Abstract
EFHC1 encodes a ciliary protein that has been linked to Juvenile Myoclonic Epilepsy. In ectodermal explants, derived from Xenopus laevis embryos, the morpholino-mediated down-regulation of EFHC1b inhibited multiciliated cell formation. In those ciliated cells that did form, axoneme but not basal body formation was inhibited. EFHC1b morphant embryos displayed defects in central nervous system (CNS) and neural crest patterning that were rescued by a EFHC1b-GFP chimera. EFHC1b-GFP localized to ciliary axonemes in epidermal, gastrocoele roof plate, and neural tube cells. In X. laevis there is a link between Wnt signaling and multiciliated cell formation. While down-regulation of EFHC1b led to a ~2-fold increase in the activity of the β-catenin/Wnt-responsive TOPFLASH reporter, EFHC1b-GFP did not inhibit β-catenin activation of TOPFLASH. Wnt8a RNA levels were increased in EFHC1b morphant ectodermal explants and intact embryos, analyzed prior to the on-set of ciliogenesis. Rescue of the EFHC1b MO’s ciliary axonemal phenotypes required the entire protein; in contrast, the EFHC1b morpholino’s Wnt8a, CNS, and neural crest phenotypes were rescued by a truncated form of EFHC1b. The EFHC1b morpholino’s Wnt8a phenotype was also rescued by the injection of RNAs encoding secreted Wnt inhibitors, suggesting that these phenotypes are due to effects on Wnt signaling, rather than the loss of cilia, an observation of potential relevance to understanding EFHC1’s role in human neural development.
Keywords: Ciliogenesis, Wnt signaling, Xenopus embryo, Myoclonic epilepsy
Introduction
Cilia are complex intracellular organelles involved in cellular motility, inter- and intracellular signaling, and the compartmentalization of cells, as in the case of photoreceptor cells. Mutations in a number of genes associated with cilia formation and function have been linked to human diseases, known collectively as ciliopathies (Brown and Witman, 2014; Ferkol and Leigh, 2012; Hildebrandt et al., 2011). Interpreting the pathological mechanisms of such mutations is complicated by the fact that a number of proteins associated with cilia have other, quite distinct functions. For example Chibby (Cby), a protein associated with the basal body region of cilia also acts as a cytoplasmic regulator of β-catenin-mediated intercellular Wnt signaling (Shi et al., 2014; Takemaru et al., 2003) while Cetn2, another basal body region associated protein, is an integral component of the DNA damage repair complex (Araki et al., 2001) and appears to play a role in the regulation of gene expression (Cunningham et al., 2014; Shi et al., 2015). It is therefore necessary to identify the range of possible functions a protein might have in order to consider its various roles in normal development and tissue function, as well as the pathological effects of variant alleles.
Here we consider the roles of one such ciliary component, EFHC1. EFHC proteins contain three conserved DM10 “domains of unknown function”. In EFHC1 there is a putative Ca2+ binding EF-hand domain in the C-terminal region. A paralogous gene, EFHC2, encodes a protein without a C-terminal EF hand domain. Conserved forms of EFHC proteins have been found in organisms with flagella, including Chlamydomonas (Rib72)(Ikeda et al., 2003), Tetrahymena (Kilburn et al., 2007), and throughout the vertebrates (Ikeda et al., 2005). Both EFHC1 and EFHC2 are reported to be associated with tektin filaments in Chlamydomonas cilia (Setter et al., 2006). Defhc1.1, a Drosophila homolog of EFHC1 has been reported to regulate microtubule stability at neuromuscular junctions (Rossetto et al., 2011); no function has yet been assigned to a second EFHC paralog. In vertebrates, EFHC1 is present in sperm flagella and tracheal cilia (Ikeda et al., 2005; Suzuki et al., 2008). After some confusion associated with antibody specificity, the expression of EFHC1 in the mouse central nervous system (CNS) appears to be primarily within the choroid plexus and ciliated ependymal cells (Yamakawa and Suzuki, 2013).
In humans, mutations in EFHC1 have been linked to Juvenile Myoclonic Epilepsy (JME; OMIM 254770)(Suzuki et al., 2004; von Podewils et al., 2015) while mutations in EFHC2 have been described and tentatively associated with JME (Gu et al., 2005) and neurological syndromes such as autism (Blaya et al., 2009; Weiss et al., 2007). Homozygous EFCH1 null mice are viable and fertile, but display increased brain ventricle volume (hydrocephalus) as well as a number of ciliopathy-associated phenotypes, including a seizure disorder. Cilia are still present in EFCH1 null mice but their motility appears to be reduced (Suzuki et al., 2009). As EFHC1 seems to be expressed in CNS and may be required during brain development, it is necessary to understand its role during vertebrate development to fully appreciate its roles in pathogenic processes. Here we describe our characterization of EFHC1 expression, protein localization, and the effects of down-regulating EFHC1 protein levels in the context of intact Xenopus laevis embryos and ectodermal explants. The results reveal an unexpected role for EFHC1 in the regulation of Wnt signaling and the identification of distinct domains of EFHC1 in ciliary localization and Wnt signaling regulation.
Results
Embryos and explants of the vertebrate tetrapod Xenopus laevis have proven to be excellent systems in which to study complex cellular processes and intercellular interactions, including ciliogenesis (Brooks and Wallingford, 2015; Werner and Mitchell, 2012). In our case, we have used ectodermal explants and intact embryos, as described previously (Shi et al., 2014), to characterize cilia-dependent and -independent functions of EFHC1. A BLAST search of the 9.1 version of the X. laevis genome reveals two EFHC1-related chromosomal scaffolds: the first (9.1: chr5L:151037124-151037414, E value: 2e−146) contains the EFHC1b gene while the second (9.1: chr5S:130244405-130244629, E value 7e−70) appears to contain the EFHC1a gene, but the coding region and intronexon structure of EFHC1a remain to be determined. Blast analysis indicates the morpholino designed to inhibit the translation of the EFHC1b RNA matches a second sequence in the X. laevis genome at 23 out of 25 positions (data not shown); this sequence is in the region of the putative EFHC1a gene and includes what appears to be a translation start site, and so may also inhibit translation of the EFHC1a RNA, although this has not been verified experimentally. Our studies focus on the EFHC1b gene.
In contrast to mouse, human, and T. thermophilia EFHC1 polypeptides, the X. laevis EFHC1b polypeptide does not appear to have an EF-hand motif in its C-terminal domain. This motif is also missing from the single EFHC1 gene found in the diploid species X. tropicalis (FIG. S1A). A BLAST-based search indicates that a number of other vertebrates have EFHC1-like proteins that are missing the C-terminal EF-hand motif, these include the zebrafish Danio rerio, the White-tailed eagle Haliaeetus albicilla, the Downy woodpecker Picoides pubescens, the American anole Anolis carolinensis, and the Burmese python Python bivittatus. We note that paralogous EFHC2 genes found in mouse, human, Tetrahymena, and both Xenopus species also lack an apparent C-terminal EF-hand motif. EFHC proteins appear to be present widely, but are apparently absent in C. elegans (King, 2006)(FIG. S1B).
