Abstract
Background & objectives:
Pre-extensively drug resistant (pre-XDR) and extensively drug resistant tuberculosis (XDR-TB) have been areas of growing concern, and are posing threat to global efforts of TB control. The present study was planned to study the presence of pre-XDR and XDR Mycobacterium tuberculosis and their genotypes in clinical isolates obtained from previously treated cases of pulmonary TB.
Methods:
A total of 219 isolates obtained from previously treated cases of pulmonary TB were subjected to first-line (streptomycin, isoniazid, rifampicin and ethambutol) and second-line (ofloxacin, kanamycin, capreomycin and amikacin) drug susceptibility testing on solid Lowenstein-Jensen medium by proportion method. Genotyping was done for pre-XDR and XDR-TB isolates using 12 loci Mycobacterial Interspersed Repetitive Units-Variable Number Tandem Repeats (MIRU-VNTR).
Results:
Multi-drug resistance was observed in 39.7 per cent (87/219) isolates. Pre-XDR and XDR M. tuberculosis isolates amongst 87 multi-drug resistant (MDR) TB isolates were 43 (49.4%) and 10 (11.4%), respectively. Two most dominant genotypes among pre-XDR and XDR M. tuberculosis isolates were Beijing and Delhi/CAS types.
Interpretation & conclusions:
Resistance to second-line anti-tubercular drugs should be routinely assessed in areas endemic for TB. Similar genotype patterns were seen in pre-XDR and XDR-TB isolates. Beijing and Delhi/CAS were predominant genotypes.
Keywords: MDR-TB, MIRU-VNTR, Mycobacterium tuberculosis, pre-XDR-TB, XDR-TB
Mult-drug resistant tuberculosis (MDR-TB) is a known phenomenon, but recently extensively drug resistant (XDR) and pre-extensively drug resistant (pre-XDR) TB have become areas of growing concern, and are posing threat to global efforts of TB control. XDR-TB is defined as resistance to atleast isoniazid (INH) and rifampicin (RIF) (i.e. MDR-TB), as well as further resistance to any fluoroquinolone (FQ) and a second-line injectable drug (kanamycin, amikacin or capreomycin)1. In 2012, according to World Health Organization (WHO), XDR-TB has been identified in 92 countries globally2. The exact prevalence of XDR-TB in most regions of the world is currently unknown. Though XDR-TB has been reported from India3,4,5, but the extent and magnitude of XDR-TB is yet to be determined. Efforts to expand surveillance to second-line anti-TB drugs are underway. Pre-XDR-TB is defined as TB with resistance to isoniazid and rifampicin and either a FQ or a second-line injectable agent but not both6. An incidence of FQ resistant MDR-TB is increasing globally but unfortunately, till date limited data are available on prevalence of pre-XDR-TB worldwide and in India7. Due to constraint in resource and high TB burden, resistance to FQs and second-line drugs is not tested routinely in developing world. Exposure to FQs and injectable aminoglycoside or both for the treatment of bacterial infection other than TB may contribute to evolution of resistance to these agents. In a country like India where TB is endemic and FQs and aminoglycosides are routinely used it may be important to know the prevalence of resistance to FQs and aminoglycosides in Mycobacterium tuberculosis isolates. Since pre-XDR and XDR-TB isolates are emerging globally, their molecular genotyping will help to study TB epidemiology as it identifies the prevalent genotypes within a defined geographical location, enhances understanding of TB transmission dynamics and helps to evaluate outbreaks of TB8. Therefore, the present study was conducted to study the presence and genotypes of pre-XDR and XDR M. tuberculosis in clinical isolates obtained from cases of pulmonary TB in north India.
Material & Methods
During May 2012 to June 2013, a total of 341 sputum samples from equal number of previously treated cases (having received > 4 wk of anti-TB drugs in the past) of pulmonary TB were received for demonstration of acid fast bacilli (AFB)9 and mycobacterial culture10 at TB laboratory, department of Microbiology, King George's Medical University, Lucknow, Uttar Pradesh, India, which is an intermediate reference laboratory for Revised National Tuberculosis Control Programme (RNTCP). Total 219 isolates of M. tuberculosis were obtained from 341 samples and subjected to first-line [streptomycin (SM), INH, RIF and ethambutol (EMB)] and second-line [ofloxacin (OFX), kanamycin (KM), capreomycin (CAP) and amikacin (AM)] drug susceptibility testing (DST) on solid Lowenstein-Jensen medium by one per cent proportion method9,10. Critical concentrations were 0.2 μg/ml for INH, 4 μg/ml for SM, 40 μg/ml for RIF, 2 μg/ml for EMB, 2 μg/ml for OFX, 30 μg/ml for KM, 40 μg/ml for CAP, 40 μg/ml for AM11,12. A standard strain M. tuberculosis H37Rv was used as the quality control strain for each new batch of medium throughout the study. Pure powder forms of all drugs were obtained from Sigma-Aldrich, USA.
