Abstract
Aims: DNA-based carrier screening is a standard component of donor eligibility protocols practiced by U.S. sperm banks. Applicants who test positive for carrying a recessive disease mutation are typically disqualified. The aim of our study was to examine the utility of a range of screening panels adopted by the industry and the effectiveness of the screening paradigm in reducing a future child's risk of inheriting disease. Methods: A cohort of 27 donor applicants, who tested negative on an initial cystic fibrosis carrier test, was further screened with three expanded commercial carrier testing panels. These results were then compared to a systematic analysis of the applicants' DNA using next-generation sequencing (NGS) data. Results: The carrier panels detected serious pediatric disease mutations in one, four, or six donor applicants. Because each panel screens distinct regions of the genome, no single donor was uniformly identified as carrier positive by all three panels. In contrast, systematic NGS analysis identified all donors as carriers of one or more mutations associated with severe monogenic pediatric disease. These included 30 variants classified as “pathogenic” based on clinical observation and 66 with a high likelihood of causing gene dysfunction. Conclusion: Despite tremendous advances in variant identification, understanding, and analysis, the vast majority of disease-causing mutation combinations remain undetected by commercial carrier screening panels, which cover a narrow, and often distinct, subset of genes and mutations. The biological reality is that all donors and recipients carry serious recessive disease mutations. This challenges the utility of any screening protocol that anchors donor eligibility to carrier status. A more effective approach to reducing recessive disease risk would consider joint comprehensive analysis of both donor and recipient disease mutations. This type of high-resolution recessive disease risk analysis is now available and affordable, but industry practice must be modified to incorporate its use.
Introduction
Like all prospective parents, those who rely on donor sperm want to conceive healthy children. The donor selection process gives these parents the opportunity to minimize risk of recessive disease inheritance by avoiding donors who carry mutations that are genetically incompatible with the reproducing parent. Since recessive disease risk is highly specific to particular reproductive pairings, a high-risk donor for one recipient is likely to be a low-risk donor for most other recipients. As a result, avoidance of high-risk donors is a clinical strategy that is both optimal and necessarily personalized.
The opportunity to minimize a future child's risk of disease, and the value placed on it by the reproductive marketplace, is reflected in the marketing claims of commercial sperm banks, which emphasize the rigor of donor screening protocols. In addition to a three-generation family history analysis (a protocol whose primary utility is the surfacing of risk for dominant and X-linked conditions), all sperm banks perform some degree of carrier screening in their applicant selection process (Sims et al., 2010). Donor applicants who test positive for a disease-associated mutation are typically disqualified.
The extent of DNA-based genetic screening, however, varies widely among sperm banks. At the federal level, sperm donation is treated as a category of tissue transplantation, and donor eligibility protocols are designed to minimize health risks to the recipient, not the recipient's future child (Centola, 2010). Stepping in to fill this gap, the major societies representing health professionals in reproductive medicine (e.g., the American Society for Reproductive Medicine [ASRM], the American College of Medical Genetics and Genomics [ACMG], and the American College of Obstetricians and Gynecologists [ACOG]) have recognized the importance of carrier screening in preconception and prenatal care (Edwards et al., 2015). All recommend global donor screening to detect selected mutations in the CFTR gene, responsible for cystic fibrosis (CF), the most prevalent recessive disease among individuals with northern European ancestry. ACMG also recommends deletion screening of the SMN1 gene associated with spinal muscular atrophy (SMA), another disease prevalent in northern European populations, and most sperm banks comply (Sims et al., 2010; Landaburu et al., 2013).
Before the emergence of next-generation sequencing (NGS) as a clinical tool, the expansiveness of carrier screening was limited by both a lack of gene-specific knowledge and the high cost of commercial testing, which ranged from several hundred to several thousand dollars per gene. Today, the cost of sequencing hundreds of genes on a commercial NGS panel is less than a thousand dollars, undermining the economic rationale for restricting any screening protocol to select groups of people or genes.