EFHC1b expression and localization in X. laevis
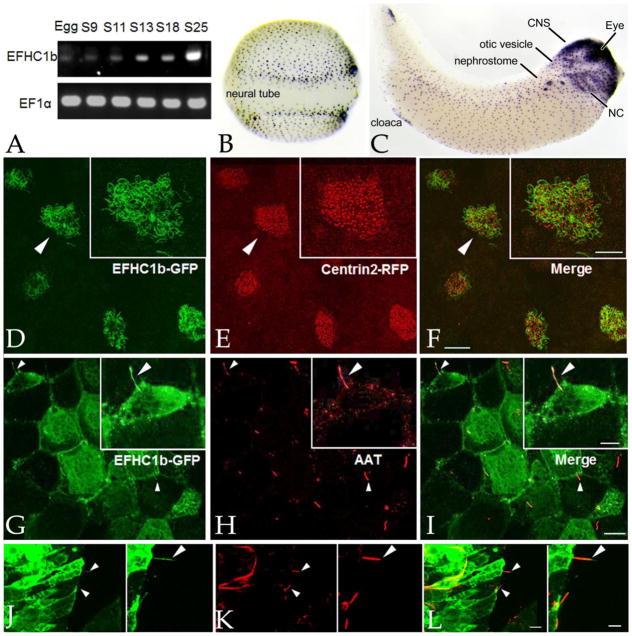
Our RT-PCR analyses confirms data from Yanai et al (Yanai et al., 2011), as accessed through XenBase (Karpinka et al., 2014). EFHC1b RNA is supplied maternally and its level increases dramatically at the onset of gastrulation (stage 10)(FIG. 1A). In situ hybridization studies reveal that EFHC1b RNA is present at high levels in the multiciliated cells (MCCs) of the embryonic epidermis of neurula stage (stage18) embryos (FIG. 1B); by tailbud stage (stage 33), EFHC1b is expressed in a variety of other tissues including the eye, otic vesicle, nephrostome, cloaca, neural crest derivatives, and anterior CNS (FIG. 1C).
Figure 1. EFHC1b is associated with various types of cilia.
A: An RT-PCR analysis during early development (stages indicated) revealed that EFHC1b RNA levels increase at the onset of gastrulation, confirming previous data from Yanai et al accessed through the Xenbase website. EF1α RNA was used as an internal control. In situ hybridization reveals the presence of EFHC1b RNA in the ciliated cells of the embryonic epidermis in stage 18 embryos (B) and in a number of different tissues in tailbud stage (stage 33) embryos (C)(CNS: central nervous system; NC: neural crest). D–F: RNAs encoding EFHC1b-GFP and Cetn2-RFP were injected into fertilized eggs and ectodermal explants were generated and analyzed when intact embryos reached stage 24; EFHC1b-GFP (D) was localized to the axonemes and excluded from the basal body regions of the cilia in multiciliated cells (E,F)(see figure 2). Scale bar of main image: 20μm; scale bar for inserts: 10 μm. G–I: The gastrocoele roof plate regions of EFHC1b-GFP RNA injected, stage 19 embryos were dissected and probed with antibodies against GFP (G) and acetylated-α-tubulin (AAT)(H,I-merged image). Individual GRP cilia are indicated by arrowheads. Scale bar of main image: 15μm; scale bar for inserts: 8μm. J–L: At stage 26, a section through the neural tube region of an EFHC1b-GFP RNA injected embryos were stained for EFHC1b-GFP (J) and AAT (K,L-merged image); arrowheads indicate primary cilia. Other AAT structures may be neuronal axons or radial glia. Scale bar of main image: 10μm; scale bar for inserts: 5μm.
In the green algae Chlamydomonas reinhardtii (Ikeda et al., 2005) and the ciliate T. thermaphila (Kilburn et al., 2007) orthologs of vertebrate EFHC1 localize to both the basal body and axonemal domains of cilia. Vertebrate EFHC1 has been reported to localize to the axonemes of sperm flagella, and the cilia of lung tracheal and CNS ependymal ventricle cells (Ikeda et al., 2005). Previous reports of EFHC1’s association with centrosomal and mitotic spindle microtubules have been questioned based on studies using EFHC1 null mice (Yamakawa and Suzuki, 2013). In the absence of a well-characterized antibody against X. laevis EFHC1b, we generated plasmids encoding a EFHC1b-GFP chimera for use in localization and morpholino-rescue studies; EFHC1b-GFP rescued all of the phenotypic effects associated with morpholino-mediated down regulation of EFHC1b (see below).
Ectodermal explants isolated from mid-blastula stage embryos rapidly (within 18 hours) differentiate to form epidermal tissue characterized by multiciliated cells surrounded by non-ciliated, mucus secreting cells and ionocytes (Dubaissi and Papalopulu, 2011). We generated ectodermal explants from fertilized eggs injected with EFHC1b-GFP RNA and analyzed them when control embryos reached stage 24. EFHC1b-GFP appeared to accumulate to higher concentrations in multiciliated cells as compared to the surrounding non-ciliated cells and was associated with the axonemal regions of cilia (FIG. 1D), but not apparently with the basal body regions, as visualized through the binding of centrin-2-RFP (Cetn2-RFP) expressed from injected RNA (FIG. 1E,F) or by staining with an anti-Xenopus Centrin (Cetn) antibody (FIG. 2H–H”). Analysis of dissected regions and sections revealed that EFHC1b-GFP was associated with the motile cilia found on gastrocoel roof plate cells (FIG. 1G–I) and the non-motile primary cilia on cells of the ventral surface of the neural tube (FIG. 1J–L); in both cases there was a high level of cytoplasmic accumulation of the chimeric polypeptide. Exogenous EFHC1b-GFP was also seen at junctional regions between cells in the gastrocoel roof plate.
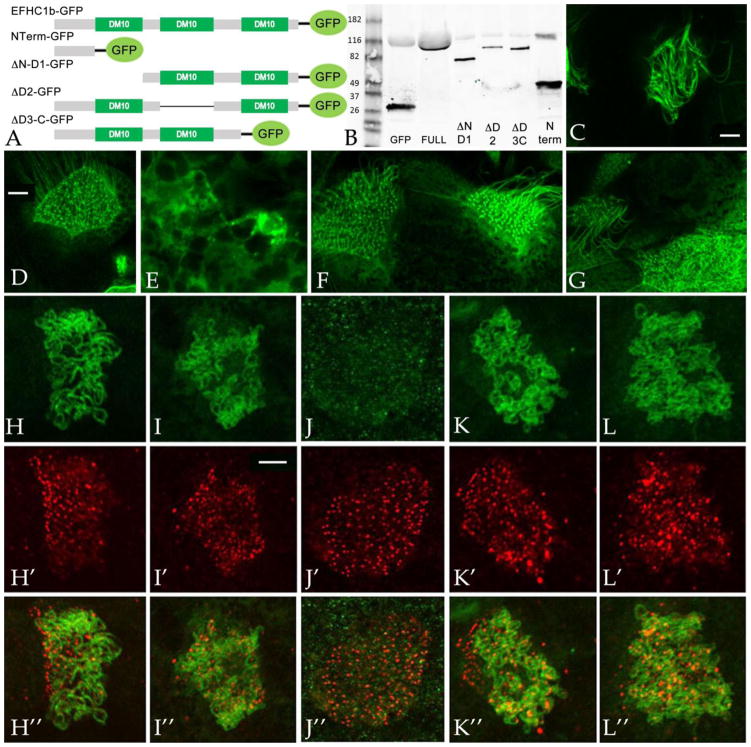
Figure 2. The N-terminus of EFHC1b is required for its axonemal localization.
