Abstract
Background & objectives:
Studies have shown that immunohistochemical (IHC) staining using epidermal growth factor receptor (EGFR) mutation specific antibodies, is an easy and cost-effective, screening method compared with molecular techniques. The purpose of present study was to assess the percentage positivity of IHC using EGFR mutation specific antibodies in lung biopsy samples from patients with primary lung adenocarcinoma (ADC).
Methods:
Two hundred and six biopsies of primary lung ADC were subjected to EGFR mutation specific antibodies against del E746-A750 and L858R. Detection of EGFR mutation done by high resolution melting analysis (HRM) was used as gold standard. A concordance was established between molecular and IHC results. Frequency of IHC positivity was assessed.
Results:
Of the 206 patients, 129 were male and 77 were female patients, with a mean age of 54.1 yr. Fifty five (26.6%) patients (36 men; 19 women) showed positivity for IHC of del E746-A750 (33) and L858R (22). HRM results were available in 14 patients which showed EGFR mutations in correspondence with del E746-750 or L858R in 64.2 per cent cases. Positive cases on HRM were further confirmed by DNA sequencing and fragment analysis. Three patients showed exon20 variation. Two cases were negative for mutation. The genotype of del E746-750 mutation was more common than L858R. A concordance was established between molecular mutation and IHC in 85.7 per cent cases.
Interpretation & conclusions:
In this preliminary study from India mutation specific IHC was used for assessment of mutation status of EGFR. Although the number tested was small, a good concordance was observed between molecular EGFR mutation and IHC expression. IHC methodology is a potentially useful tool to guide clinicians for personalized treatment in lung ADC, especially where facilities for molecular analysis are not readily available and for use in small biopsies where material is scant for molecular tests.
Keywords: Adenocarcinoma, epidermal growth factor receptor (EGFR), exon 19, exon 21, immunohistochemical (IHC), mutation specific, non-small cell lung carcinomas (NSCLC)
Lung cancer has emerged as a common cancer in urban India and is a leading cause of cancer related mortality1,2,3. Non-small cell lung carcinomas (NSCLC) are the most common type with adenocarcinoma (ADC) being the most common subtype4,5. With the emergence of selective tyrosine kinase inhibitors (TKI) targeting epidermal growth factor receptor (EGFR) there is a persistent need to identify the subset of NSCLCs harbouring specific EGFR mutations6. Hence, molecular testing of patients with adenocarcinoma of lung for selection of personalized therapy is standard of care in clinical practice7.
Activating mutations in tyrosine kinase domain of EGFR †have been reported in 10-50 per cent of NSCLCs, associated commonly with adenocarcinoma histology, female sex, East Asian ethnicity and never smokers8. About 90 per cent of activating EGFR mutations include deletions of exon 19 and point mutations of exon 216,8. Studies from India have reported EGFR mutations in approximately 20 to 50 per cent of cases studied in which mutation was detected by molecular methods which include PCR based methods and DNA sequencing3,9,10,11,12,13,14,15. Though there are various methods to detect most common EGFR deletion and point mutations, there is no approved gold standard method. The molecular mutation detection methods such as DNA sequencing and real time polymerase chain reaction (RT-PCR) are limited by their low sensitivity, procedure complexity, increased cost and turnaround time16,17. Moreover, these methods cannot be incorporated into the routine diagnostic laboratory especially in low resource settings such as in India. For these reasons, immunohistochemistry (IHC) for EGFR mutation specific antibodies has emerged as a relevant alternative for determining EGFR status in NSCLC18,19.
We hypothesized that EGFR mutation specific IHC can be utilized as a routine method in a clinical diagnostic laboratory to detect the two most common mutations for identifying patients for personalized treatment of EGFR TKI. Here we report the IHC results of 206 biopsy samples obtained from patients of ADC of lung which were scored taking into account the intensity and area of staining.
Material & Methods
Between January 2013 and June 2014, we prospectively analyzed 206 lung biopsies diagnosed as NSCLC with subtype adenocarcinoma based on morphology and immunohistochemistry (TTF-1 positive, p40 negative) in the department of Pathology, All India Institute of Medical Sciences, New Delhi. Approval from ethics committee of the All India Institute of Medical Sciences, New Delhi, was obtained prior to the study. Informed written consent from each patient was taken at the time of biopsy. The diagnosis was made according to International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification for lung adenocarcinomas20.
