Wann et al., 1999). Briefly, an aliquot of an overnight culture was normalized to an OD600 of 0.05 and then grown to the late-exponential/post-exponential phase for optimal capsule production. After the cells had grown to the appropriate phase, they were pelleted with centrifugation, treated with RNAprotect bacteria reagent (Qiagen), and stored at − 80 °C until analysis was carried out. The samples were thawed on ice and total RNA was extracted as previously described (Sahukhal & Elasri, 2014; Samanta & Elasri, 2014). qRT-PCR was performed and the relative fold change in gene expression was calculated using gyrA as the internal control (Goerke et al., 2000; Sahukhal & Elasri, 2014; Samanta & Elasri, 2014).
Absolute quantification of the msaB transcript
Absolute quantification of the msaB transcript was performed by the method described by Chini et al. (2007). Briefly, the msaB and gyrA genes were amplified from chromosomal DNA using primers external to the primers used for qRT-PCR (Table 2). These amplicons were purified, and their concentrations were measured using a NanoDrop spectrophotometer. The corresponding concentrations were converted to copies μl− 1 by a method described elsewhere (Sambrook et al., 1989). Tenfold serial dilutions (ranging from 102 to 106) of these amplicons were used as templates for qRT-PCR. Standard curves were generated by plotting cycle threshold (C t) values against the log of the copy numbers [log starting quantity (SQ)]. SQs of ‘unknown’ wild-type samples (early, mid-, late- and post-exponential cDNA of Mu50 and UAMS-1) were calculated by plotting the respective C t values on the standard curve. Copy numbers were measured by raising 10 to the power of the SQ (10SQ). Copy numbers of msaB were normalized to those of gyrA and plotted. The process was repeated in triplicate independently.
In vitro capsule production assay
Total capsule production was determined with a dot-blotting method described by Luong et al. (2003) with the following modifications. In brief, 2 ml of an 18 h culture, adjusted to an OD660 of 5.0, was pelleted and resuspended in 100 μl PBS. The suspension was treated consecutively with the following enzymes at 37 °C: lysostaphin (100 μg ml− 1) for 15 min, DNase I (300 U ml− 1) for 15 min and proteinase K (100 μg ml− 1) for 1 h. The proteinase K was subsequently inactivated by heating at 75 °C for 10 min. The crude capsule preparations were assayed by immunoblotting, as described elsewhere (Luong et al., 2003).
In vitro whole blood survival assay
The survival of S. aureus in whole human blood was measured with an in vitro method (Nygaard et al., 2012) with minor modifications. In brief, heparinized venous blood samples were collected from healthy donors according to a protocol approved by the Institutional Review Board for Human Subjects at the University of Southern Mississippi. S. aureus cells were harvested in the post-exponential phase of growth (∼9 × 1010 cells) to ensure optimal capsule production. The blood (3 ml) was inoculated with 3 × 105 S. aureus cells in a 14 ml culture tube and incubated at 37 °C for 2 h with end-over-end rotation at 20 r.p.m. The samples were then diluted with sterile deionized water to lyse the blood cells and the number of c.f.u. was counted after plating on TSA.
Human neutrophil phagocytosis assay
The phagocytosis of S. aureus by freshly isolated human PMNs was measured as described elsewhere (Voyich et al., 2005), with the following modifications. In brief, PMNs (1 × 106) were combined with S. aureus (1 × 107) in a 24-well tissue culture plate and centrifuged at 380 g for 8 min. The cells were then incubated for 2 h at 37 °C with 5 % CO2. After incubation, the extracellular bacteria in the medium were plated on TSA and incubated overnight at 37 °C. The number of c.f.u. was counted and the test samples compared with the controls, which contained deionized water instead of the neutrophil suspension. This assay measures the total number of viable uningested bacteria.
Construction of cap promoter–luxAB fusions and luciferase assays
The E. coli–staphylococcal shuttle vector pCN58, which was used in this study, contains a low-copy-number staphylococcal replicon cassette (pT181copwt repC) and a promoterless reporter gene, luxAB (encoding the luciferase from Vibrio fischeri), constructed to analyse transcriptional fusions (Charpentier et al., 2004). The promoter region of the identified cap promoter, including the 10 bp inverted repeat that was identified and described by Ouyang et al. (1999), was fused to luxAB for promoter studies in the msaABCR and msaB mutants. To study the promoter–luciferase activity, overnight bacterial cultures were diluted 1 : 10 in TSB and incubated for an additional 3 h. The cells were then normalized to an OD600 of 0.05 and incubated further at 37 °C with shaking (220 r.p.m.). The bacterial cells (5 ml) were harvested in different phases of growth: OD600 of 1.0, 4.0 and 7.0, or after growth for 18 h, representing the early, mid-, late- and post-exponential growth phases, respectively. The cells were washed once with PBS and resuspended in 1 ml PBS. The cell suspensions were mixed with 100 μl 1 % (v/v) decanal in 90 % ethanol and luminescence was measured with a luminometer immediately after mixing, using 10 s measurements in the integrated data mode. The luciferase activities were recorded in relative luminescence units (RLUs) and the specific luciferase activities were calculated by dividing the RLU values by the optical density of the culture (RLU/OD600). The promoterless version of the reporter-gene plasmid (pCN58) was used as the control.
Expression and purification of MsaB and antibody production
The msaB ORF (SAV1402) of S. aureus strain Mu50 was cloned into the pH6HTN His6HaloTag T7 plasmid (Promega) as an XbaI–ApaI fragment. E. coli strain BL21(DE3) was then transformed with the resulting plasmid, pH6HTN-msaB, and the transformants were selected on LB agar plates containing ampicillin. LB broth (1 l) with ampicillin (100 μg ml− 1) was inoculated with an overnight culture (10 ml) of a positive colony. Protein expression was induced by adding IPTG at a final concentration of 0.5 mM. The cells were pelleted 4 h after induction and resuspended in PBS (pH 7.4) with a protease inhibitor cocktail, and then lysed by sonication. The cell lysate was centrifuged at 10 000 g for 30 min to remove the cell debris. The His6Halo–MsaB fusion protein was purified from the clear lysate with a nickel column (HisPur Ni-NTA resin; Thermo Scientific). The fusion protein was then cleaved with the tobacco etch virus (TEV) protease (ProTEV Plus; Promega) to remove the His6Halo tag. The cleaved protein band was excised from the polyacrylamide gel and identified with MS by a commercial company (MS Bioworks). The MsaB protein was purified from the tag with reversed-phase HPLC. After the removal of acetonitrile by vacuum centrifugation, the protein was dissolved in PBS (pH 7.4) and stored for future use. Antibodies directed against MsaB were raised in rabbit by a commercial company (Thermo Scientific).