A: Plasmids encoding GFP-tagged wild type and deleted forms of the EFHC1b were generated. B: Immunoblot analysis was carried out using an anti-GFP antibody on stage11 embryos injected with RNAs encoding these GFP-tagged polypeptides. C,H: Full length EFHC1b-GFP localizes to axonemes. NTerm-GFP (D,I), ΔD2-GFP (F,K) and ΔD3-C-GFP (G, L) were also associated with axonemes, although higher cytoplasmic levels were found compared to the full length protein. The ΔN-D1-GFP (E, J) polypeptide accumulates to detectable levels in every cell, in contrast to the other mutants and the full-length polypeptides, which appear to accumulate preferentially in ciliated cells. In double labeling studies, fertilized eggs were injected with RNAs encoding EFHC1b-GFP (H–H”), NTerm-GFP (I–I”), ΔN-D1-GFP (J–J”), ΔD2-GFP (K–K”), and ΔD3-C-GFP (L–L”); ectodermal explants were generated and analyzed when intact embryos reached stage 24. Basal bodies were visualized by anti-Xenopus centrin antibody staining (H’, I’, J’, K’, and L’; in no case was there any obvious overlap between the EFHC1 and centrin staining (overlap images in parts H”, I”, J”, K”, and L”). Scale bars: 5μm.
EFHC family proteins, both EFHC1 and EFHC2, contain three conserved DM10 “domains of unknown function”. DM10 domains have been identified in only two types of proteins: the nm23-H7 class of nucleoside disphosphate kinases and the EFHC proteins. To define the domains of EFHC1b that are necessary for ciliary localization and other functions, we generated GFP-tagged mutants that lack either the N-terminal domain and first DM10 domain (ΔN-D1-GFP, 199~552aa), the second DM10 domain (ΔD2-GFP, 1~238, 359~552aa), and the third DM10 domain together with the C-terminus (ΔD3-C-GFP, 1~416aa), as well as an N-terminal only construct (NTerm-GFP, 1~92aa)(FIG. 2A). RNAs encoding these polypeptides were injected into fertilized eggs and examined by immunoblot at stage 11 using an anti-GFP antibody; polypeptides of the expected size were found to accumulate (FIG. 2B). To monitor the interaction between these mutant polypeptides with cilia, ectodermal explants were isolated from RNA-injected embryos, allowed to develop to stage 24, when ciliogenesis is complete, and examined by confocal microscopy. The full length EFHC1b-GFP polypeptide was found localized to ciliary axonemes (FIG. 2C,H). Surprisingly, the NTerm-GFP polypeptide was also found associated with axonemes (FIG. 2D, I), as were the ΔD2-GFP (FIG. 2F,K) and the ΔD3-C-GFP (FIG. 2G, L) polypeptides. There appears to be a strong localization of basal bodies with the N-terminal construct while the ΔN-D1-GFP polypeptide was found in cytoplasmic aggregates (FIG. 2E, J). Compared to the full length EFHC1b-GFP, deleted polypeptides appear to accumulate to higher levels in the cytoplasm.
EFHC1b is required for cilia and ciliated cell formation
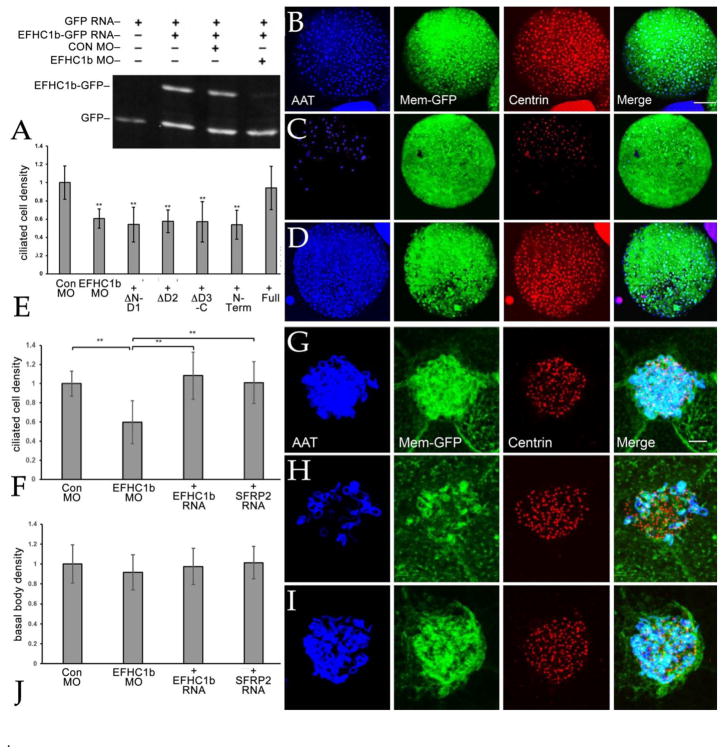
To define the functional roles of EFHC1b we had Gene Tools, LLC design and synthesize a translation blocking morpholino (MO). A BLAST search of the X. laevis genome revealed that the EFHC1b MO’s only other match (23 out of 25 positions) was a site within the likely EFHC1a gene (see above); whether EFHC1b MO inhibits the translation of EFHC1a RNA (or even if there is an expressed EFHC1a RNA) has yet to be determined. The plasmid that encodes EFHC1b-GFP contains an exact match to the sequence targeted by the EFHC1b MO and the co-injection of the EFHC1b MO with EFHC1b-GFP RNA led to a dramatic reduction in the accumulation of the EFHC1b-GFP polypeptide (FIG. 3A). To analyze the effect of down-regulating EFHC1b, fertilized eggs were injected with RNAs encoding a membrane-bound GFP together with either control or EFHC1b MOs; ectodermal explants were generated, fixed at stage 24, and stained with an anti-acetylated α-tubulin (AAT) antibody to mark axonemes and an anti-Cetn antibody to mark basal bodies. We used a semi-automated ImageJ script to quantify the numbers of ciliated cells in the injected regions of explants (Shi et al., 2014). The density of ciliated cells per explant was reduced in EFHC1b morphants (FIG. 3B–E) while the overall cell number remained the same (FIG. S3). We were surprised to find that the number of basal bodies, visualized by staining with an anti-Xenopus Cetn antibody, was unchanged while the numbers of axonemes was clearly reduced (FIG. 3G–J).