IHC analyses of EGFR mutation specific antibodies: Paraffin blocks were cut to a thickness of 4 micron for immunostaining. Conventional de-waxing and hydration treatment were undertaken using xylene and a graded series of ethanol, respectively. Antigen retrieval was performed using citric acid at pH 6.0. Intrinsic peroxidase activity was blocked by treatment with 0.3 per cent hydrogen peroxide and methanol solution for 15 min. After washing in tris-buffered saline for 15 min, two primary antibodies [delE746-A750 mutation specific monoclonal antibody (6B6) and L858R mutation specific monoclonal antibody (43B2); Cell Signaling Technology, Danvers, MA, USA] were diluted separately at 1:100 and added to the specimens fixed to the slides. The slides were incubated for two hours at room temperature with mutation specific antibodies. Slides were then incubated and analyzed by a detection kit (EnVision Plus HRP, DakoCytomation, Glostrup, Denmark) according to the manufacturer's instructions, developing peroxidase activity with 3-3diaminobenzidine. Finally, slides were counterstained with hematoxylin, dehydrated and mounted21,22.
IHC scoring: The IHC staining score was based on the staining intensity and percentage positivity (0-100%) of cells in the membrane and/or cytoplasm of tumour cells as previously described23,24.
Four grades were employed: 0, 1+, 2+, 3+. 0= no staining; 1+ =faint membrane and/or cytoplasmic staining in >10 per cent of tumour cells in x 40 objective magnification; 2+ = moderate membrane and/or cytoplasmic staining in x 10 to x 20 objective magnification; 3+ = strong membrane and/or cytoplasmic staining x 2 to x 4 objective magnification. Zero was negative; whereas 1+, 2+ and 3+ were considered as positive cases. Lung adenocarcinomas with mutation status confirmed by molecular testing were used as positive and negative controls. The intensity of the staining was assessed by two pathologists independently.
Validation of IHC with molecular detection by high resolution melting analysis (HRMA): Polymerase chain reaction (PCR) was performed to amplify exons 19 or 21 of EGFR using Light Scanner master mix (Idaho Technology, USA) on Veriti thermal cycler (Applied Biosystems, USA) using gene specific primers (Idaho). The PCR product was denatured at 95°C for 30 sec and then cooled to 28°C to promote the formation of heteroduplexes. The plate was transferred to Light Scanner (Idaho Technology) and melt analysis was performed according to manufacturer's instructions. Data were analyzed using the software (Idaho Technology). After normalization and temperature-adjustment steps, melting curve shapes were compared between the tumour samples and control samples. Human genomic DNA extracted from leukocytes was used as the negative control sample with wild-type EGFR and known sample harbouring the mutation was used as positive control. Samples having left-shifted curves or curves similar to positive controls were judged to have mutations and further validated by sequencing. The L858R mutation was further confirmed by direct sequencing and deletion E746-750 by fragment analysis (outsourced to Applied Biosystems).
Statistical analysis: Appropriate statistical analysis was done to calculate sensitivity, specificity, positive and negative predictive values. χ2 test was used for categorical variables.
Results
Of the 206 patients, 129 were male and 77 were female patients (M:F 1.6:1), with a mean age of 54.1 ± 1.4 yr (range 21-81 yr). No patients had received neoadjuvant chemotherapy or radiotherapy prior to biopsy.
IHC analysis of mutation-specific antibodies against EGFR mutants in lung adenocarcinomas: The mutation-specific antibodies against L858R point mutation in exon 21 and E746-A750 deletion in exon 19 were used to stain the tumour sections. Positive IHC staining of del E746-A750 and L858R with faint (1+; 7 cases)(Fig. 1A, B), moderate (2+; 25 cases) (Fig. 1C), and strong (3+; 23 cases) (Fig. 1D) intensity was identified in 26.6 per cent (55/206) cases. E746-A750 del (33/55; 60%) was more common than L858R (22/55; 40%). The mutant-specific antibodies showed cytoplasmic and membranous staining in positive cases. There was no positive staining in normal tissues. Three cases showed 1+ positivity for both mutation specific antibodies, on repeat IHC two of these cases showed negative results, thus all three samples were considered negative.