Chromatin immunoprecipitation (ChIP) with anti-MsaB antibody
The ChIP assay was performed as described by Sengupta et al. (2012), with minor modifications. Briefly, exponentially growing S. aureus cells were treated with 1 % formaldehyde and 10 nM sodium phosphate to facilitate the cross-linking of MsaB to its targets. After 20 min, the cross-linking reaction was quenched by the addition of 0.1 volumes 3 M glycine. The cultures were then pelleted and washed with an equal volume of 0.1 M phosphate buffer to remove excess formaldehyde. The phosphate buffer was removed by centrifugation, and the cells were resuspended in 200 μl IP buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 5 % (v/v) glycerol, 1 % Triton X-100) and broken with bead beating. The cell lysates were then diluted with 200 μl IP buffer and the cellular DNA was sheared with further bead beating. The cell debris was removed by centrifugation and the clear supernatant was diluted with 1 ml IP buffer. To 200 μl of the clear lysate, an anti-MsaB antibody (diluted 1 : 1000) was added and incubated at room temperature for 2 h. This antigen–antibody mixture was then added to prewashed protein-G-coupled magnetic beads (Thermo Scientific) and incubated under ambient conditions for 1 h. The antigen–antibody–bead complex was collected with a magnetic tube holder, washed twice with wash buffer (Tris-buffered saline, 0.05 % Tween 20, 0.5 M NaCl), and then washed with ultrapure water. The beads were resuspended in 100 μl elution buffer (50 mM Tris/HCl pH 8.0, 1 mM EDTA, 1 % SDS) and incubated at 65 °C overnight to reverse the cross-links. The DNA was extracted with a phenol/chloroform extraction method. The DNA was used as the template to detect the MsaB-bound promoter sequences with PCR amplification using promoter-specific primers. The msaABCR operon mutant was used as an internal negative control to demonstrate that the MsaB antibody enriches the cap promoter region specifically.
Electrophoretic mobility shift assay (EMSA)
Duplex DNA fragments, 22 nt in size, containing the 10 bp inverted repeat described by Ouyang et al. (1999) were synthesized and biotinylated at the 5′ end by a commercial provider (Integrated DNA Technologies). The shift assays were performed with the LightShift chemiluminescent EMSA kit (Pierce), according to the manufacturer's protocol. Briefly, the 5′-biotinylated duplex fragments were incubated with increasing concentrations of purified MsaB protein for 20 min at room temperature in 20 μl reaction buffer (1 × binding buffer, 2.5 % (v/v) glycerol, 5 mM MgCl2, 50 ng ml− 1 poly dI-dC, 0.05 % NP-40). Unlabelled DNA probe was used in 10 M excess as a specific competitive binding control in the reaction. Additionally, a DNA probe that did not contain the MsaB-binding site was used as a negative binding control in the reaction. The samples were subjected to electrophoresis in a native 6.0 % polyacrylamide gel. The proteins in the gel were then visualized using the detection module supplied with the kit, according to the manufacturer's protocol, and imaged with the ChemiDoc system (Bio-Rad) to detect any shifted band.
Western blotting of MsaB and its quantification
To measure the expression level of MsaB across the growth phases of Mu50 and UAMS-1, quantitative Western blotting was performed with whole-cell lysates. Overnight cultures were normalized to OD600 0.05 and incubated at 37 °C. The cells were harvested at OD600 (early), (mid), and (late), or after overnight incubation (post-exponential), and frozen until analysis was carried out. The pellets were resuspended in PBS with a protease inhibitor and lysed with bead beating. The crude lysates were centrifuged to remove the cell debris. The clear supernatants were collected and their protein concentrations determined with the BCA method, using the Pierce BCA protein assay kit (Life Technologies), and 25 μg from each sample was loaded into an SDS-polyacrylamide gel for separation. After the proteins were blotted onto a PVDF membrane and blocked with 5 % non-fat skimmed milk, MsaB was detected with an anti-MsaB antibody and a peroxidase-conjugated secondary antibody. The MsaB bands were quantified with ImageJ software (Schneider et al., 2012).
Altered nutrient experiments
The production of the capsule was measured under nutrient-altered conditions to test the effects of the regulation of nutrients on capsule production. Briefly, the cultures were subjected to different nutrient conditions in different growth phases. For the nutrient-depleted conditions, overnight cultures were normalized to an OD600 of 0.05, incubated at 37 °C with shaking (220 r.p.m.) and allowed to grow to early exponential phase. Cells were pelleted and then resuspended in filter-sterilized overnight-spent medium and then incubated at 37 °C with shaking (220 r.p.m.) for an additional 2 h. The capsule production assay and ChIP assay were performed as described above using mid-phase grown cells under normal conditions as controls for comparative analysis. For the nutrient-replenished conditions, overnight cultures were normalized to an OD600 of 0.05, incubated at 37 °C with shaking (220 r.p.m.) and allowed to grow to late-exponential phase, when the cells were collected by centrifugation. The pellet was resuspended in fresh growth medium (TSB) and incubated further at 37 °C with shaking (220 r.p.m.) for an additional 2 h. Capsule production was then assayed as described above using late-phase-grown cells under normal conditions as controls for comparative analysis.
Results
msaABCR operon is essential for capsule production
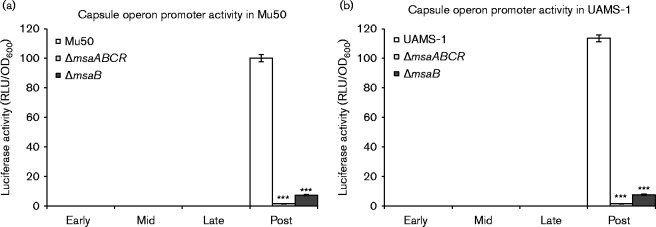
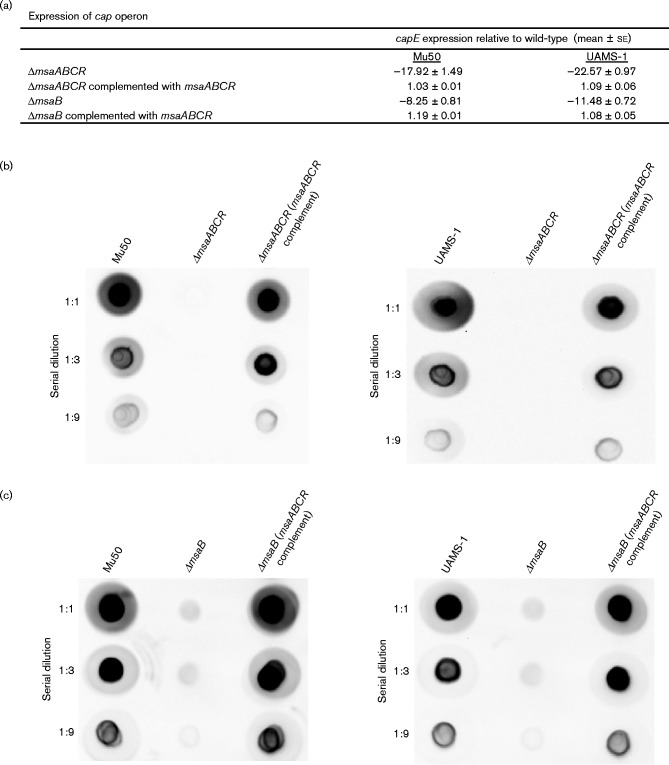
To investigate the role of the msaABCR operon in the regulation of capsule production, we deleted the msaABCR operon in the strains Mu50 and UAMS-1, which represent the two most commonly encountered clinical serotypes, CP5 and CP8, respectively. We first examined the expression of the capsule operon in both strains by measuring the expression of the cap genes in four different growth phases using a reporter fusion protein. As previously reported, both strains showed reporter activity only in the post-exponential growth phase, whereas the msaABCR deletion mutant showed no reporter activity in any of the four growth phases tested (Fig. 1) (Dassy & Fournier, 1996; Cunnion, 2001). This suggests that the deletion of the msaABCR operon abolished the activation of the cap promoter. We confirmed our findings by measuring the transcription of the capE gene, which is required for capsule production, using qRT-PCR, and by directly measuring capsule production with immunoblotting using anti-CP antibody (Fig. 2a, b). We found that msaABCR is essential for the transcription of the cap operon and the production of capsule in the post-exponential growth phase in both strains. Complementation of the deletion mutants with a functional copy of the msaABCR operon restored the transcription of the operon and the production of the capsule in both strains to levels similar to those in the wild-type (Fig. 2a, b). This confirms that the msaABCR operon is required for the production of the capsule in two different strains.