Figure 3. Depletion of EFHC1b results in deficient cilia and multiciliated cell formation.
A: The EFHC1b morpholino is efficient for EFHC1b depletion. Embryos were injected with RNAs encoding GFP (150 pgs/embryo) and EFHC1b-GFP match (100 pgs/side) and either control or EFHC1b MO (10 ngs/embryo) and analyzed at stage 11. EFHC1b MO reduced EFHC1b-GFP protein levels, as visualized using an anti-GFP antibody. To examine the effects of reducing EFHC1b levels on the number of ciliated cells (B–D)(scale bar: 100 μm), the basal body density (J), and the number of ciliary axonemes per cell (G–I) (scale bar: 5 μm), both blastomeres of two-cell embryos were injected with membrane-GFP RNA together with either control MO, EFHC1b MO, EFHC1b MO plus EFHC1b-GFP RNA, or EFHC1b MO plus SFRP2 RNA. Membrane-GFP was visualized using an anti-GFP antibody, while anti-acetylated α-tubulin and anti-centrin antibodies were used to visualized cilia and basal bodies, respectively. Quantitation of the EFHC1b morpholino's effect on the number of ciliated cells per cap (E, F) and the basal body density (J)(calculated by dividing the total basal body number per cell by the cell size) are shown as mean ± S.D. All the conditions were normalized to control MO (set as 1). N=20 to 25 explants per experimental condition. Significant differences between conditions are marked by horizontal bars (** for p<0.01)
EFHC1b plays a role in Wnt signaling
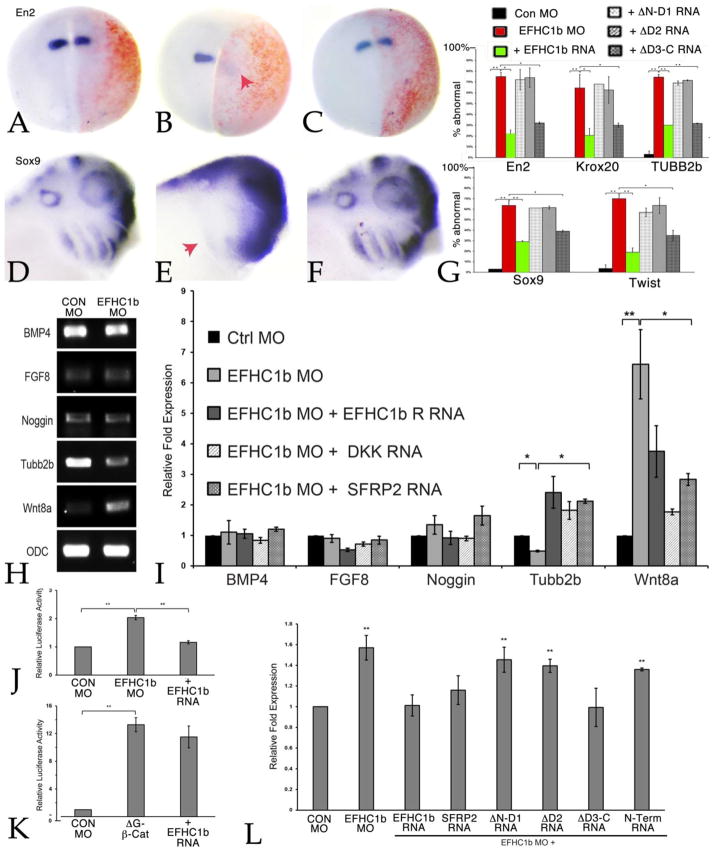
We examined EFHC1b MO effects in intact embryos by injecting one cell of two-cell embryos, together with either GFP or β-galactosidase RNAs as lineage markers. Embryos were sorted at stage 11 and fixed and examined at either stage 18 or 25 by in situ hybridization. EFHC1b morphants displayed defects in the expression of the CNS markers Krox20, Engrailed-2 (En2), and neural β-tubulin (Tubb2) (FIG.4A–C, G, FIG. S4A) and the neural crest markers Sox9 and Twist (FIG. 4D–F, G, FIG. S4B). We found dramatically reduced expression of all five genes tested in EFHC1 morphants. Overall morphology the EFHC1b morphant embryos was distorted (see FIG. S5).
Figure 4. EFHC1b plays a role in Wnt signaling.
In situ hybridization shows defects (red arrows) in central nervous system (En2) patterning (A, B) and neural crest (Sox9) migration (D, E) in EFHC1b morphants at stage 18 and stage 25, respectively. These phenotypes were rescued by co-injection of EFHC1b-GFP-rescue RNA (C, F). Embryos were injected with either control MO, EFHC1b MO, or EFHC1b MO together with EFHC1b-GFP RNA. All embryos (A–C) were injected with RNA encoding β-galactosidase (red) as a lineage tracer. The loss of neural patterning markers (En2, Krox20, Tubb2b) as well as neural crest markers (Sox9, Twist1) was also rescued by ΔD3-C-GFP (G). The percentage of embryos that were “abnormal”, that is displayed reduced marker gene expression, was calculated from two independent experiments (N=55 to 60 embryos per condition). H: RNA levels in control and EFHC1b morpholino explants were analyzed at stage 18 using RT-PCR; EFHC1b morphant explants displayed decreased levels of Tubb2 RNA and increased levels of Wnt8a RNA. Levels of BMP4, Noggin and FGF8 RNA were unchanged. I: qPCR analyses (N=3) of control and EFHC1b morphant explants co-injected with EFHC1b-GFP-rescue, Dkk1, or SRFP2 RNAs. Both EFHC1b-GFP-rescue and the two Wnt signaling inhibitors returned all RNAs to control levels. J, K: Embryos were injected with TOPFLASH and FOPFLASH (control) plasmid DNAs (100 pgs/embryo) together with ΔG-β-catenin RNA (100 pgs/embryo) either alone or together with GFP or EFHC1b-GFP (100 pgs/embryo) RNAs or control, EFHC1b MO (10 ngs/embryo) and EFHC1b MO with SFRP2 RNA. The Y-axis indicates the fold-increase relative to the control TOPFLASH/FOPFLASH value (set equal to 1, N=3). L: qPCR analyses (N=3) of control and EFHC1 morphant explants co-injected with EFHC1b-GFP-rescue, SRFP2, or EFHC1b mutant RNAs on Wnt8 RNA. Besides the EFHC1b-GFP and the Wnt inhibitor SFRP2, injection of ΔD3-C-GFP RNA (100 pgs/embryo) also returned the Wnt8 RNA to control levels. Significant differences between conditions are marked by horizontal bars; in each case, data are represented as mean ± SD, *for p<0.05 and ** for p<0.01.