In response to these technological advances, a number of sperm banks have expanded their global screening protocol with panels offered by companies such as Good Start Genetics (GSG), which covers 23 recessive disease genes (e.g., Xytex Cryo International, CryoGam Colorado, NW Cryobank, and Manhattan Cryobank), or Counsyl (e.g., MidWest Sperm Bank), which covers ∼100 genes (Lazarin et al., 2013; Perreault-Micale et al., 2015). The two largest sperm banks in the United States, California Cryobank and Fairfax Cryobank, still limit global donor screening by DNA analysis to just two genes (associated with CF and SMA), with expanded screening for donors who identify as members of a few select ethnic groups such as Ashkenazi Jewish, Irish, or French Canadian. However, this ethnicity-based paradigm is challenged by an increasingly multiethnic populace (Nazareth et al., 2015) and a growing body of evidence that many so-called ethnic diseases reach far beyond the boundaries of specific communities (Lazarin et al., 2013; Lim et al., 2015). Sperm banks do not generally assume any responsibility for genetic education or genetic testing of clients receiving donor sperm (Isley and Callum, 2013).
In this study, we report a comparative mutation analysis of 27 provisionally qualified sperm donors who had tested negative on an expanded CF-carrier screen. This purported CF-negative cohort was screened further with the GeneVu panel offered by GSG. We also performed proxy screens based on public records describing the targeted mutation panels commercialized by Counsyl and 23andMe. Finally, we performed a systematic NGS analysis of the complete coding regions of 479 well-characterized recessive pediatric disease genes. A comparative analysis of screening results demonstrated a rate of disease-causing variant detection that scaled with the level of variant coverage of each panel: the greater the number of genes tested and depth of the DNA analysis (i.e., number of targeted base pairs), the higher the number of carriers detected. Our systematic analysis of both clinical and molecular data demonstrates that every member of the donor cohort carries at least one clinically characterized and/or clinically relevant pediatric disease-associated genetic variant.
Materials and Methods
Subjects and specimen collection
Till 2014, the participating donor sperm bank used the CF-specific CFplus genetic test from Integrated Genetics, Inc. as the carrier-screening component of its donor qualification process. The CFplus screen targets 94 disease-causing mutations and fulfills the ACOG recommendation for carrier screening. Donor applicants who tested positive were eliminated from further consideration.
In 2014, the participating sperm bank replaced the CFplus test with the GeneVu carrier-screening panel offered by GSG. During a transitionary period, 27 provisionally qualified donors who had tested negative on the CF-specific CFplus carrier screen were rescreened with the GeneVu panel. These 27 twice-screened individuals make up the cohort of subjects used for this study. The cohort is an ethnically diverse group of self-selected donor applicants (Table 1). The participating sperm bank is representative of the larger donor bank community and does not recruit, include, or exclude applicants based on unusual metrics.
Table 1.
Donors in Cohort
| No. | Ancestrya | Panel resultsb | NGSc |
|---|---|---|---|
| 1 | Latin American | DHCR7:c.964-1G>C (CSY); IDUA:p.Q70* (CSY) | 8 |
| 2 | Latin American | HBA:g.-3.7 (GSG) | 8 |
| 3 | African American | 7 | |
| 4 | Latin American | 7 | |
| 5 | W. European | 7 | |
| 6 | E. European/W. European/S. European | GBA:p.N409S (GSG, CSY) | 6 |
| 7 | W. European/S. European | 6 | |
| 8 | W. European | 5 | |
| 9 | East Asian | 5 | |
| 10 | South Asian | SMN1:g.delExon7 (GSG) | 5 |
| 11 | East Asian | SMN1:g.delExon7 (GSG) | 5 |
| 12 | African American | 4 | |
| 13 | European/Native American | 4 | |
| 14 | S. European | 4 | |
| 15 | African American | HBA:g.-3.7 (GSG) | 3 |
| 16 | W. European | 3 | |
| 17 | S. European/E. European/W. European | BLM:p.M1T (GSG) | 3 |
| 18 | W. European/Latin American | 3 | |
| 19 | E. European | 3 | |
| 20 | Asiand | 3 | |
| 21 | W. European | ACADM:p.K333E (CSY, TAM) | 2 |
| 22 | W. European/Native American | 2 | |
| 23 | W. European/S. European | 2 | |
| 24 | W. European | PYGM:p.R50* (CSY) | 2 |
| 25 | S. European | 1 | |
| 26 | W. European/E. European | 1 | |
| 27 | Latin American | 1 |
Self-identified.
HGVS nomenclature.
Total number of DCV and HLGD variants detected by NGS.
Self-reported ancestry not available. Race/ethnicity taken from GSG report used as indication of ancestry.