Fig. 1.
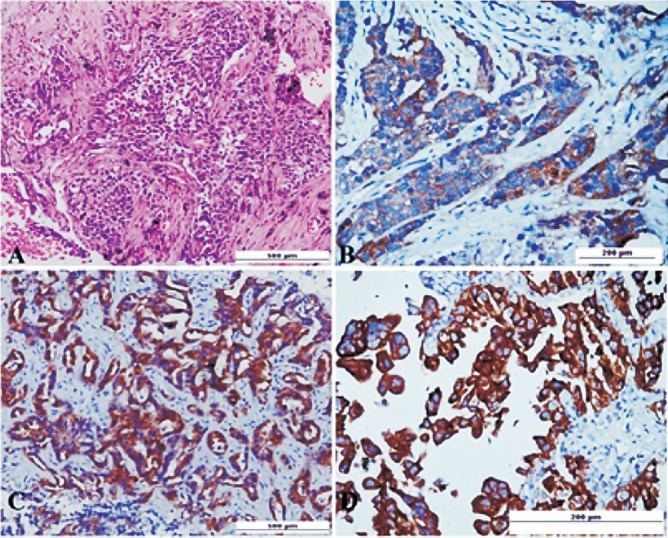
(A). Photomicrograph of biopsy from the mass lesion in lung shows an invasive adenocarcinoma H&E x 20. (B) EGFR del E746-A750 IHC shows weak cytoplasmic staining without membrane accentuation scored as 1+ original magnification x10. (C) EGFR L858R IHC shows moderate intensity of cytoplasmic staining with rare membrane reactivity scored as 2+ original magnification x20. (D) EGFR del E746-A750 IHC shows strong cytoplasmic positivity with diffuse membrane staining scored as 3+ original magnification x10.
Concordance of IHC with molecular analysis: HRMA results were available in 14 patients which showed EGFR mutations in correspondence with del E746-750 (Fig. 2) or L858R (Fig. 3) in 64.2 per cent (9/14) cases. Three patients showed exon 20 variation. Two cases were negative for mutation. The findings of IHC were in concordance with the results of molecular analysis in 85.7 per cent (12/14) of tested biopsy samples. There was no significant difference in positive EGFR mutation ratio determined by molecular testing and IHC. Based on molecular testing, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of immunostaining results using the two antibodies in 14 samples are presented in the Table. Discordant molecular and IHC results were found in two cases, which were positive for L858R point mutation in exon 21 by HRM and DNA sequencing (Fig. 4) but negative by IHC. E746-750 deletion of exon 19 was further confirmed by fragment analysis (Fig. 5). Three cases positive for exon 20 variation (2361 G>A and 2462 T>A in one of the cases) and two cases negative for any mutation did not show positivity by IHC.
Fig. 2.
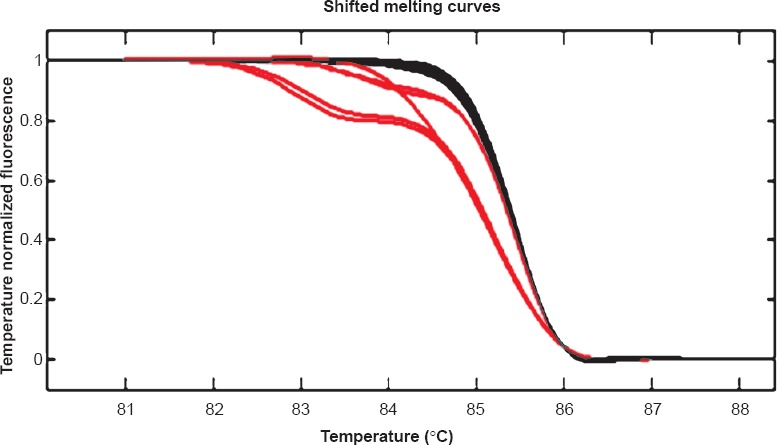
Melt Analysis of exon 19: Left shifted curves (red) are patient harbouring deletion, Right sided curves (grey) are wild type.
Fig. 3.