Fig. 1.
cap operon promoter activity was measured in the wild-types, msaABCR mutants and msaB mutants of cap5 (a) and cap8 (b) in different growth phases (early, mid-, late- and post-exponential phases). The vector pCN58 containing luxAB without a promoter was used as a negative control. These results represent the means of three independent experiments. Error bars represent se. Student's unpaired t-test was used to compare the results of the wild-types to their respective mutants. ***P ≤ 0.001.
Fig. 2.
Capsule production assays. qRT-PCR was used to measure the relative fold changes in cap5E and cap8E in the mutants and the complemented mutants compared with the wild-type (a). Total capsule production was assessed in the representative CP5 and CP8 strains, including their respective mutants and complements of msaABCR (b) and msaB (c). Samples were serially diluted as indicated on the left and dot-blotted directly onto the membrane. The blots were processed using CP5- and CP8-specific antibodies. These results are representative of three independent experiments for each sample set.
The msaABCR operon is not yet fully defined. Although we have learned a substantial amount about the phenotypes with which the operon is associated, we do not have a clear picture of the contributions made by each gene yet. We have analysed the structure of the msaABCR operon and found that it encodes three functional non-coding RNAs (msaA, msaC and msaR) that are essential for the function of the operon (Sahukhal & Elasri, 2014). Additionally, we have found that MsaB is the only protein encoded within the transcript of the msaABCR operon. To determine whether the three non-coding RNAs (msaA, msaC and msaR) act in concert to regulate the expression of MsaB, which likely functions as a transcription factor (Sahukhal & Elasri, 2014), we examined the contribution of msaB to the regulation of capsule production. We deleted the msaB region alone and compared the phenotype of the resulting mutant with that of the msaABCR mutant. The deletion of the msaB gene alone caused a similar decline in the transcription and production of the capsule as observed in the msaABCR mutant (Fig. 2a). However, importantly, the msaB mutants showed reduced capsule production, whereas the msaABCR mutants showed no detectable capsule production (Fig. 2c).
Deletion of msaABCR operon leads to reduced survival in blood
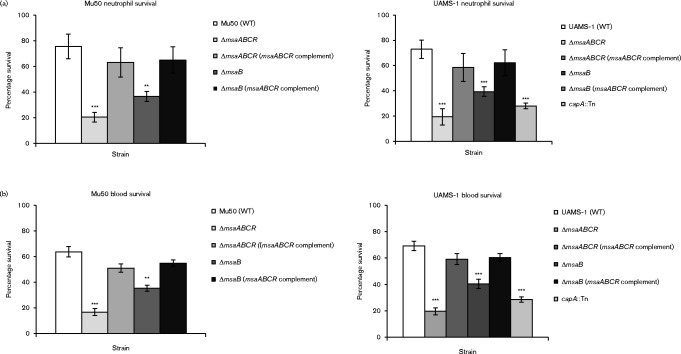
Several studies have shown that the production of the capsule by S. aureus plays a role in its evasion of either phagocytosis or its elimination by components of the host immune system (O'Riordan & Lee, 2004; Thakker et al., 1998; Luong & Lee, 2002; Watts et al., 2005; Nilsson et al., 1997). We measured the susceptibility of the msaABCR and msaB mutants to phagocytosis by human PMNs in vitro. In both serotypes, the deletion of the msaABCR operon and msaB led to a significant increase in phagocytosis (Fig. 3a). We also examined the effect of the deletion of the msaABCR operon and msaB on bacterial clearance from heparinized whole human blood. We added post-exponentially grown cultures of both strains and their corresponding msaABCR and msaB deletion mutants to heparinized whole human blood that had been freshly collected from volunteers. The wild-type strains of both serotypes survived significantly better than both the msaABCR and msaB mutants (Fig. 3b). Complementation of the mutants with a functional copy of the msaABCR operon restored their survival to levels approaching those of the wild-type. When comparing the results of the mutation of the msaABCR operon and the msaB region alone, we noticed that even though both mutations lead to significant differences in clearance in neutrophils and whole blood the results of the msaB region alone are not as significant as for the whole msaABCR operon. We speculate that this is likely due to the low levels of capsule production displayed by the msaB mutant. These findings suggest that additional elements in the msaABCR operon, such as the three non-coding RNAs (msaA, msaC and msaR), play a role in the regulation of capsule production. Additionally, we compared the results from both experimental models to the UAMS-1 capA : : Tn mutant. We observed that the mutation of the msaABCR operon, msaB and capA all resulted in a similar increase in clearance in both models used. These results suggest that the msaABCR operon is important in the bacterial resistance to phagocytosis and serum immune factors. Due to these phenotypes being associated with capsule production (O'Riordan & Lee, 2004; Thakker et al., 1998; Luong & Lee, 2002; Watts et al., 2005; Nilsson et al., 1997), we concluded that the increased susceptibility to phagocytosis and serum killing in the msaABCR and msaB deletion mutants is likely due to the lack of capsule production. This is supported by our finding that there was no significant difference between the msaABCR and the capA : : Tn mutants in survival assays (Fig. 3). However, given the pleiotropic effect of deletion of the msaABCR operon, we cannot rule out factors other than the lack of capsule production in the reduced survival of the msaABCR mutant. Indeed, we have also shown in several of our studies that the msaABCR operon plays a role in regulation of many virulence factors, biofilm development and antibiotic resistance (Sambanthamoorthy et al., 2006, 2008; Samanta & Elasri, 2014; Sahukhal & Elasri, 2014; Sahukhal et al., 2015). Further studies are needed to examine the putative contribution of other factors affected by the msaABCR operon.