Genotyping: DNA extraction of mycobacteria was performed according to method described by Aslan et al13. Genotyping was performed using 12 loci Mycobacterial Interspersed Repetitive Units-Variable Number Tandem Repeats (MIRU-VNTR), for pre-XDR and XDR M. tuberculosis isolates14. Primer sequences used to study various MIRU loci were according to that mentioned by Supply et al15. Primers were obtained from Bioserve biotech, Hyderabad. PCR products were separated using an agarose gel and were analysed by gel document system (Alfa Imager HP3400, California, USA). The sizes of amplicons were used to determine the number of repetitions by reference to a table (available at http://www.miruvntrplus.org). The results are expressed as a numerical code representing the number of MIRU-VNTRs in each of the 12 genomic loci. Via MIRU-VNTRplus database, study isolates were compared with the reference strains for the assignment of MTB genotype16 and dendrogram of the typing method was generated using UPGMA analysis (www.miruvntrplus.org).
A cluster was defined as two or more isolates with identical genetic patterns. Clusters were assumed to have arisen from recent transmission, and the clustering rate was used to determine recent transmission of MTB17. Clustering rate was calculated using the formula: (number of clustered strains - number of clusters)/total number of strains17. The allelic diversity of MIRU loci was calculated using the Hunter-Gaston Discriminatory Index (HGDI)18 and was classified as highly discriminant (> 0.6), moderately discriminant (≥ 0.3) and poorly discriminant (< 0.3)19.
Results
Of the 219 isolates subjected to first-line DST, 69 (31.5%) were susceptible to all four first-line drugs (SM, INH, RIF and EMB), 87 (39.7%) were MDR, 22 (10%) were poly-resistant, 41 (18.7%) were monoresistant and 38 (14.3%) were resistant to all four first-line drugs tested (Table I). These 219 isolates were also subjected to second-line DST, of which 129 (58.9%) were susceptible to all four second-line drugs (OFX, KM, CAP and AM); 72 (32.8%) isolates were resistant to OFX. CAP, KM and AM resistance was seen in 22 (10%), 14 (6.3%) and 10 (4.5%) isolates, respectively (Table II). Of the 87 MDR-TB isolates, 34 (39%) were susceptible to all the four second-line drugs tested. Resistance to OFX was detected in 44 (50.5%) isolates. Resistance to KM, CAP and AM was found in 8 (9.1%), 15 (17.2%) and 7 (8%) isolates, respectively. The pre-XDR-TB (amongst MDR-TB) was seen in 43 (49.4%) isolates. Total 34 of 87 (39%) isolates were OFX resistant pre-XDR. KM, CAP and AM resistant pre-XDR isolates were one, three and two, respectively. Presence of XDR-TB amongst MDR-TB was seen in ten isolates (11.4%) (Table II).
Table I.
First-line anti-tuberculosis drug resistance among M. tuberculosis isolates from previously treated cases of pulmonary tuberculosis
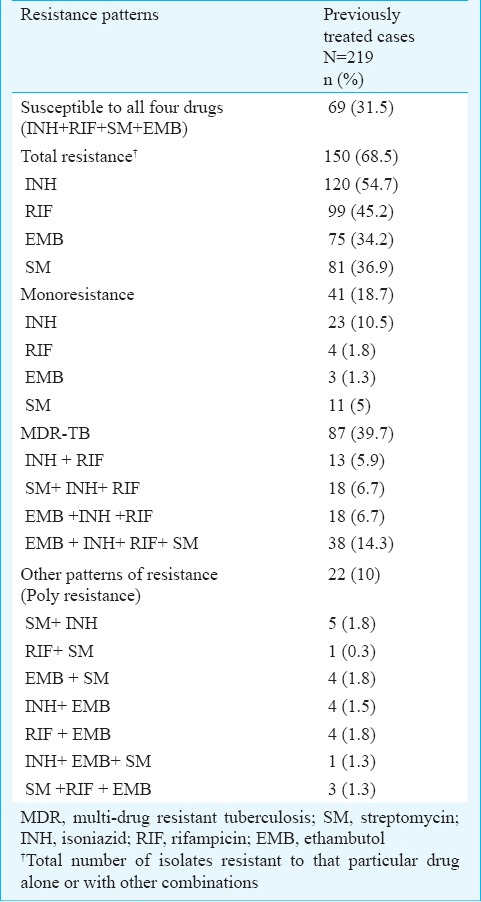
Table II.