When analyzing EFHC1 morphants we noted the loss of Tubb2b expression in neural ectoderm, as monitored by in situ hybridization (Fig. S3A) and RT-PCR (see below). This led us to explore changes in gene expression; Wnt8a, BMP4, FGF8 and Noggin RNA levels were examined by both standard and quantitative RT-PCR. The result revealed an increase in Wnt8a RNA and a decrease in Tubb2 RNA levels in ectodermal explants; in contrast, BMP4 and Noggin RNAs were unaltered (FIG. 4H,I). Moreover, a small increase in Wnt8a RNA was observed in intact EFHC1b morphant embryos analyzed at stage 11 (FIG. 4L), well before cilia formation, which occurs around stage 17–18 (Chu and Klymkowsky, 1989; Steinman, 1968).
Basal body and cilia-associated proteins may function in developmental processes by regulating signaling pathways. Cby is a basal-body associated protein that acts as an inhibitor of β-catenin mediated Wnt signaling (Li et al., 2008; Takemaru et al., 2003; Takemaru et al., 2009). In X. laevis Wnt8a RNA levels are increased in Cby morphant gastrula stage embryos and there is a dramatic increase in the activity of the β-catenin-responsive TOPFLASH reporter (Shi et al., 2014). In EFHC1 morphants, we found an increase in TOPFLASH activity that was returned to control levels by EFHC1b RNA (FIG. 4J). In contrast to the situation with Cby (Shi et al., 2014), EFHC1b-GFP RNA did not inhibit the ability of a stabilized form of β-catenin (ΔG-β-catenin) to activate TOPFLASH (FIG. 4K).
ΔD3-C mutant rescues the Wnt signaling related phenotype
We found that all EFHC1b MO phenotypes were rescued by the injection of EFHC1b-GFP-rescue RNA, in which the EFHC1b MO’s target sequence has been altered to minimize complementarity without changing the final polypeptide’s sequence (FIG. 3D,F,I; FIG.4 G,I,L). In contrast, none of the EFHC1b MO’s effects on ciliated cell density or ciliary axoneme formation (FIG. 3E) were rescued by any of the deletion constructs. In contrast, EFHC1b MO’s effects on CNS patterning, neural crest markers, and Wnt8a RNA levels were rescued by ΔD3-C-GFP (FIG. 4G, L).
Autocrine-paracrine effects of EFHC1b loss
The increased level of Wnt8a RNA observed in EFHC1b morphant explants and embryos and the ability of ΔD3-C-GFP to rescue this increase, as well as the EFHC1b MO’s CNS and neural crest phenotypes suggests that these later stage phenotypes are due at least in part to effects on Wnt signaling rather than effects on cilia. To test this hypothesis, we co-injected EFHC1b morphant embryos with RNAs encoding the secreted Wnt inhibitors SFRP2 (Bradley et al., 2000) or Dickkopf (Dkk) (Glinka et al., 1998); both inhibited the changes in Wnt8a and Tubb2 RNA levels observed in EFHC1b morphant ectodermal explants (FIG. 4I) as well as the change in Wnt8a RNA levels observed in EFHC1b early gastrula stage embryos (FIG. 4L). SFRP2 rescued the reduction in ciliated cell density (FIG. 3F), but not the reduction in axonemes per ciliated cells observed in EFHC1b morphant embryonic explants (FIG. S2). We have not yet been able to define the exact stage where the development (and ectodermal insertion) of ectodermal multiciliated cells is arrested in EFHC1b morphant explants.
Discussion
While mutations in EFHC1 have been implicated in JME (de Nijs et al., 2013), it remains unclear exactly what cellular mechanisms are involved. The situation is further complicated by the observation that JME-associated mutations in EFHC1 have been found in apparently normal individuals (Bai et al., 2009; Pal and Helbig, 2015; Subaran et al., 2015) and different EFHC1 alleles appear to act through distinctly different mechanisms (Stogmann et al., 2006). While clearly a component of cilia (Ikeda et al., 2005), EFHC1 has been reported to associate with non-ciliary proteins, including the redox-sensitive TRPM2 channel (Katano et al., 2012) and two transcription factors, the β-catenin-binding HMG box protein TCF4 and the Zinc finger transcription factor ZBED1 (Sahni et al., 2015). Given a recent report of TRPM2 induced amyloid-β-induced neurovascular dysfunction (Park et al., 2014), it is possible that EFHC1-TRPM2 interactions may alter brain development and function. Mutations in EFHC1 have also been reported to lead to increased apoptosis (Katano et al., 2012) through an as yet undefined mechanism.
Our observations indicate that in X. laevis EFHC1b is necessary for axoneme, but not basal body formation in ectodermal explants. This axonemal effect appears to be structural, requires the intact polypeptide, and cannot be rescued by inhibiting the EFHC1b MO’s effects on Wnt signaling. This is in contrast to the phenotypes observed in EFHC1−/− mice (Suzuki et al., 2009) and Tetrahymena (manuscript in preparation) which have cilia, although in mice (but not Tetrahymena) cilia beat frequency has been found to be reduced. Whether these differences are due to complementary effects of EFHC2 or other ciliary proteins remains to be resolved. In this light, it is interesting to consider a recent paper that found that differences between mutant and morphant phenotypes can arise from genetic compensation effects in mutants (Rossi et al., 2015).
What is new and significant in our findings is the observation that reducing EFHC1b levels alters β-catenin-mediated Wnt signaling, as monitored by the activation of the TOPFLASH reporter, and leads to an increase in Wnt8a RNA, and that some aspects of the EFHC1 morpholino phenotype can be rescued by the expression of extracellular inhibitors of Wnt signaling. This effect is distinct from that described previously for the basal body associated protein Cby (Shi et al., 2014). How the absence of EFHC1 influences Wnt8a RNA levels is unclear and is the subject of on-going studies. It is tempting to suggest, based on the reported interaction between EFHC1 and the transcription factor TCF4 (Sahni et al., 2015), that the effect is direct but this remains to be demonstrated, but interactions between cilia and Wnt signaling have been found in a number of contexts (Wallingford and Mitchell, 2011). Effects on other cellular processes, such as the disruption of signaling systems associated with the inhibition of proteosome activity, described for other ciliopathy-associated mutations (Liu et al., 2014), could impact Wnt signaling. Gerdes et al (Gerdes et al., 2007) found that disruption of basal bodies led to the stabilization of β-catenin and to an increase in Wnt signaling. Basal bodies appear to be intact in EFHC1b morphants. That said, it is clear that early embryonic effects, arising from the absence of EFHC1b, lead to effects on later embryonic stages in X. laevis, specifically the patterning of the CNS and the formation of the neural crest. Such effects, if they occur in response to EFHC1 mutations during human development, could produce subtle and pathogenically significant effects on brain organization and function.