Fig. 3.
Survival assays were performed using two different methods. Bacterial cells were mixed with freshly isolated human neutrophils to measure their survival rates against phagocytosis (a). Bacterial cells were mixed with heparinized human whole blood to measure their survival rates in the presence of complementing proteins (b). Results are presented as percentages of surviving bacterial cells relative to the control, which contained water instead of neutrophils (a) or by comparing the direct count of bacteria added to the total inoculum with the total c.f.u. enumerated, corrected by any dilution factor (b). Results represent the means of three independent experiments. Error bars represent se. Student's unpaired t-test was used to compare the results of wild-types to their respective mutants. The results are displayed with asterisks using the following P value cut-offs: ** P ≤ 0.01; ***, P ≤ 0.001.
MsaB binds to the 10 bp regulatory region within the cap promoter
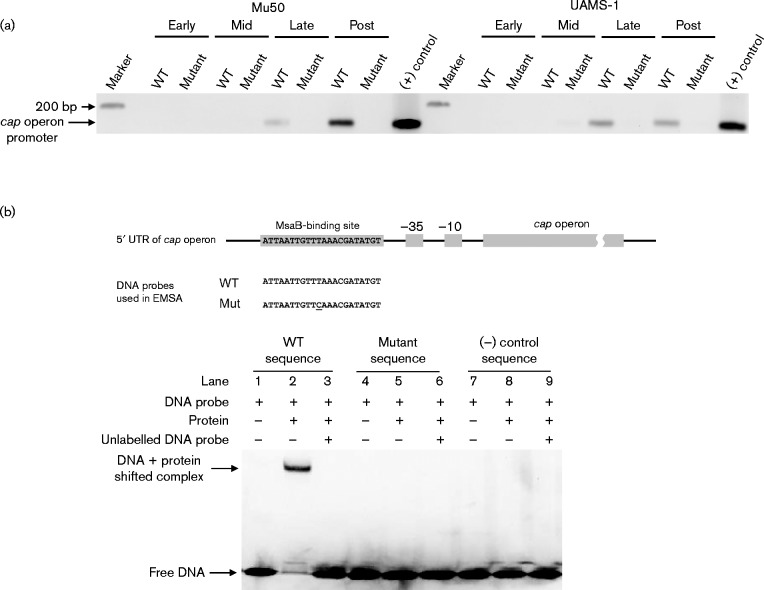
Detailed analysis of the cap operon promoter identified the tentative − 35 and − 10 sequences, and led to the conclusion that it is highly regulated (Ouyang et al., 1999). Several DNA-binding proteins have been shown to directly interact with the cap promoter region, but only one regulator, RbsR, has been shown to directly interact with the 10 bp inverted repeat (Zhao et al., 2010; Majerczyk et al., 2010; Sun et al., 2012; Ding et al., 2014; Jutras et al., 2013; Lei & Lee, 2015). We investigated the regulatory relationship between the msaABCR and cap operons by testing whether the putative DNA-binding protein, MsaB, interacts with the cap promoter, using a ChIP assay (Fig. 4a). The MsaB protein bound to the promoter region of the cap operon in both serotypes (Fig. 4a). Previously, Ouyang et al. (1999) identified a 10 bp inverted repeat directly upstream from the promoter region, which was proposed as a binding site for an unknown activator. We tested whether MsaB binds specifically to this 10 bp binding domain. We synthesized a 22 nt DNA duplex containing the 10 bp inverted repeat and tested its binding to purified MsaB using EMSAs (Fig. 4b). We observed a shift in the band when MsaB and the probe containing the 10 bp inverted repeat were mixed, indicating a specific interaction between MsaB and the 10 bp inverted repeat region (Fig. 4b). Unlabelled competitor DNA was added in 10-fold molar excess as a specific competitor and was found to reduce the formation of the protein–DNA complex. Furthermore, no retarded protein–DNA complex was detected when a random DNA probe that did not contain the 10 bp inverted repeat was used as a control. We further tested the specificity of this binding event by mutating 1 nt in the 10 bp region (T to C) and showed that this change abolished its binding to MsaB (Fig. 4b). This confirms the previous findings of Ouyang et al. (1999), who performed a full analysis on the individual nucleotides within this repeat. This nucleotide was found to be one of the critical nucleotides involved in the regulatory functions of the repeat and proposed to play an important role in the binding of a putative activator. Taken together, these results show that the 10 bp inverted repeat is an MsaB-binding site that is required for transcriptional activation of the cap operon by MsaB.
Fig. 4.
(a) ChIP using an anti-MsaB antibody was performed to determine whether the promoter region of the cap operon binds to MsaB from whole-cell extracts in different growth phases (early, mid-, late- and post-exponential phases) from two strains that produce the two clinically relevant capsule serotypes (Mu50 for CP5 and UAMS-1 for CP8). Primers specific for the cap promoter region were used to amplify the DNA after immunoprecipitation. Lanes are labelled as follows: WT represents whole-cell extract from the wild-type strain and anti-MsaB; mutant is the negative control, representing the whole-cell extract from the msaABCR deletion mutant and anti-MsaB; (+) control represents the PCR product amplified from the genomic DNA of the tested strain. (b) Map of the cap promoter region showing the 10 bp repeat region that binds MsaB. DNA probes used in the EMSAs are also shown. EMSAs showed that purified MsaB bound to the DNA probe containing the 10 bp repeat region. All of these results are representative of at least three independent experiments.
MsaB binding to the cap promoter is dependent on nutrient availability
The cap operon is expressed in the late- and post-exponential growth phases, but not in the early and mid-exponential growth phases during in vitro growing conditions. We have shown that MsaB specifically binds to the 10 bp regulatory region upstream from the cap promoter. Based on the time points that capsule is produced in our in vitro analysis and the known regulatory functions of capsule production, we investigated the role of MsaB in the temporal regulation of capsule production and how it might interact with other regulators.