Prevalence of second-line anti-tuberculosis drug resistance among M. tuberculosis isolates from previously treated cases of pulmonary tuberculosis
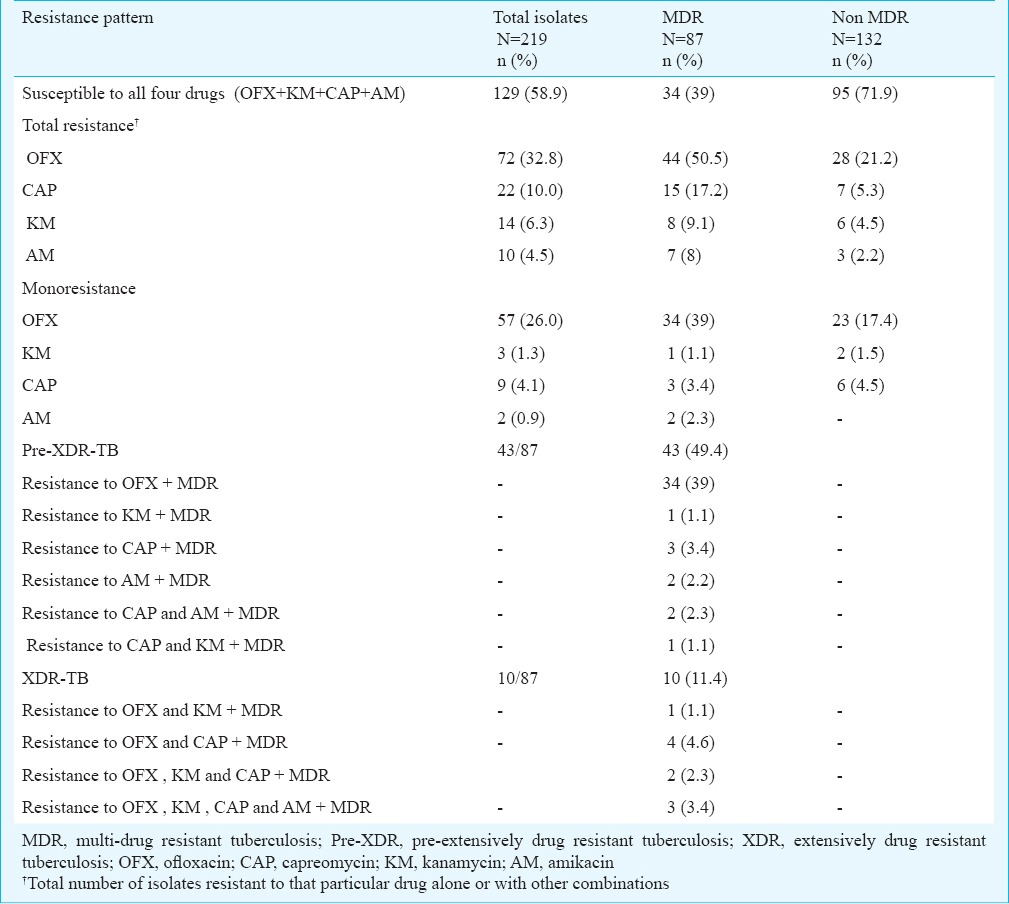
Of the 132 non-MDR-TB isolates, resistance to OFX only was found in 23 and resistance to an aminoglycoside only was seen in eight isolates (Table II). Six non MDR-TB isolates were resistant to more than one second-line drug.