Materials and methods
Embryos, their manipulation and analysis
X. laevis embryos were injected, explants were generated, and staged following standard procedures (Nieuwkoop and Faber, 1967; Shi et al., 2011; Shi et al., 2014; Sive et al., 2000). At the two-cell stage, embryo injections were directed equatorially; for explant studies injections were targeted to the animal hemisphere. Animals were held in IACUC approved facilities. As an injection tracer, we routinely included RNAs (150 pgs/embryo) encoding β-galactosidase, green fluorescent protein (GFP) or GFP-CAAX, a membrane-associated form of GFP. In the case of GFP/GFP-CAAX RNA injections, embryos were examined at stage 10–11 by fluorescent microscopy to confirm the accuracy of injection. RNA isolation, RT-PCR and qPCR analyses were carried out as described previously (Shi et al., 2014; Zhang et al., 2003; Zhang et al., 2006). cDNA synthesis was performed using 1 μg purified RNA and a Verso cDNA kit (Thermo Scientific) following manufacturer’s directions. Real-time (quantitative) PCR was carried out using a Mastercycler Epgradient Realplex device (Eppendorf). PCR reactions were set up using DyNAmo SYBR Green qPCR kits (Finnzymes). Each sample was normalized to the expression level of ornithine decarboxylase (ODC) or elongation factor 1-alpha (EF1α). The cycling conditions used were: 95°C for 5 minutes; then 40 cycles of 95°C for 15 seconds, 56°C for 15 seconds, 60°C for 30 seconds. The ΔΔCT method was used to calculate real-time PCR results. The primers used for RT-PCR analysis were EF1α: [U 5'-TGG TGA CAG CAA GAA TGA CC-3' D 5'-AAC TTG CAA GCA ATG TGA GC-3']; ODC: [U 5'-CAG CTA GCT GTG GTG TGG-3' D 5'-CAA CAT GGA AAC TCA CAC-3']; Wnt8a [U 5'-TGA TGC CTT CAC TTC TGT GG-3' D 5'-TCC TGC AGC TTC TTC TCT CC -3']; BMP4 [U 5'-TGG TGG ATT AGT CTC GTG TCC -3' D 5'-TCA ACC TCA GCA GCA TTC C -3']; Noggin [U 5'-AGT TGC AGA TGT GGC TCT -3' D 5'-AGT CCA AGA GTC TCA GCA -3’]; FGF8 [U 5’-TGG TGA CCG ACC AAC TAA GC D 5’-CGA TTA ACT TGG CGT GTG g-3’]; Tubb2 [U 5’-CCA GGC TTT GCC CCA TTA AC D 5’-GCT ACT GTG AGG TAG CGT CC].
Morpholinos and plasmids
The EFHC1b MO was designed and synthesized by Gene Tools, Inc. The control morpholino used has been described previously (Zhang and Klymkowsky, 2009). For ectodermal explant experiments 20 ngs/embryo of morpholino was injected per embryo; for whole embryo experiments 10 ngs/embryo of morpholino was injected into either one of two cells, at the two-cell stage, or 10 ngs/blastomere was injected into both cells of a two-cell embryo. We generated plasmids that encode EFHC1b-GFP chimeras that either matched (pCS2-EFHC1b-GFP-match) or maximally mismatched (pCS2-EFHC1b-GFP-rescue) the EFHC1b MO. Both plasmids were sequenced to confirm the absence of mutations introduced in the course of their construction. Other plasmids used include those that encode membrane-bound GFP, GFP-CAAX, supplied by Kristen Kwan (U. Utah), ΔG-β-catenin (Merriam et al., 1997), Dickkopf-1 (Glinka et al., 1998), SFRP2 (Bradley et al., 2000), and RFP-tagged centrin plasmids from Sergie Sokol (Mt. Sinai School of Medicine) and John Wallingford (U. Texas). 200 pgs/embryo of RNA was injected for most of the experiments otherwise stated in the text. The details of the TOPFLASH/FOPFLASH assay have been described previously (Zhang et al., 2006). Statistical analyses were based on values expressed as mean ± standard deviation. All conditions were normalized to control MO, which was set to 1. Comparisons between EFHC1 MO alone or together with rescue RNA versus control MO were analyzed by unpaired student’s t-test. p <0.05 was considered as significant in all analyses.
In situ hybridization studies and immunoblot
Anti-sense probes for EFHC1b, Sox9, Twist1, Krox20, En2, and Tubb2 RNAs were synthesized using digoxigenin-UTP following standard methods. For in situ hybridization studies, embryos were co-injected with β-galactosidase RNA (50 pg/embryo); β-galactosidase activity was visualized in fixed embryos using a brief Red-Gal (Research Organics) reaction. In situ hybridization images were captured using a Nikon D5000 camera on a Wild microscope. Immunoblot analysis was carried out as described previously using a mouse anti-GFP antibody (Covance, Princeton, NJ)(for FIG. 2B) or a rabbit anti-GFP antibody (for FIG.3A). Secondary antibodies were anti-rabbit Alexa680 (Invitrogen) and anti-mouse IR800, which were detected on a LI-COR Odyssey infrared imager (LI-COR, Lincoln, NE).
Immunocytochemistry and confocal microscopy
Whole-mount immunocytochemistry was carried out as described by Dent et al (Dent et al., 1989). Gastrocoele and neural tube cilia were visualized as described in Shi et al (Shi et al., 2014). Images were collected using a Zeiss 510 Confocal Laser Scanning Microscope. The mouse monoclonal anti-acetylated α-tubulin antibody 6-B-11 (Sigma) was used to visualized ciliated cells (Chu and Klymkowsky, 1989). Rabbit anti-Xenopus centrin antibody was supplied by Sergei Sokol (Kim et al., 2012); a chicken anti-GFP antibody was purchased from Immunology Consultants Lab, Inc. Quantitative confocal microscopy was carried using a customized ImageJ script written by Domenico Galati, as described previously (Shi et al., 2014).
Supplementary Material
A: Schematic showing the protein domains and motifs within EFHC1 and EFHC2. B: A phylogenetic tree of EFHC1 and EFHC2 protein families across different species. The tree was built by using Phylogeny.fr [S1].
Embryos were injected with membrane-GFP RNA together with either control MO, EFHC1b MO plus SFRP2 RNA. Membrane-GFP was visualized using an anti-GFP antibody, while anti-acetylated α-tubulin and anti-centrin antibodies were used to visualized cilia and basal bodies, respectively. Defect in cilia formation on individual multiciliated cells was not rescued by co-injection of SFRP2 RNA. Scale bar: 5μm.
Embryos were injected with control MO, EFHC1b MO, and EFHC1b MO plus EFHC1b RNA, in each case using a membrane-GFP as a tracer and cell boundary marker. For each explant, three 100μm x 100μm regions were examined to determine the overall cell number. Relative total cell numbers are shown as mean ± S.D. All the conditions were normalized to control MO (set as 1). N=20~25 per experimental condition.
In situ hybridization shows reduced expression (red arrows) of the CNS markers Krox20 and Tubb2b (A) and the neural crest marker Twist (B) in EFHC1b morphants at stage 18 and stage 25, respectively. These phenotypes were rescued by co-injection of EFHC1b-GFP-rescue RNA. Embryos were injected with either control MO, EFHC1b MO, or EFHC1b MO together with EFHC1b-GFP RNA. All embryos were injected with RNA encoding β-galactosidase RNA (red) as a lineage tracer.