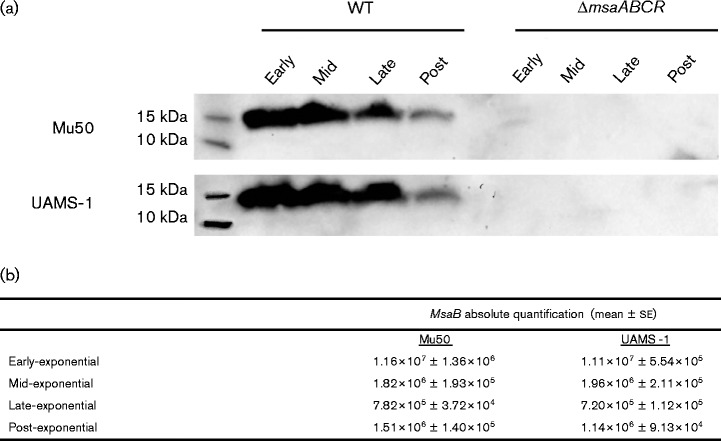
We measured the expression of MsaB in four growth phases (early, mid-, late- and post-exponential) with a Western analysis using an anti-MsaB antibody. We found that MsaB was produced in all four growth phases in both Mu50 and UAMS-1 (Fig. 5a), although the expression of MsaB decreased in the post-exponential growth phase. Additionally, we measured the transcript level of msaB in all four of the growth phases in both strains. The absolute values of msaB were calculated normalizing the unknown C t values to standard values plotted on a standard curve (Fig. 5b). We used a ChIP analysis to examine the binding capacity of MsaB in all four growth phases and found that MsaB binds to the cap promoter only during the late- and post-exponential growth phases (Fig. 4a). This indicates that despite the presence of MsaB in the early and mid-exponential growth phases, it is not able to bind to the cap promoter. This may be attributable to a change in the affinity of MsaB for the 10 bp binding site or the presence of another regulator that interferes with the binding of MsaB during the early and mid-exponential growth phases. Other regulators have been shown to repress capsule production by binding to the cap promoter. Based on the growth phase dependence of capsule production, we next tested whether nutrient availability influenced binding of MsaB to the cap promoter region and capsule production. Cells were inoculated in nutrient-replenished medium in the late phase of growth and allowed to grow for an additional 2 h; the growth was compared to cells grown under normal conditions in the late phase of growth. We observed a significant decline in the production of capsule relative to that in cells grown under normal conditions (Fig. 6a). We also observed similar results in transcriptional downregulation ( − 6.01-fold in Mu50; − 4.74-fold in UAMS-1) of capA under these conditions. Cells were also tested in nutrient-depleted medium during the early-exponential phase of growth after an additional 2 h and compared to cells grown under normal conditions to the mid-exponential phase of growth. We found that capsule was produced in the nutrient-depleted growth conditions, whereas the control cells grown in normal medium produced no capsule (Fig. 6b). This correlated with the upregulation of transcription of capA under the same conditions (2.87-fold in Mu50; 2.57-fold in UAMS-1). As a result of these findings, we used a ChIP analysis to test whether MsaB binds to the cap promoter in these nutrient-depleted conditions. Indeed, we found that MsaB bound to the cap promoter in the cells grown in nutrient-depleted conditions, differing from cells measured during the mid-exponential growth phase under normal growing conditions (Fig. 6c). These results suggest that nutrient availability has a direct effect on the ability of MsaB to bind the cap promoter and activate capsule production. These data suggest that MsaB protein is involved in sensing nutrient levels directly or that MsaB can only bind to the cap promoter when other nutrient-dependent regulators are not bound, which occurs when nutrient levels are low. Further studies are needed to investigate any direct links of MsaB-sensing nutrients, and the possibility of an interplay between MsaB and other nutrient-sensing regulators in controlling capsule production.
Fig. 5.
(a) Western blot of Mu50 and UAMS-1 whole-cell lysates probed with an anti-MsaB antibody. Cells were harvested in the early, mid-, late- and post-exponential phases of growth and lysed, and 25 μg protein was loaded in each lane. These results are representative of three independent experiments for each sample set. (b) Absolute quantification of msaB in the four growth phases. Amplicons of msaB were converted to copies μl− 1, and then serially diluted and used as templates for qRT-PCR. Standard curves were generated by plotting C t values against the log of the copy numbers (log SQ). SQs of ‘unknown’ samples were calculated by plotting the respective C t values on the standard curve. Copy numbers were measured by raising 10 to the power of the SQ (10SQ). These results are representative of triplicate independently treated samples.
Fig. 6.
Total capsule production was assessed in the representative CP5 and CP8 strains, including the wild-types, the respective mutants and the controls, in nutrient-replenished medium (a) and nutrient-depleted medium (b). Samples were serially diluted as indicated on the left and dot-blotted directly onto the membrane. The blots were processed using CP5- and CP8-specific antibodies. (c) ChIP using an anti-MsaB antibody was performed to determine whether the promoter region of the cap operon binds to MsaB from whole-cell extract under nutrient-depleted conditions in the mid-exponential phase of growth. As in previously described ChIP reactions, primers specific for the cap promoter region were used to amplify the DNA after immunoprecipitation. Lanes are labelled as follows: Mu50 (nutrient depleted) and UAMS-1 (nutrient depleted) represent whole-cell extracts from wild-type CP5 and CP8 strains, respectively, and anti-MsaB antibody; ΔmsaABCR mutant represents whole-cell extract from an msaABCR deletion mutant and anti-MsaB antibody; (+) control represents PCR product amplified from the genomic DNA of the tested strain. These results are representative of three independent experiments.
Discussion
The capsule is a very important virulence factor in S. aureus and facilitates its evasion of phagocytosis during infection (O'Riordan & Lee, 2004). This is particularly important during certain types of infection, such as bacteraemia or septicaemia, when the pathogen must survive in blood (Thakker et al., 1998; Luong & Lee, 2002; Watts et al., 2005; Nilsson et al., 1997; Kampen et al., 2005). In this study, we have demonstrated that the msaABCR operon is essential for capsule production in the two most clinically relevant capsule serotypes of S. aureus, CP5 and CP8. Importantly, we have shown that the regulation of the cap operon by the msaABCR operon is similar in both serotypes. This is an important consideration because future therapeutic agents are being developed against the capsule and it is therefore important to understand whether strain differences are an important factor that must be taken into account (Fattom et al., 1993; Lattar et al., 2014; Bagnoli et al., 2012). However, we cannot rule out possible differences in the regulation of capsule production in other strains because we have only tested two representative strains. Although capsule production is very important in the survival of S. aureus during different phases of infection, very little is known about the mechanism that regulates the cap operon. Several global regulators have been shown to affect the production of the capsule, indicating that several environmental and host signals must be integrated to fine-tune this process (Luong et al., 2002, 2003, 2011; Luong & Lee, 2006; Chen et al., 2007; Dassy et al., 1993; Gupta et al., 2013).