CSY, Counsyl; DCV, defined clinical variants; GSG, Good Start Genetics; HLGD, high likelihood of gene dysfunction; NGS, next-generation sequencing; TAM, 23andMe.
All subjects provided written consent for genetic testing. The participating sperm bank provided deidentified saliva samples and subject data, including self-identified ethnicity. Anonymity was reinforced with the assignment of an unrelated identification number to each subject. We did not seek IRB approval because this investigation did not involve human subjects as defined by the Common Rule (45 CFR 46.101).
Next-generation sequencing
Purified DNA from saliva samples was obtained and processed as previously described (Larson et al., 2015). Sample enrichment and library preparation were achieved on the Illumina TruSight One (TSO) platform, which targets complete coding regions from 4813 genes having known clinical phenotypes (Illumina, 2016). Enriched libraries were sequenced on the MiSeq system according to the manufacturer's instructions (Illumina). Raw sequence files (FASTQ) were aligned to the GRCh37/hg19 human reference sequence with the Burrows-Wheeler Alignment tool (Li and Durbin, 2010). Variant calling, quality control, and local realignment were performed with the Genome Analysis Toolkit (GATK) version 3.2 (Van der Auwera, 2013).
Variant analysis was confined to a panel of 479 well-characterized TSO-covered genes associated with severe recessive monogenic pediatric disease (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/gtmb). The analysis panel includes genes curated by the Kingsmore group (Bell et al., 2011; Kingsmore, 2012) and additional recessive disease genes included on any of the commercial carrier screens examined in this study. Genes that did not generate consistent high-quality sequencing data for all canonical exons in all subjects were not included in our analytic panel. Analytic validation of all variant calls in all included genes was demonstrated with Phred quality scores of at least 30 (99.9% confidence) as determined with the GATK toolkit.
The comprehensive panel includes the 23 genes on the GSG GeneVu panel, the 98 genes on the Counsyl (CSY) standard Family Prep Screen (Lazarin et al., 2013), and the 35 genes on the 23andMe (TAM) Personal Genome Service. Genes on the X chromosome were excluded from all analyses.
Commercial carrier screening
The participating sperm bank provided deidentified GSG GeneVu carrier test reports for the CF-prescreened provisional donors. The GeneVu test uses NGS to detect variant genotypes at a targeted set of clinically defined chromosomal positions as well as novel protein truncation mutations in 19 autosomal genes with a relatively high incidence of recessive disease in individuals of Ashkenazi Jewish descent (Perreault-Micale et al., 2015). GeneVu also reports carrier status for three additional genetic diseases (alpha- and beta-thalassemia and SMA) tested with non-NGS methods. Six provisional donors were scored positive on the GeneVu test and were disqualified by the participating sperm bank.
Proxy carrier screening
We used the publicly reported targeted DNA variant content of the CSY Family Prep Screen 1.0 and the TAM Personal Genome Service to develop analytic tools that act as proxies for these genetic tests in conjunction with a subject's DNA sequence data. Variants included in each mutation panel (417 in the Family Prep Screen 1.0 and only 100 in the Personal Genome Service) were identified from the respective vendor's website (both accessed on November 10, 2015). Our proxies for these screens evaluate exactly the same disease-causing variants that are targeted by the original screens.
Two additional commercial panels were chosen for inclusion in our study based on widespread use or availability in their respective markets. The CSY Family Prep Screen represents the most common physician-ordered carrier test and the TAM Personal Genome Service is the only FDA-approved direct-to-consumer carrier test. These two tests have some overlap, with 33 genes and 84 variants common to both panels. The CSY and TAM panels also cover 19 and 12, respectively, of the same genes as the GSG test.
Systematic variant analysis
Our systematic analysis began with the identification of all variants in the 479 autosomal genes included in our panel (Supplementary Table S1). For the purpose of this study, we pursued a conservative approach in identifying variants with a very strong likelihood of causing gene dysfunction and, by extrapolation, recessive disease symptoms in individuals with two dysfunctional alleles. A clear limitation of such a stringent variant identification process is that an unknown number of disease-associated variants will likely be missed. We use the term “dysfunctional” throughout this article to signify variants that are predicted to be disease causing in homozygotes and/or compound heterozygotes.