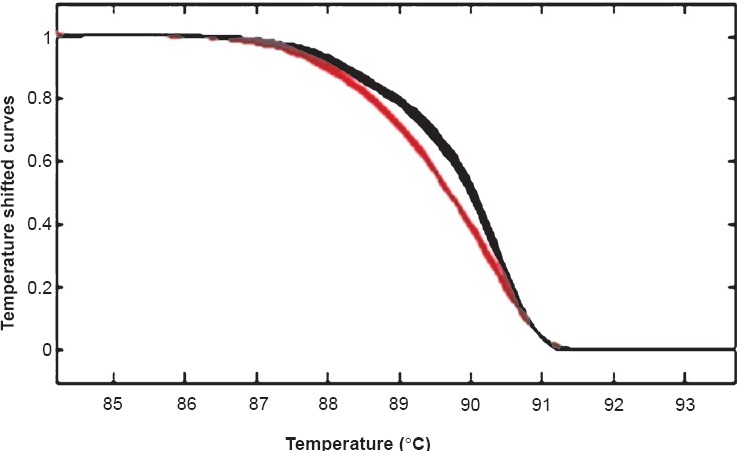
Melt Analysis of exon 21: Left shifted curves (red) are patient harbouring mutation, Right sided curves (grey) are wild type.
Table.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of two mutation specific antibodies.
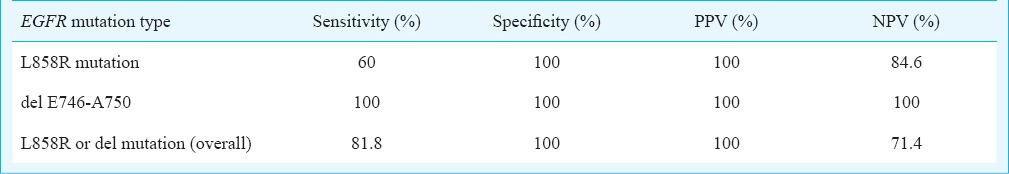
Fig. 4.
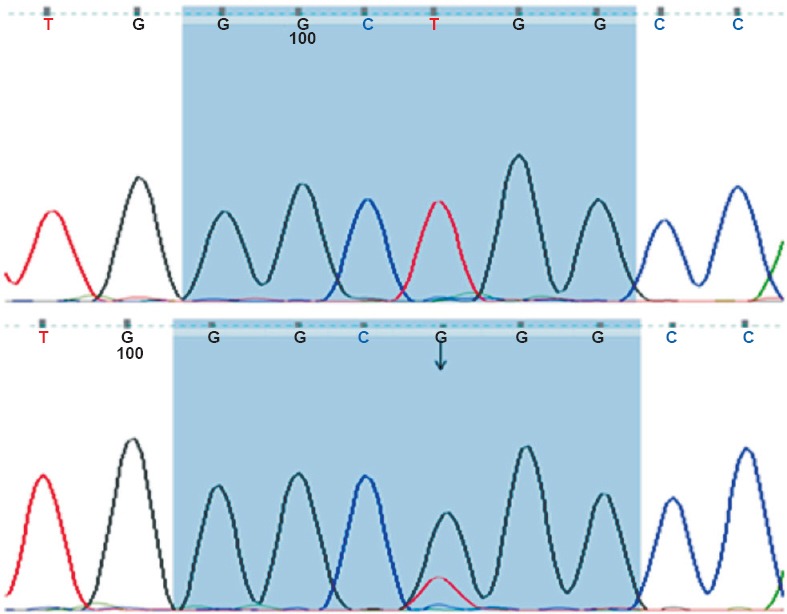
Sequencing chromatograms of exon 21: Upper panel; Wild type and Lower panel; Arrow indicating L858R mutation.
Fig. 5.
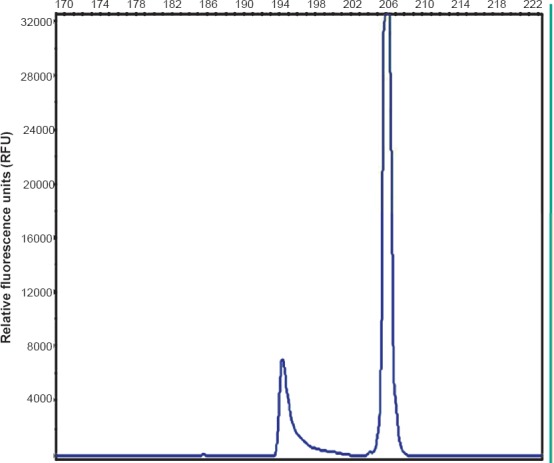
Fragment analysis of exon 19: Smaller peak depicting the deletion.