Of the 43 pre-XDR M. tuberculosis isolates, 24 (55.8%) were Beijing genotype, 15 (34.8%) were Delhi/CAS, two (4.6%) were Latin American and Mediterranean (LAM) and two were not assigned any genotype (orphan) (Fig. 1). Of the 10 XDR isolates, six were Beijing and four were Delhi/CAS (Fig. 2). Among pre-XDR M. tuberculosis isolates, a total of 12 distinct patterns were identified which included seven clusters each with 2-15 isolates. In total, 38 (88.3%) isolates clustered and clustering rate was 72 per cent. Among XDR M. tuberculosis isolates, two clusters each with 2-4 isolates were found (Fig. 2) and clustering rate was 40 per cent. Analysis of the allelic diversity of the 12 MIRU-VNTR loci revealed that MIRU loci 10, 40, 31, 26, 16 and 23 were highly discriminatory; loci 4, 24, 27 and 39 were moderately discriminatory; and loci 2 and 20 were poorly discriminatory.
Fig. 1.
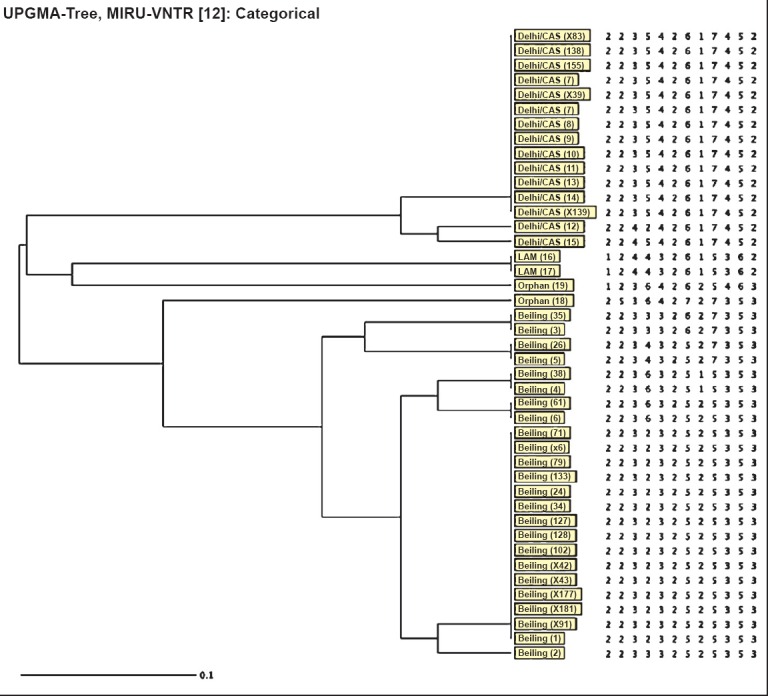
Dendogram showing MIRU-VNTR (Mycobacterial Interspersed Repetitive Units - Variable Number Tandem Repeats) analysis of pre-XDR M. tuberculosis isolates (n=43). LAM, Latin American and Mediterranean.
Fig. 2.
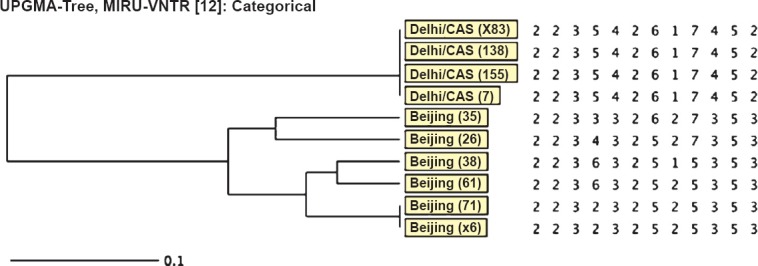
Dendogram showing MIRU-VNTR (Mycobacterial Interspesred Repetitive Units - Variable Number Tandem Repeats) analysis of XDR M. tuberculosis isolates (n=10).
Discussion
In the present study multi-drug resistance was found in 39.7 per cent isolates. Presence of pre-XDR and XDR-TB among MDR-TB isolates was 49.4 and 11.4 per cent, respectively. The percentage of pre-XDR and XDR-TB among MDR-TB isolates was slightly higher than reported in a previous study from our laboratory during 2007-20097. In earlier study only two second-line drugs OFX and KM were tested. Inclusion of two more second-line drugs, CAP and KM in test panel may have resulted in increase of the number of pre-XDR and XDR isolates. The prevalence rate of pre-XDR-TB among MDR-TB patients was reported to be 12.1 per cent in Poland, 16.7 per cent in Nigeria, 18 per cent in California, 31 per cent in China and 51.4 per cent in Philippines6,20,21,22,23. Majority of the pre-XDR-TB isolates were resistant to OFX. The OFX resistant pre-XDR-TB in our study was 39 per cent, which was higher than reported in other studies ranging from 7.7 to 27.6 per cent7,20,24. KM resistance pre-XDR-isolates in our study was 1.1 per cent, similar to that found in earlier studies varying from 0.6 to 14.6 per cent7,20,24. AM resistance in pre-XDR isolates in our study was 2.2 per cent, similar to Poland (2.6%) and China (2.4%)20,21.