Both blastomeres of two-cell embryos were injected with either 20 ngs/ embryo total (A) or EFHC1 morpholino (B); embryos that passed successfully through gastrulation were fixed at stage 35. Compared to uninjected (not shown) or control MO injected embryos (A), EFHC1 MO injected embryos typically displayed a noticeable kink and were visibly smaller (B).
Significance statement.
EFHC1 is a highly conserved ciliary protein. In humans mutations in EFHC1 have been linked to Juvenile Myoclonic Epilepsy. Using embryos and ectodermal explants from the clawed frog X. laevis, we find that down-regulation of EFHC1 leads to up-regulation of intracellular Wnt signaling. Blocking Wnt signaling rescues central nervous system defects associated with EFHC1 down-regulation. Defects in Wnt signaling could play a role in producing the defects observed in the human condition.
Highlights.
In Xenopus, EHFC1 localizes to the axonemal region of cilia.
The N-terminal domain of EHFC1 appears to be necessary and sufficient for axonemal localization
Reduction of EFHC1 levels leads to the up-regulation of Wnt RNA levels and signaling.
Wnt effects in EFHC1 morphants lead to central nervous system and neural crest defects
Acknowledgments
We thank Sergei Sokol for anti-centrin antibody, and Sergei Sokol and John Wallingford for RFP-tagged centrin plasmids. The work was supported primarily through a collaborative supplement to the NIH 3R01GM074746 - 06W1 grant to Mark Winey and Mike Klymkowsky.
Footnotes
Author contributions
Together YZ, JS, and MWK designed the Xenopus studies, which were carried out primarily by YZ and JS; YZ analyzed the original experimental data; YZ and MWK wrote the manuscript; all authors were involved in overall final data analysis and manuscript preparation.
Competing Interests: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- Bai D, Bailey JN, Duron RM, Alonso ME, Medina MT, Martinez-Juarez IE, Suzuki T, Machado-Salas J, Ramos-Ramirez R, Tanaka M, et al. DNA variants in coding region of EFHC1: SNPs do not associate with juvenile myoclonic epilepsy. Epilepsia. 2009;50:1184–1190. doi: 10.1111/j.1528-1167.2008.01762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaya C, Moorjani P, Salum GA, Gonçalves L, Weiss LA, Leistner-Segal S, Manfro GG, Smoller JW. Preliminary evidence of association between EFHC2, a gene implicated in fear recognition, and harm avoidance. Neuroscience letters. 2009;452:84–86. doi: 10.1016/j.neulet.2009.01.036. [DOI] [PubMed] [Google Scholar]
- Bradley L, Sun B, Collins-Racie L, LaVallie E, McCoy J, Sive H. Different activities of the frizzled-related proteins frzb2 and sizzled2 during Xenopus anteroposterior patterning. Dev Biol. 2000;227:118–132. doi: 10.1006/dbio.2000.9873. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. In vivo investigation of cilia structure and function using Xenopus. Methods in cell biology. 2015;127:131–159. doi: 10.1016/bs.mcb.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Witman G. Cilia and diseases. BioScience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DTW, Klymkowsky MW. The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Dev Biol. 1989;136:104–117. doi: 10.1016/0012-1606(89)90134-6. [DOI] [PubMed] [Google Scholar]
- Cunningham CN, Schmidt CA, Schramm NJ, Gaylord MR, Resendes KK. Human TREX2 components PCID2 and centrin 2, but not ENY2, have distinct functions in protein export and co-localize to the centrosome. Experimental cell research. 2014;320:209–218. doi: 10.1016/j.yexcr.2013.11.015. [DOI] [PubMed] [Google Scholar]
- de Nijs L, Wolkoff N, Grisar T, Lakaye B. Juvenile myoclonic epilepsy as a possible neurodevelopmental disease: Role of EFHC1 or Myoclonin1. Epilepsy & Behavior. 2013;28:S58–S60. doi: 10.1016/j.yebeh.2012.06.034. [DOI] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- Dubaissi E, Papalopulu N. Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Disease models & mechanisms. 2011;4:179–192. doi: 10.1242/dmm.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkol TW, Leigh MW. Ciliopathies: the central role of cilia in a spectrum of pediatric disorders. The Journal of pediatrics. 2012;160:366–371. doi: 10.1016/j.jpeds.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Liu Y, Zaghloul NA, Leitch CC, Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nature genetics. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Gu W, Sander TL, Heils A, Lenzen KP, Steinlein OK. A new EF-hand containing gene EFHC2 on Xp11. 4: Tentative evidence for association with juvenile myoclonic epilepsy. Epilepsy research. 2005;66:91–98. doi: 10.1016/j.eplepsyres.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. Journal of Biological Chemistry. 2003;278:7725–7734. doi: 10.1074/jbc.M210751200. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Ikeda K, Enomoto M, Park MK, Hirono M, Kamiya R. The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 2005;579:819–822. doi: 10.1016/j.febslet.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic acids research. 2014 doi: 10.1093/nar/gku956. gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katano M, Numata T, Aguan K, Hara Y, Kiyonaka S, Yamamoto S, Miki T, Sawamura S, Suzuki T, Yamakawa K, et al. The juvenile myoclonic epilepsy-related protein EFHC1 interacts with the redox-sensitive TRPM2 channel linked to cell death. Cell Calcium. 2012;51:179–185. doi: 10.1016/j.ceca.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KD, Lake BB, Haremaki T, Weinstein DC, Sokol SY. Rab11 regulates planar polarity and migratory behavior of multiciliated cells in Xenopus embryonic epidermis. Developmental Dynamics. 2012;241:1385–1395. doi: 10.1002/dvdy.23826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM. Axonemal protofilament ribbons, DM10 domains, and the link to juvenile myoclonic epilepsy. Cell motility and the cytoskeleton. 2006;63:245–253. doi: 10.1002/cm.20129. [DOI] [PubMed] [Google Scholar]
- Li FQ, Mofunanya A, Harris K, Takemaru KI. Chibby cooperates with 14-3-3 to regulate β-catenin subcellular distribution and signaling activity. The Journal of cell biology. 2008;181:1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Tsai IC, Morleo M, Oh EC, Leitch CC, Massa F, Lee BH, Parker DS, Finley D, Zaghloul NA, et al. Ciliopathy proteins regulate paracrine signaling by modulating proteasomal degradation of mediators. The Journal of clinical investigation. 2014;124:0–0. doi: 10.1172/JCI71898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam JM, Rubenstein AB, Klymkowsky MW. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the end of Metamorphosis. Amsterdam, New York: Amsterdam Publishing Company; 1967. republished in 1994 by Garland Publishing. [Google Scholar]