In seven positive cases (6 del19 and 1 L858R) which were in concordance with molecular mutation, IHC score was 2+ (3 cases) and 3+ (4 cases).
Discussion
In the present study 26.6 per cent positivity was observed for the two most frequently found targeted mutations by IHC analysis that was within the range of prior studies3,9,10,11,12,13,14,15. A frequency on the lower side in the present study can be explained by the method of detection that is IHC which detects only two most common mutations. Clinically male predominance was observed similar to earlier reports in Indian and Asian origin of patients10,11,25.
In routine clinical practice the most frequently received specimen for the diagnosis of lung cancer is either a small biopsy or a cytology sample. Some of these limited samples, at times, fail molecular testing mostly due to scant cellularity. Moreover, limitations of the use of molecular testing in low resource settings include high costs of tests and reagents as well as availability of experts for the interpretation of the results.
Routine testing requires the development of an efficient, cost-effective and practical test that can be standardized and performed by diagnostic pathology laboratories, to triage specimens and expedite treatment decisions by a shorter diagnosis time. Immunohistochemistry has been used in routine clinical pathology practice due to its reduced turnaround time, cost-effectiveness, easy availability and rapid interpretation by pathologists21,22,26,27. These advantages make IHC a useful test that can be used to screen for EGFR mutations in NSCLCs. Immunohistochemistry for total EGFR is not recommended for selection of tyrosine kinase inhibitor therapy because it has been shown to correlate poorly or not at all with the presence of EGFR mutations12.
The present study showed that mutation-specific antibodies against L858R point mutation in exon 21 and E746-A750 deletion in exon 19 were reasonably useful tools for rapid screening to detect the two most common forms of targetable EGFR mutations. Clone 43B2 detected L858R mutations with a sensitivity of 60 per cent and PPV of 100 per cent and, clone 6B6 detected the 15 base-pair exon-19 deletions with a sensitivity of 100 per cent and PPV of 100 per cent.
In earlier studies, IHC with the EGFR L858R mutated antibody has confirmed excellent sensitivity and specificity relative to mutation testing19,26,28,29. The EGFR exon 19 mutated-specific antibody showed good sensitivity and specificity for cases with the 15-bp deletion in exon 19 but reduced sensitivity was observed for exon 19 deletions of other sizes26,27,28,29. Though we had small number of cases, EGFR exon 19 deletion was detected in all six cases indicating 15-bp deletion. Exon 21 L858R mutation was not detected in the two cases possibly due to defective antigen retrieval, mutational heterogeneity or technical error30.
One major drawback for the use of IHC to detect EGFR mutations is that these are specific only to their target mutations (E746-A750 deletion in exon 19 and L858R in exon 21). Other mutations such as TKI resistance exon 20 mutations cannot be detected by these two antibodies. Though suboptimal sensitivity for L858R mutation precludes use of these antibodies as ascreening tool23, these may potentially be used at the time of diagnosis due to their high specificity and positive predictive value (though based on low number of cases), and for the obvious advantages described earlier especially in Indian setting. Moreover, IHC is useful to overcome some of the obstacles that are unavoidable in molecular testing which include low cellularity in small biopsies and contamination of tumour by normal tissue. The high specificity observed for both antibodies was similar to the earlier studies, where specificity of exon19 mutations ranged from 92 to 100 per cent and for exon21 mutations from 97 to 100 per cent26,27,28. IHC with EGFR mutation specific antibodies can be used as an initial screen to identify most patients who are candidates for TKI, if IHC scoring cut-offs are set stringently to ensure a high positive predictive value.
The pattern of IHC positivity is important; only homogeneous moderate to strong cytoplasmic and membranous staining correlated with the presence of the mutation23,31. Allo et al31 have observed small yet significant false positive rate especially in small biopsies29,31. It has been noted that false positive cases are associated with 1+ staining31. However, further studies are needed to establish the exact rate of false positivity associated with these antibodies. It has been recommended that 2+ and 3+ cases can be considered true positives whereas 1+ cases should undergo molecular testing28,31.