Previously, we have shown that the msaABCR operon is involved in virulence, biofilm development and antibiotic resistance (Sambanthamoorthy et al., 2006, 2008; Sahukhal & Elasri, 2014; Samanta & Elasri, 2014; Sahukhal et al., 2015). In this study, we found that MsaB, encoded by the msaABCR operon, binds directly to a previously identified 10 bp inverted regulatory repeat located immediately upstream from the identified cap promoter region. We have shown that MsaB is an activator of the cap operon that binds to the cap promoter region in the late- and post-exponential phases of growth. Since MsaB is produced throughout all growth phases, we hypothesize that either MsaB is not in its active form in all these phases or its binding site is not accessible when other regulators are bound to this region. Our findings suggest that nutrient availability is an important factor in MsaB binding to this region and activating capsule production. One possible mechanism for regulation of the cap operon by MsaB is that in early phases of growth, when nutrients and metabolic factors are high, nutrient-sensing regulators (cap repressors) such as CodY or CcpE bind to this region and block MsaB from binding and activating cap. Nutrient-and metabolic-sensing regulators such as CodY and CcpE respond to nutrient levels, which change with the depletion of nutrients and metabolic factors as the cells progress through their growth phases. Indeed, in Bacillus subtilis, CodY was shown to have a high affinity for its binding site under high-nutrient conditions, such as in the early-exponential growth phase, and a low affinity under low-nutrient conditions, such as in the post-exponential growth phase (Ratnayake-Lecamwasam et al., 2001; Shivers & Sonenshein, 2004). When nutrient availability declines in later phases of growth, these repressors putatively become unbound, allowing the MsaB-binding site to be available and to activate transcription. Our results suggest that in the absence of MsaB, the cap operon is expressed at the basal level and therefore the fine-tuning by other regulators (e.g. CodY) is necessary for the full production of the capsule under low-nutrient conditions. Because several global regulators have been shown to modulate the production of the capsule, we speculate that other environmental stimuli are involved in the repression or activation of the cap operon. It will be interesting to investigate how these signals are integrated with the proposed MsaB regulatory mechanism (activation) of capsule production. Additionally, MsaB itself may be sensing nutrient levels in an undetermined mechanism altering its ability to bind to this region. Considering these different possibilities, we plan to test these two possible regulatory mechanisms and to examine their relevance to pathogenesis.
The msaABCR operon regulates capsule production very tightly because the deletion of the operon leads to undetectable levels of the capsule. We believe that this is mainly attributable to the specific binding of the MsaB protein to the 10 bp inverted repeat upstream from the cap promoter. This region was analysed in detail by Ouyang et al. (1999), who showed that several nucleotides are essential for its binding. Indeed, with respect to the prior work of Ouyang et al. (1999) the mutation of a single nucleotide (T to C) led to the loss of binding of MsaB to this repeat region (Fig. 4b). We chose this particular mutation to test the specificity of MsaB binding to its target region because this mutation is found in the epidemic USA300 lineage. The USA300 strains do not produce a capsule even though they carry the cap operon with a sequence similar to those of Mu50 and UAMS-1, except for the T-to-C mutation in the MsaB-binding region (Montgomery et al., 2008; Boyle-Vavra et al., 2015). We propose that this mutation may be the main reason that USA300 strains do not produce capsule, but without further analysis we cannot rule out other causes. It is also unclear at this point whether the lack of capsule production contributes to the heightened virulence of this lineage (Montgomery et al., 2008; Boyle-Vavra et al., 2015). Additionally, several strains that are classified as capsule nontypable or capsule nonproducers have also been shown to contain mutations in the MsaB-binding region, and we also propose that these strains do not produce capsule because MsaB cannot activate the cap operon (Cocchiaro et al., 2006). Of note, RbsU has also been found to bind to this region. At this time, it is unknown whether any interactions take place between RbsU and MsaB in the binding to this region and we intend to investigate this further.
The deletion of the msaABCR operon led to a significant reduction in the transcription of the cap operon and a reduction in capsule production to undetectable levels. However, the msaB deletion mutant showed some residual capsule production. At this point, it is unclear why we observed any residual activity, because neither the msaABCR nor the msaB mutant expresses MsaB. Furthermore, the msaB mutants had reduced survival in the presence of neutrophils and whole blood relative to that of the wild-type, but not reduced to the same level as the msaABCR mutants. It is not clear whether the difference in the survival rates of the msaB and msaABCR deletion mutants is significant to the relative virulence of the two strains. However, these findings suggest that another element in the operon plays an additional role in the regulation of capsule production. Also, it is likely that the operon and MsaB regulator have additional functions other than the production of capsule that are involved in the survival against host immune components. Our previous work on the msaABCR operon showed that MsaB is the only protein-coding gene in the operon. Therefore, we hypothesize that one of the three non-coding RNAs (msaA, msaC or msaR) has a role in regard to the regulation of capsule production (Sahukhal & Elasri, 2014). We plan to investigate this possibility in order to fully understand this regulatory process. This hypothesis is further supported by the observation that the msaB mutants cannot be complemented by the reintroduction of msaB alone, but are only complemented by the reintroduction of the whole msaABCR operon.
MsaB is predicted to be part of the cold shock protein family, carrying the DNA-binding cold shock domain, based on a protein homology analysis (Rost et al., 2004). Members of this family have been shown to be either constitutively expressed or induced by stresses, including nutrient starvation during the late-exponential or stationary phase of growth (Graumann et al., 1997). Additionally, MsaB also appears to contain GTP-binding regions, which may respond to nutrient availability (Chen et al., 2012). The findings of our study suggest that nutrient availability or metabolic sensors may play a key role in capsule production, and we propose that MsaB is one of the factors that are directly sensitive to nutrient levels or other signals required for the production. We intend to investigate this possibility in the future. Taken together, these findings support the mechanism of an interplay between several regulators including the fine-tuning of binding between transcriptional repressors and the activator MsaB, which controls the production of the capsule in response to growth and nutrient availability.
Acknowledgements
This work was funded by the National Institutes of Health (grant number 1R15AI099922, to M. O. E.) and the Mississippi INBRE, with an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (grant number P20GM103476). We gratefully acknowledge Dr Ali Fattom and Dr Chia Lee for providing the capsule type 5 and capsule type 8 antibodies used in this work. We are also very grateful to all of the numerous volunteers who donated their blood for use in the experiments.
Abbreviations:
- ChIP
chromatin immunoprecipitation
- CP
capsular polysaccharide
- EMSA
electrophoretic mobility shift assay
- NARSA
Network on Antimicrobial Resistance in Staphylococcus aureus
- PMN
polymorphonuclear neutrophil
- qRT-PCR
quantitative reverse transcriptase PCR
- RLU
relative luminescence unit
- SQ
starting quantity.