- Pal D, Helbig I. Commentary: Pathogenic EFHC1 mutations are tolerated in healthy individuals dependent on reported ancestry. Epilepsia. 2015 doi: 10.1111/epi.12906. [DOI] [PubMed] [Google Scholar]
- Park L, Wang G, Moore J, Girouard H, Zhou P, Anrather J, Iadecola C. The key role of transient receptor potential melastatin-2 channels in amyloid-β-induced neurovascular dysfunction. Nature communications. 2014:5. doi: 10.1038/ncomms6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto MG, Zanarella E, Orso G, Scorzeto M, Megighian A, Kumar V, Delgado-Escueta AV, Daga A. Defhc1.1, a homologue of the juvenile myoclonic gene EFHC1, modulates architecture and basal activity of the neuromuscular junction in Drosophila. Hum Mol Genet. 2011;20:4248–4257. doi: 10.1093/hmg/ddr352. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Sahni N, Yi S, Taipale M, Bass J, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter P, Malvey-Dorn E, Steffen W, Stephens R, Linck R. Tektin interactions and a model for molecular functions. Experimental cell research. 2006;312:2880–2896. doi: 10.1016/j.yexcr.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Shi J, Severson C, Yang J, Wedlich D, Klymkowsky MW. Snail2 controls BMP/Wnt mesodermal induction of neural crest. Development. 2011;138:3135–3145. doi: 10.1242/dev.064394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Galati N, Winey M, Klymkowsky MW. Chibby functions in Xenopus ciliary assembly, embryonic development, and the regulation of gene expression. Dev Biol. 2014;395:287–298. doi: 10.1016/j.ydbio.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhou Y, Vonderfecht T, Winey M, Klymkowsky MW. Centrin-2 (Cetn2) mediated regulation of FGF/FGFR gene expression in Xenopus. Scientific Reports. 2015;5:10283. doi: 10.1038/srep10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Steinman RM. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. American Journal of Anatomy. 1968;122:19–55. doi: 10.1002/aja.1001220103. [DOI] [PubMed] [Google Scholar]
- Stogmann E, Lichtner P, Baumgartner C, Bonelli S, Assem-Hilger E, Leutmezer F, Schmied M, Hotzy C, Strom TM, Meitinger T, et al. Idiopathic generalized epilepsy phenotypes associated with different EFHC1 mutations. Neurology. 2006;67:2029–2031. doi: 10.1212/01.wnl.0000250254.67042.1b. [DOI] [PubMed] [Google Scholar]
- Subaran R, Conte J, Stewart W, Greenberg D. Pathogenic EFHC1 mutations are tolerated in healthy individuals dependent on reported ancestry. Epilepsia. 2015 doi: 10.1111/epi.12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, Hara Y, Nishida M, Numata T, Medina MT, Takeuchi T, et al. Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet. 2004;36:842–849. doi: 10.1038/ng1393. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Inoue I, Yamagata T, Morita N, Furuichi T, Yamakawa K. Sequential expression of Efhc1/myoclonin1 in choroid plexus and ependymal cell cilia. Biochem Biophys Res Commun. 2008;367:226–233. doi: 10.1016/j.bbrc.2007.12.126. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyamoto H, Nakahari T, Inoue I, Suemoto T, Jiang B, Hirota Y, Itohara S, Saido TC, Tsumoto T, et al. Efhc1 deficiency causes spontaneous myoclonus and increased seizure susceptibility. Hum Mol Genet. 2009;18:1099–1109. doi: 10.1093/hmg/ddp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- Takemaru KI, Fischer V, Li FQ. Fine-tuning of nuclear β-catenin by Chibby and 14-3-3. Cell Cycle. 2009;8:210–213. doi: 10.4161/cc.8.2.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Podewils F, Kowoll V, Schroeder W, Geithner J, Wang Z, Gaida B, Bombach P, Kessler C, Felbor U, Runge U. Predictive value of EFHC1 variants for the long-term seizure outcome in juvenile myoclonic epilepsy. Epilepsy & Behavior. 2015;44:61–66. doi: 10.1016/j.yebeh.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes & development. 2011;25:201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Purcell S, Waggoner S, Lawrence K, Spektor D, Daly MJ, Sklar P, Skuse D. Identification of EFHC2 as a quantitative trait locus for fear recognition in Turner syndrome. Human molecular genetics. 2007;16:107–113. doi: 10.1093/hmg/ddl445. [DOI] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Understanding ciliated epithelia: the power of Xenopus. Genesis. 2012;50:176–185. doi: 10.1002/dvg.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K, Suzuki T. Re-evaluation of myoclonin1 immunosignals in neuron, mitotic spindle, and midbody—Nonspecific? Epilepsy & Behavior. 2013;28:S61–S62. doi: 10.1016/j.yebeh.2012.06.032. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zhang C, Carl TF, Trudeau ED, Simmet T, Klymkowsky MW. An NF-kappaB and slug regulatory loop active in early vertebrate mesoderm. PLoS ONE. 2006;1:e106. doi: 10.1371/journal.pone.0000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Klymkowsky MW. Unexpected functional redundancy between Twist and Slug (Snail2) and their feedback regulation of NF-kappaB via Nodal and Cerberus. Dev Biol. 2009;331:340–349. doi: 10.1016/j.ydbio.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Schematic showing the protein domains and motifs within EFHC1 and EFHC2. B: A phylogenetic tree of EFHC1 and EFHC2 protein families across different species. The tree was built by using Phylogeny.fr [S1].
Embryos were injected with membrane-GFP RNA together with either control MO, EFHC1b MO plus SFRP2 RNA. Membrane-GFP was visualized using an anti-GFP antibody, while anti-acetylated α-tubulin and anti-centrin antibodies were used to visualized cilia and basal bodies, respectively. Defect in cilia formation on individual multiciliated cells was not rescued by co-injection of SFRP2 RNA. Scale bar: 5μm.
Embryos were injected with control MO, EFHC1b MO, and EFHC1b MO plus EFHC1b RNA, in each case using a membrane-GFP as a tracer and cell boundary marker. For each explant, three 100μm x 100μm regions were examined to determine the overall cell number. Relative total cell numbers are shown as mean ± S.D. All the conditions were normalized to control MO (set as 1). N=20~25 per experimental condition.
In situ hybridization shows reduced expression (red arrows) of the CNS markers Krox20 and Tubb2b (A) and the neural crest marker Twist (B) in EFHC1b morphants at stage 18 and stage 25, respectively. These phenotypes were rescued by co-injection of EFHC1b-GFP-rescue RNA. Embryos were injected with either control MO, EFHC1b MO, or EFHC1b MO together with EFHC1b-GFP RNA. All embryos were injected with RNA encoding β-galactosidase RNA (red) as a lineage tracer.
Both blastomeres of two-cell embryos were injected with either 20 ngs/ embryo total (A) or EFHC1 morpholino (B); embryos that passed successfully through gastrulation were fixed at stage 35. Compared to uninjected (not shown) or control MO injected embryos (A), EFHC1 MO injected embryos typically displayed a noticeable kink and were visibly smaller (B).