As with any immunostain, possibilities for false-negative and false-positive results still exist, and confirmation of an equivocal immunostain can be done by molecular analysis. In addition, due to their low NPV all specimens negative with these two mutation-specific monoclonal antibodies should also be considered valid for molecular testing.
There are two different clones available for these two antibodies; 6B6 for del exon 19 and 43B2 for L858R mutation (used in the present study) and SP125 (del exon 19) and SP111 (L858R mutation). In the present study equivalent performance of these clones has been observed as previously reported18,31.
In conclusion, this is a preliminary study of IHC analysis of EGFR mutation specific antibodies in 206 Indian patients of ADC of lung to examine the usefulness of this test in routine setting. One limitation of the present study was lack of information about clinical responsiveness of mutation positive cases due to prospective nature of the study. Secondly, the number of samples tested by molecular methods was less for establishment of accurate concordance. We propose IHC as an initial, rapid and cost-effective tool especially in our diagnostic set-up and peripheral laboratory services where facilities for molecular analysis are limited. It is also useful to avoid the challenges of mutation testing in small low tumour cellularity specimens. However, more studies are needed to validate these results further in large number of patients and to evaluate their utility in patient management.
Acknowledgment
Authors acknowledge the financial support received from the Research Section, All India Institute of Medical Sciences, New Delhi.
Footnotes
Conflicts of Interest: None.
References
- 1.Ferlay J SH, Bray F, Forman D, Mathers C, Parkin DM. Lyon, France: International Agency for Research on Cancer; [accessed on July 2, 2013]. GLOBOCAN 2008 v2.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 10. Available from: http://globocan.iarc.fr . [Google Scholar]
- 2.Dikshit R, Gupta PC, Ramasundarahettige C, Gajalakshmi V, Aleksandrowicz L, Badwe R, et al. Cancer mortality in India: a nationally representative survey. Lancet. 2012;379:1807–16. doi: 10.1016/S0140-6736(12)60358-4. [DOI] [PubMed] [Google Scholar]
- 3.Noronha V, Dikshit R, Raut N, Joshi A, Pramesh CS, George K, et al. Epidemiology of lung cancer in India: focus on the differences between non-smokers and smokers: a single-centre experience. Indian J Cancer. 2012;49:74–81. doi: 10.4103/0019-509X.98925. [DOI] [PubMed] [Google Scholar]
- 4.Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–9. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
- 5.Malik PS, Sharma MC, Mohanti BK, Shukla NK, Deo S, Mohan A, et al. Clinico-pathological profile of lung cancer at AIIMS: a changing paradigm in India. Asian Pac J Cancer Prev. 2013;14:489–94. doi: 10.7314/apjcp.2013.14.1.489. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137:828–60. doi: 10.5858/arpa.2012-0720-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–62. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 9.Sahoo R, Harini VV, Babu VC, Patil Okaly GV, Rao S, Nargund A, et al. Screening for EGFR mutations in lung cancer, a report from India. Lung Cancer. 2011;73:316–9. doi: 10.1016/j.lungcan.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Chougule A, Prabhash K, Noronha V, Joshi A, Thavamani A, Chandrani P, et al. Frequency of EGFR mutations in 907 lung adenocarcioma patients of Indian ethnicity. PLoS One. 2013;8:e76164. doi: 10.1371/journal.pone.0076164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choughule A, Noronha V, Joshi A, Desai S, Jambhekar N, Utture S, et al. Epidermal growth factor receptor mutation subtypes and geographical distribution among Indian non-small cell lung cancer patients. Indian J Cancer. 2013;50:107–11. doi: 10.4103/0019-509X.117023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta J. Molecular epidemiology of epidermal growth factor receptor mutations in lung cancers in Indian population. Indian J Cancer. 2013;50:102–6. doi: 10.4103/0019-509X.117019. [DOI] [PubMed] [Google Scholar]