References
- Arbeit R. D., Karakawa W. W., Vann W. F., Robbins J. B. (1984). Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus Diagn Microbiol Infect Dis 2 85–91 10.1016/0732-8893(84)90002-6 . [DOI] [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection Plasmid 55 58–63 10.1016/j.plasmid.2005.05.005 . [DOI] [PubMed] [Google Scholar]
- Bagnoli F., Bertholet S., Grandi G. (2012). Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials Front Cell Infect Microbiol 2 16 10.3389/fcimb.2012.00016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Dunman P., Kormanec J., Macapagal D., Murphy E., Mounts W., Berger-Bachi B., Projan S. (2004). Microarray-based analysis of the Staphylococcus aureus σB regulon J Bacteriol 186 4085–4099 10.1128/JB.186.13.4085-4099.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Vavra S., Li X., Alam M. T., Read T. D., Sieth J., Cywes-Bentley C., Dobbins G., David M. Z., Kumar N., other authors (2015). USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus MBio 6 e02585-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner S., Monteil H., Prevost G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications FEMS Microbiol Rev 28 183–200 10.1016/j.femsre.2003.09.003 . [DOI] [PubMed] [Google Scholar]
- Charpentier E., Anton A. I., Barry P., Alfonso B., Fang Y., Novick R. P. (2004). Novel cassette-based shuttle vector system for gram-positive bacteria Appl Environ Microbiol 70 6076–6085 10.1128/AEM.70.10.6076-6085.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Luong T. T., Lee C. Y. (2007). The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus J Bacteriol 189 7343–7350 10.1128/JB.01079-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Mizianty M. J., Kurgan L. (2012). Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors Bioinformatics 28 331–341 10.1093/bioinformatics/btr657 . [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y. Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus FEMS Immunol Med Microbiol 40 1–9 10.1016/S0928-8244(03)00309-2 . [DOI] [PubMed] [Google Scholar]
- Chini V., Foka A., Dimitracopoulos G., Spiliopoulou I. (2007). Absolute and relative real-time PCR in the quantification of tst gene expression among methicillin-resistant Staphylococcus aureus: evaluation by two mathematical models Lett Appl Microbiol 45 479–484 10.1111/j.1472-765X.2007.02208.x . [DOI] [PubMed] [Google Scholar]
- Cocchiaro J. L., Gomez M. I., Risley A., Solinga R., Sordelli D. O., Lee J. C. (2006). Molecular characterization of the capsule locus from non-typeable Staphylococcus aureus Mol Microbiol 59 948–960 10.1111/j.1365-2958.2005.04978.x . [DOI] [PubMed] [Google Scholar]
- Cunnion K. M., Lee J. C., Frank M. M. (2001). Capsule production and growth phase influence binding of complement to Staphylococcus aureus Infect Immun 69 6796–6803 10.1128/IAI.69.11.6796-6803.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassy B., Fournier J. M. (1996). Respiratory activity is essential for post-exponential-phase production of type 5 capsular polysaccharide by Staphylococcus aureus Infect Immun 64 2408–2414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassy B., Hogan T., Foster T. J., Fournier J. M. (1993). Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide by Staphylococcus aureus J Gen Microbiol 139 1301–1306 10.1099/00221287-139-6-1301 . [DOI] [PubMed] [Google Scholar]
- David M. Z., Daum R. S. (2010). Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic Clin Microbiol Rev 23 616–687 10.1128/CMR.00081-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Liu X., Chen F., Di H., Xu B., Zhou L., Deng X., Wu M., Yang C. G., Lan L. (2014). Metabolic sensor governing bacterial virulence in Staphylococcus aureus Proc Natl Acad Sci U S A 111 E4981–E4990 10.1073/pnas.1411077111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattom A., Schneerson R., Watson D. C., Karakawa W. W., Fitzgerald D., Pastan I., Li X., Shiloach J., Bryla D. A., Robbins J. B. (1993). Laboratory and clinical evaluation of conjugate vaccines composed of Staphylococcus aureus type 5 and type 8 capsular polysaccharides bound to Pseudomonas aeruginosa recombinant exoprotein A Infect Immun 61 1023–1032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattom A. I., Sarwar J., Ortiz A., Naso R. (1996). Staphylococcus aureus capsular polysaccharide (CP) vaccine and CP-specific antibodies protect mice against bacterial challenge Infect Immun 64 1659–1665 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Campana S., Bayer M. G., Doring G., Botzenhart K., Wolz C. (2000). Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro Infect Immun 68 1304–1311 10.1128/IAI.68.3.1304-1311.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P., Wendrich T. M., Weber M. H., Schroder K., Marahiel M. A. (1997). A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures Mol Microbiol 25 741–756 10.1046/j.1365-2958.1997.5121878.x . [DOI] [PubMed] [Google Scholar]
- Gupta R. K., Alba J., Xiong Y. Q., Bayer A. S., Lee C. Y. (2013). MgrA activates expression of capsule genes, but not the α-toxin gene in experimental Staphylococcus aureus endocarditis J Infect Dis 208 1841–1848 10.1093/infdis/jit367 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras B. L., Chenail A. M., Rowland C. L., Carroll D., Miller M. C., Bykowski T., Stevenson B. (2013). Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins PLoS One 8 e66683 10.1371/journal.pone.0066683 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen A. H., Tollersrud T., Lund A. (2005). Staphylococcus aureus capsular polysaccharide types 5 and 8 reduce killing by bovine neutrophils in vitro Infect Immun 73 1578–1583 10.1128/IAI.73.3.1578-1583.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakawa W. W., Vann W. F. (1982). Capsular polysaccharides of Staphylococcus aureus Semin Infect Dis 4 285–293 .6232086 [Google Scholar]
- Karakawa W. W., Sutton A., Schneerson R., Karpas A., Vann W. F. (1988). Capsular antibodies induce type-specific phagocytosis of capsulated Staphylococcus aureus by human polymorphonuclear leukocytes Infect Immun 56 1090–1095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattar S. M., Noto Llana M., Denoel P., Germain S., Buzzola F. R., Lee J. C., Sordelli D. O. (2014). Protein antigens increase the protective efficacy of a capsule-based vaccine against Staphylococcus aureus in a rat model of osteomyelitis Infect Immun 82 83–91 10.1128/IAI.01050-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Park J. S., Shepherd S. E., Carey V., Fattom A. (1997). Protective efficacy of antibodies to the Staphylococcus aureus type 5 capsular polysaccharide in a modified model of endocarditis in rats Infect Immun 65 4146–4151 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M. G., Lee C. Y. (2015). RbsR activates capsule but represses the rbsUDK operon in Staphylococcus aureus J Bacteriol 197 3666–3675 10.1128/JB.00640-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections N Engl J Med 339 520–532 10.1056/NEJM199808203390806 . [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (2011). How Staphylococcus aureus adapts to its host N Engl J Med 364 1987–1990 10.1056/NEJMp1100251 . [DOI] [PubMed] [Google Scholar]
- Luong T. T., Lee C. Y. (2002). Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence Infect Immun 70 3389–3395 10.1128/IAI.70.7.3389-3395.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Lee C. Y. (2006). The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway Microbiology 152 3123–3131 10.1099/mic.0.29177-0 . [DOI] [PubMed] [Google Scholar]