The actual incidence and prevalence of XDR-TB in India is not available. A few scattered reports reveal the prevalence ranging from 2.4-33.3 per cent3,25,26. Percentage of MDR-TB (39.7%) was slightly higher than reported in our previous studies7,27. The high level of resistance to FQ (i.e OFX in our study) agreed well with the extensive use of FQs for the treatment of new TB cases and its irrational use for other bacterial infections. Even aminoglycosides are commonly used antibiotics in various infections. Their injudicious use has led to emergence of resistance to the second-line injectable agents.
The limitation of our study was that only 12 loci were used instead of 24 loci MIRU for genotyping and molecular epidemiological studies of M. tuberculosis. Moreover, this initial MIRU-12 set was replaced by the standard MIRU-15 set, and subsequently, standard MIRU-24 loci set28 has been proposed for optimal discrimination of closely related strains. Various studies have defined a minimal set of 12 loci for genotyping Beijing strains which made up more than 90 per cent of the isolates investigated from Asia29,30,31. The Beijing family was the predominant genotype in pre-XDR and XDR M. tuberculosis isolates in this study. Zhao et al22 from China found that 90.9 per cent of the XDR isolates were Beijing genotype. A study from New Delhi also revealed predominance of Beijing genotype in XDR M. tuberculosis isolates32. A study conducted in Kanpur, north India, found that the MDR-TB was significantly higher in Beijing strains than others33. Beijing genotype was initially found in China but these strains have spread to different geographic regions of the world34. The second predominant genotype in pre-XDR and XDR M. tuberculosis isolates in our study was Delhi/CAS. The Delhi/CAS is localized in Middle East and Central Asia, preferentially in India35. A study from Delhi showed that Delhi/CAS was predominant genotype among M. tuberculosis clinical isolates36. A study conducted in Kerala revealed that ancestral East-African Indian (EAI) lineage comprised the majority of circulating MTB genotypic clones37.
Not much data are available on genotypes of pre-XDR/ XDR MTB isolates from Indian subcontinent. Our study showed predominance of Beijing and Delhi/CAS genotypes. High clustering rate among genotypes found in our study implies high transmission rate. This suggests that the same genotypes are being transmitted and pre-XDR/ XDR-isolates are not associated with emergence of new genotype. Population based studies over long periods are needed to understand epidemiology of pre-XDR and XDR-TB. Studies on molecular epidemiology of drug resistant M. tuberculosis should be undertaken to detect chains of transmission and improve TB control activities.
In view of high resistance to OFX, KM, CAP and AM among MDR-TB isolates in our study, it is a necessity that second-line DST of M. tuberculosis isolates should be performed routinely. Documenting the emergence of pre-XDR and XDR-TB requires a laboratory-based diagnosis that relies on first- and second-line drug susceptibility testing. As access to treatment with second-line drugs increases, standardized methods, improved diagnostics, and quality assurance for second-line drug susceptibility testing are urgently needed to enable reliable testing and design of appropriate treatment regimen. Emphasis needs to be given on identification of pre-XDR-TB patients among MDR-TB patients and treatment monitoring to ensure cure and halting the progression to XDR-TB.
Footnotes
Conflicts of Interest: None.
References
- 1.Report of the meeting of the WHO Global Task Force on XDR-TB. Geneva, Switzerland: WHO; 2009. [accessed on February 2, 2013]. World Health Organization. Geneva, Switzerland, 9-10 October, 2006 WHO/HTM/TB/2007.375. Available from: http://whqlibdoc.who.int/hq/2007/who_htm_tb_2007375_eng.pdf . [Google Scholar]
- 2.World Health Organization. Global Tuberculosis Report 2013. [accessed on February 5, 2014]. Available from: www.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf .
- 3.Mondal R, Jain A. Extensively drug resistant Mycobacterium tuberculosis, India. Emerg Infect Dis. 2007;13:1429–30. doi: 10.3201/eid1309.070443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porwal C, Kaushik A, Makkar N, Banavaliker JN, Hanif M, Singla R, et al. Incidence and risk factors for extensively drug-resistant tuberculosis in Delhi region. PLoS One. 2013;8(2):e55299. doi: 10.1371/journal.pone.0055299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michael JS, John TJ. Extensively drug-resistant tuberculosis in India: a review. Indian J Med Res. 2012;136:599–604. [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee R, Allen J, Westenhouse J, Oh P, Elms W, Desmond E, et al. Extensively drug-resistant tuberculosis in California, 1993-2006. Clin Infect Dis. 2008;47:450–7. doi: 10.1086/590009. [DOI] [PubMed] [Google Scholar]
- 7.Jain A, Dixit P, Prasad R. Pre-XDR & XDR in MDR and ofloxacin and kanamycin resistance in non-MDR M. tuberculosis isolates. Tuberculosis. 2012;92:404–6. doi: 10.1016/j.tube.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Agonafir M, Lemma E, Wolde-Meskel D, Goshu S, Santhanam A, Girmachew F, et al. Phenotypic and genotypic analysis of multidrug-resistant tuberculosis in Ethiopia. Int J Tuberc Lung Dis. 2010;14:1259–65. [PubMed] [Google Scholar]
- 9.Manual for laboratory technician. New Delhi: Central TB Division, Directorate General of Health Services, Ministry of Health and Family Welfare. New Delhi, India; [accessed on February 2, 2014]. Revised National Tuberculosis Control Programme (RNTCP) Available from: http://www.tbcindia.nic.in/pdfs/Module%20for%20Laboratory%20Technician.pdf . [Google Scholar]
- 10.Revised National Tuberculosis Control Programme. Training manual for Mycobacterium tuberculosis culture & drug susceptibility testing, April 2009, Central TB Division, Directorate General of Health Services, Ministry of Health and Family Welfare, New Delhi, India. [accessed on February 2, 2014]. Available from: http://tbcindia.nic.in/pdfs/Training%20manual%20M%20tuberculosis%20C%20DST.pdf .
- 11.Geneva, Switzerland: WHO; 2001. [accessed on February 2, 2014]. World Health Organization. Guidelines for drug susceptibility testing for second-line anti-tuberculosis drugs for DOTS-plus. WHO/CDS/TB/2001.288. Available from: http://whqlib.doc.who.int/hq/2001/WHO_CDS_TB_2001288.pdf . [Google Scholar]
- 12.Tuberculosis Research Centre (now National Institute for Research in Tuberculosis), Chennai. Standard operating procedure for mycobacteriology laboratory. November 2010. Chennai (India) [accessed on February 2, 2014]. Available from: http://www.nirt.res.in/pdf/bact/SOP.pdf .
- 13.Aslan G, Tezcan S, Serin MS, Emekdas G. Genotypic anaylysis of isoniazid and rifampin resistance in drug-resistant clinical Mycobacterium tuberculosis complex isolates in Southern Turkey. Jpn J Infect Dis. 2008;61:255–60. [PubMed] [Google Scholar]
- 14.Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, et al. High-resolution minisatellite based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA. 2001;98:1901–6. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human mini-satellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–77. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 16.Weniger TH, Krawczyk J, Supply PH, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:1–6. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabyonga L, Kateete DP, Katabazi FA, Odong PR, Whalen CC, Dickman KR, et al. Determination of circulating Mycobacterium tuberculosis strains and transmission patterns 346 INDIAN J MED RES, MARCH 2016among pulmonary TB patients in Kawempe municipality, Uganda, using MIRU-VNTR. BMC Res Notes. 2011;4:280. doi: 10.1186/1756-0500-4-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun YJ, Bellamy R, Lee AS, Ng ST, Ravindran S, Wong SY, et al. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J Clin Microbiol. 2004;42:1986–93. doi: 10.1128/JCM.42.5.1986-1993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozińska M, Brzostek A, Krawiecka D, Rybczyńska M, Zwolska Z, Augustynowicz-Kopeć E. MDR, pre-XDR and XDR drug-resistant tuberculosis in Poland in 2000-2009. Pneumonol Alergol Pol. 2011;79:278–87. [PubMed] [Google Scholar]