- 13.Doval DC, Azam S, Batra U, Choudhury KD, Talwar V, Gupta SK, et al. Epidermal growth factor receptor mutation in lung adenocarcinoma in India: A single center study. J Carcinog. 2013;12:12. doi: 10.4103/1477-3163.114970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noronha V, Prabhash K, Thavamani A, Chougule A, Purandare N, Joshi A, et al. EGFR mutations in Indian lung cancer patients: clinical correlation and outcome to EGFR targeted therapy. PLoS One. 2013;8:e61561. doi: 10.1371/journal.pone.0061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veldore VH, Rao RM, Kakara S, Pattanayak S, Tejaswi R, Sahoo R, et al. Epidermal growth factor receptor mutation in non-small-cell lung carcinomas: a retrospective analysis of 1036 lung cancer specimens from a network of tertiary cancer care centers in India. Indian J Cancer. 2013;50:87–93. doi: 10.4103/0019-509X.117013. [DOI] [PubMed] [Google Scholar]
- 16.Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7:36–9. doi: 10.1016/s1525-1578(10)60006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams C1, Pontén F, Moberg C, Söderkvist P, Uhlén M, Pontén J, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–71. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo AN, Park TI, Jin Y, Sun PL, Kim H, Chang H, et al. Novel EGFR mutation-specific antibodies for lung adenocarcinoma: highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry. Lung Cancer. 2014;83:316–23. doi: 10.1016/j.lungcan.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Wang X, Xue L, Xu N, Ye X, Zeng H, et al. The use of mutation-specific antibodies in predicting the effect of EGFR-TKIs in patients with non-small-cell lung cancer. J Cancer Res Clin Oncol. 2014;140:849–57. doi: 10.1007/s00432-014-1618-2. [DOI] [PubMed] [Google Scholar]
- 20.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X, Liu B, Xu H, Yu B, Shi S, Zhang J, et al. Immunostaining with EGFR mutation-specific antibodies: a reliable screening method for lung adenocarcinomas harboring EGFR mutation in biopsy and resection samples. Hum Pathol. 2013;44:1499–507. doi: 10.1016/j.humpath.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Kane S, Wu J, Benedettini E, Li D, Reeves C, et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res. 2009;15:3023–8. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 23.Bondgaard AL, Høgdall E, Mellemgaard A, Skov BG. High specificity but low sensitivity of mutation-specific antibodies against EGFR mutations in non-small-cell lung cancer. Mod Pathol. 2014;27:1590–8. doi: 10.1038/modpathol.2014.67. [DOI] [PubMed] [Google Scholar]
- 24.Wen YH, Brogi E, Hasanovic A, Ladanyi M, Soslow RA, Chitale D, et al. Immunohistochemical staining with EGFR mutation-specific antibodies: high specificity as a diagnostic marker for lung adenocarcinoma. Mod Pathol. 2013;26:1197–203. doi: 10.1038/modpathol.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–62. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozu Y, Tsuta K, Kohno T, Sekine I, Yoshida A, Watanabe S, et al. The usefulness of mutation-specific antibodies in detecting epidermal growth factor receptor mutations and in predicting response to tyrosine kinase inhibitor therapy in lung adenocarcinoma. Lung Cancer. 2011;73:45–50. doi: 10.1016/j.lungcan.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara A, Azuma K, Sumi A, Taira T, Nakashima K, Aikawa E, et al. Identification of non-small-cell lung cancer with activating EGFR mutations in malignant effusion and cerebrospinal fluid: rapid and sensitive detection of exon 19 deletion E746-A750 and exon 21 L858R mutation by immunocytochemistry. Lung Cancer. 2011;74:35–40. doi: 10.1016/j.lungcan.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Kato Y, Peled N, Wynes MW, Yoshida K, Pardo M, Mascaux C, et al. Novel epidermal growth factor receptor mutation-specific antibodies for non-small cell lung cancer: immunohistochemistry as a possible screening method for epidermal growth factor receptor mutations. J Thorac Oncol. 2010;5:1551–8. doi: 10.1097/JTO.0b013e3181e9da60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12:169–76. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurada A, Lara-Guerra H, Liu N, Shepherd FA, Tsao MS. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J Thorac Oncol. 2008;3:527–9. doi: 10.1097/JTO.0b013e318168be93. [DOI] [PubMed] [Google Scholar]
- 31.Allo G, Bandarchi B, Yanagawa N, Wang A, Shih W, Xu J, et al. Epidermal growth factor receptor mutation-specific immunohistochemical antibodies in lung adenocarcinoma. Histopathology. 2014;64:826–39. doi: 10.1111/his.12331. [DOI] [PubMed] [Google Scholar]