- Luong T., Sau S., Gomez M., Lee J. C., Lee C. Y. (2002). Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA Infect Immun 70 444–450 10.1128/IAI.70.2.444-450.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Newell S. W., Lee C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus J Bacteriol 185 3703–3710 10.1128/JB.185.13.3703-3710.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Sau K., Roux C., Sau S., Dunman P. M., Lee C. Y. (2011). Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain Newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS J Bacteriol 193 686–694 10.1128/JB.00987-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., Bodi K., Sonenshein A. L. (2010). Direct targets of CodY in Staphylococcus aureus J Bacteriol 192 2861–2877 10.1128/JB.00220-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S., Goerke C., Wolz C., Seidl K., Homerova D., Schulthess B., Kormanec J., Berger-Bachi B., Bischoff M. (2007). σB and the σB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus Infect Immun 75 4562–4571 10.1128/IAI.00392-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Adem P. V., Lee J. C., Husain A. N., Clasen J., Daum R. S. (2008). Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia J Infect Dis 198 561–570 10.1086/590157 . [DOI] [PubMed] [Google Scholar]
- Nanra J. S., Buitrago S. M., Crawford S., Ng J., Fink P. S., Hawkins J., Scully I. L., McNeil L. K., Aste-Amézaga J. M., other authors (2013). Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus Hum Vaccin Immunother 9 480–487 10.4161/hv.23223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth J., Lee J. C. (1995). Antibodies to capsular polysaccharides are not protective against experimental Staphylococcus aureus endocarditis Infect Immun 63 375–380 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. M., Lee J. C., Bremell T., Ryden C., Tarkowski A. (1997). The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis Infect Immun 65 4216–4221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Geisinger E. (2008). Quorum sensing in staphylococci Annu Rev Genet 42 541–564 10.1146/annurev.genet.42.110807.091640 . [DOI] [PubMed] [Google Scholar]
- Nygaard T. K., Pallister K. B., Dumont A. L., Dewald M., Watkins R. L., Pallister E. Q., Malone C., Griffith S., Horswill A. R., other authors (2012). Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection PLoS One 7 e36532 10.1371/journal.pone.0036532 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan K., Lee J. C. (2004). Staphylococcus aureus capsular polysaccharides Clin Microbiol Rev 17 218–234 10.1128/CMR.17.1.218-234.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., Sau S., Lee C. Y. (1999). Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus J Bacteriol 181 2492–2500 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pané-Farré J., Jonas B., Förstner K., Engelmann S., Hecker M. (2006). The σB regulon in Staphylococcus aureus and its regulation Int J Med Microbiol 296 237–258 10.1016/j.ijmm.2005.11.011 . [DOI] [PubMed] [Google Scholar]
- Pohl K., Francois P., Stenz L., Schlink F., Geiger T., Herbert S., Goerke C., Schrenzel J., Wolz C. (2009). CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression J Bacteriol 191 2953–2963 10.1128/JB.01492-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutrel B., Gilbert F. B., Lebrun M. (1995). Effects of culture conditions on production of type 5 capsular polysaccharide by human and bovine Staphylococcus aureus strains Clin Diagn Lab Immunol 2 166–171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M., Serror P., Wong K. W., Sonenshein A. L. (2001). Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels Genes Dev 15 1093–1103 10.1101/gad.874201 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. (2004). The PredictProtein server Nucleic Acids Res 32 W321–W326 10.1093/nar/gkh377 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahukhal G. S., Elasri M. O. (2014). Identification and characterization of an operon, msaABCR, that controls virulence and biofilm development in Staphylococcus aureus BMC Microbiol 14 154 10.1186/1471-2180-14-154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahukhal G. S., Batte J. L., Elasri M. O. (2015). msaABCR operon positively regulates biofilm development by repressing proteases and autolysis in Staphylococcus aureus FEMS Microbiol Lett 362 1–10 10.1093/femsle/fnv006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D., Elasri M. O. (2014). The msaABCR operon regulates resistance in vancomycin-intermediate Staphylococcus aureus strains Antimicrob Agents Chemother 58 6685–6695 10.1128/AAC.03280-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambanthamoorthy K., Smeltzer M. S., Elasri M. O. (2006). Identification and characterization of msa (SA1233), a gene involved in expression of SarA and several virulence factors in Staphylococcus aureus Microbiology 152 2559–2572 10.1099/mic.0.29071-0 . [DOI] [PubMed] [Google Scholar]
- Sambanthamoorthy K., Schwartz A., Nagarajan V., Elasri M. O. (2008). The role of msa in Staphylococcus aureus biofilm formation BMC Microbiol 8 221 10.1186/1471-2180-8-221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Concentration of DNA solution. In Molecular Cloning: a Laboratory Manual, p. C1 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sau S., Bhasin N., Wann E. R., Lee J. C., Foster T. J., Lee C. Y. (1997a). The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes Microbiology 143 2395–2405 10.1099/00221287-143-7-2395 . [DOI] [PubMed] [Google Scholar]
- Sau S., Sun J., Lee C. Y. (1997b). Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus J Bacteriol 179 1614–1621 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis Nat Methods 9 671–675 10.1038/nmeth.2089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M., Jain V., Wilkinson B. J., Jayaswal R. K. (2012). Chromatin immunoprecipitation identifies genes under direct VraSR regulation in Staphylococcus aureus Can J Microbiol 58 703–708 10.1139/w2012-043 . [DOI] [PubMed] [Google Scholar]
- Shivers R. P., Sonenshein A. L. (2004). Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids Mol Microbiol 53 599–611 10.1111/j.1365-2958.2004.04135.x . [DOI] [PubMed] [Google Scholar]
- Somerville G. A., Proctor R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci Microbiol Mol Biol Rev 73 233–248 10.1128/MMBR.00005-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompolinsky D., Samra Z., Karakawa W. W., Vann W. F., Schneerson R., Malik Z. (1985). Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types J Clin Microbiol 22 828–834 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Ji Q., Jones M. B., Deng X., Liang H., Frank B., Telser J., Peterson S. N., Bae T., He C. (2012). AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus J Am Chem Soc 134 305–314 10.1021/ja2071835 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker M., Park J. S., Carey V., Lee J. C. (1998). Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model Infect Immun 66 5183–5189 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchscherr L., Loffler B., Buzzola F. R., Sordelli D. O. (2010). Staphylococcus aureus adaptation to the host and persistence: role of loss of capsular polysaccharide expression Future Microbiol 5 1823–1832 10.2217/fmb.10.147 . [DOI] [PubMed] [Google Scholar]
- Voyich J. M., Braughton K. R., Sturdevant D. E., Whitney A. R., Said-Salim B., Porcella S. F., Long R. D., Dorward D. W., Gardner D. J., other authors (2005). Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils J Immunol 175 3907–3919 10.4049/jimmunol.175.6.3907 . [DOI] [PubMed] [Google Scholar]
- Wann E. R., Dassy B., Fournier J. M., Foster T. J. (1999). Genetic analysis of the cap5 locus of Staphylococcus aureus FEMS Microbiol Lett 170 97–103 10.1111/j.1574-6968.1999.tb13360.x . [DOI] [PubMed] [Google Scholar]
- Watts A., Ke D., Wang Q., Pillay A., Nicholson-Weller A., Lee J. C. (2005). Staphylococcus aureus strains that express serotype 5 or serotype 8 capsular polysaccharides differ in virulence Infect Immun 73 3502–3511 10.1128/IAI.73.6.3502-3511.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Xue T., Shang F., Sun H., Sun B. (2010). Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence Infect Immun 78 3506–3515 10.1128/IAI.00131-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]