- 21.Qi YC, Ma MJ, Li DJ, Chen MJ, Lu QB, Xiu-Jun Li, et al. Multidrug-resistant and extensively drug-resistant tuberculosis in multi-ethnic region, Xinjiang Uygur Autonomous Region, China. PLoS One. 2012;7:e32103. doi: 10.1371/journal.pone.0032103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, Li X, Xu P, Shen X, Gui X, Wang L, et al. Transmission of MDR and XDR tuberculosis in Shanghai, China. PLoS One. 2009;4:e4370. doi: 10.1371/journal.pone.0004370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimaldo ER, Tupasi TE, Rivera AB, Quelapio MI, Cardano RC, Derilo JO, et al. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant Mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int J Tuberc Lung Dis. 2001;5:546–50. [PubMed] [Google Scholar]
- 24.Olusoji D, Eltayeb O, Olanrewaju O, Olapade GD. Pre-extensive drug resistant tuberculosis (Pre-XDR-TB) among MDR-TB patents in Nigeria. Global Adv Res J Microbiol. 2013;2:22–5. [Google Scholar]
- 25.Sharma SK, George N, Kadhiravan T, Saha PK, Mishra HK, Hanif M. Prevalence of extensively drug-resistant tuberculosis among patients with multidrug-resistant tuberculosis: a retrospective hospital-based study. Indian J Med Res. 2009;130:392–5. [PubMed] [Google Scholar]
- 26.Singh S, Sankar MM, Gopinath K. High rate of extensively drug-resistant tuberculosis in Indian AIDS patients. AIDS. 2007;21:2345–7. doi: 10.1097/QAD.0b013e3282f125c9. [DOI] [PubMed] [Google Scholar]
- 27.Jain A, Mondal R, Prasad R, Singh K, Ahuja RC. Prevalence of multidrug resistant Mycobacterium tuberculosis in Lucknow, Uttar Pradesh. Indian J Med Res. 2008;128:300–6. [PubMed] [Google Scholar]
- 28.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murase Y, Mitari S, Sugawara I, Kato S, Maeda S. Promising loci of variable number of tandem repeats for typing Beijing family Mycobacterium tuberculosis. J Med Microbiol. 2008;57:873–80. doi: 10.1099/jmm.0.47564-0. [DOI] [PubMed] [Google Scholar]
- 30.Dong H, Shi L, Zhao X, BaSang, Lv B, Xiaomin Yang, et al. Genetic diversity of Mycobacterium tuberculosis isolates from Tibetans in Tibet, China. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0033904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou A, Nawaz M, Xue X, Karakousis PC, Yao Y, Xu J. Molecular genotyping of Mycobacterium tuberculosis in Xi’an, China using MIRU-VNTR typing. Int J Tuberc Lung Dis. 2011;15:517–22. doi: 10.5588/ijtld.10.0495. [DOI] [PubMed] [Google Scholar]
- 32.Arora J, Bhalla M, Sidiq Z, Lal P, Behera D, Rastogi N, et al. Predominance of Beijing genotype in extensively drug resistant Mycobacterium tuberculosis isolates from a tertiary care hospital in New Delhi, India. Int J Mycobacteriol. 2013;2:109–13. doi: 10.1016/j.ijmyco.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Purwar S, Chaudhari S, Katoch VM, Sampath A, Sharma P, Upadhyay P, et al. Determination of drug susceptibility patterns and genotypes of Mycobacterium tuberculosis isolates from Kanpur district, North India. Infect Genet Evol. 2011;11:469–75. doi: 10.1016/j.meegid.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Glynn JR, Whiteley J, Bifani PJ, Kremer K, Van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002;8:843–9. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhanu NV, van Soolingen D, van Embden JD, Dar L, Pandey RM, Seth P. Predominace of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinb) 2002;82:105–12. doi: 10.1054/tube.2002.0332. [DOI] [PubMed] [Google Scholar]
- 36.Singh UB, Suresh N, Bhanu NV, Arora J, Pant H, Sinha S, et al. Predominant tuberculosis spoligotypes, Delhi, India. Emerg Infect Dis. 2004;10:1138–42. doi: 10.3201/eid1006.030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph B BV, Soman S, Radhakrishnan I, Hill V, Dhanasooraj D, Kumar RA, et al. Molecular epidemiology of Mycobacterium tuberculosis isolates from Kerala, India using IS6110-RFLP, spoligotyping and MIRU-VNTRs. Infect Genet Evol. 2013;16:157–64. doi: 10.1016/j.meegid.2013.01.012. [DOI] [PubMed] [Google Scholar]