Variants were processed through a series of clinical and analytic filters. We began with the assumption that individual variants present in greater than 5% of cohort members are very unlikely to cause disease in homozygous individuals. Based on this assumption, variants present in more than two donors were excluded. Remaining variants were cross-referenced to genome positions in the NIH-sponsored ClinVar database. ClinVar records contain clinical assertions from the published biomedical literature and submissions from genetic testing laboratories (Landrum et al., 2014). Each clinical assertion is assigned a ClinVar classification represented by an integer number, for example, 0: variant of uncertain significance, 2: benign, 3: likely benign, 4: likely pathogenic, and 5: pathogenic. Variants are also assigned a ClinGen star rating according to their review status. The ClinGen rating was not taken into consideration since only a minority of variants had been properly reviewed at this point in time (Rehm et al., 2015).
An individual variant can be assigned multiple classifications if multiple reports provide conflicting clinical assertions. Donor variants that appear in the ClinVar catalogue (November 2, 2015 release) with any benign, likely benign, or uncertain clinical assertion were excluded. Those with an unconflicted pathogenic or likely pathogenic classification comprise our “defined clinical variants” (DCV) list.
We next used three well-established computational models (PolyPhen2, PROVEAN, and CADD) to distinguish variants subject to strong negative selection during evolution. Based on our stringent approach to dysfunctional variant identification, we excluded all missense mutations (other than start codon changes) with a protein damage likelihood below 0.99 according to the PolyPhen2 protein modeling tool (Adzhubei et al., 2010). Missense mutations were further filtered by requiring a CADD score for deleteriousness above 99.9% of all potential genome variants (i.e., a Phred-scaled score of at least 30) (Kircher et al., 2014) and/or a PROVEAN score higher than 90% of all potential coding region variants (i.e., a score less than −4.1) (Choi et al., 2012). Missense mutations retained at the end of the filtering process were included in a non-ClinVar “high likelihood of gene dysfunction” (HLGD) group.
The remaining non-ClinVar nonmissense variants were evaluated for a predicted molecular effect on RNA splicing or polypeptide translation. Variants were added to the HLGD group if they (1) cause premature protein termination through a nonsense substitution or an out-of-frame indel, (2) fail to initiate translation through a modification of the methionine start codon, or (3) are very likely to interrupt splicing through a single base substitution in the first or second intronic base abutting an exon.
Finally, we used a previously described analysis of NGS read depth over SMN1, SMN2, and control genes to determine SMA carrier status of each donor (Larson et al., 2015). GSG reports of alpha-thalassemia carriers were confirmed through a comparison of NGS read depth at HBA1 and HBA2. All other variants evaluated by the GSG and proxy panels are included in our systematic sequence analysis.
Results
Commercial carrier screens are inconsistent in identifying donors with disease variants
Five independent carrier assessments were conducted for a selected set of sperm donor applicants (Materials and Methods section). In the first assessment, applicants were prescreened for CF-associated mutations and those who tested positive were excluded from further consideration by the participating donor bank. Twenty-seven applicants who tested negative were provisionally qualified as donors and were included as subjects in this study.
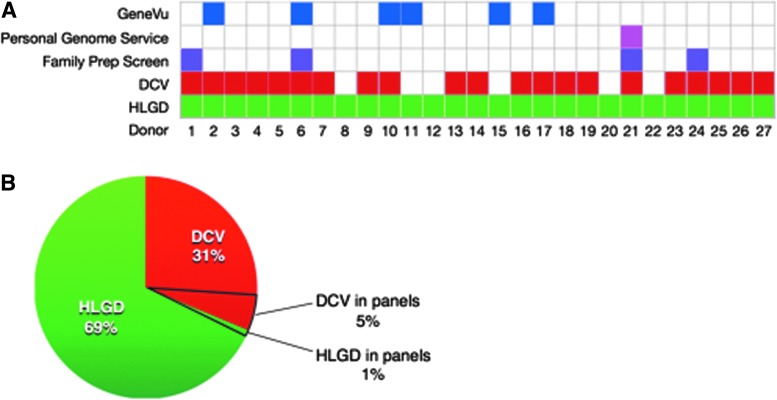
The second assessment was based on GeneVu test reports provided by GSG to the participating donor bank. These reports flagged six provisional donors as carriers (Fig. 1A and Table 1). Two of these carriers were identified with NGS data: one is heterozygous for a major GBA variant associated with Gaucher disease and the second is heterozygous for a novel BLM variant predicted to be associated with Bloom syndrome. Based on non-NGS tests, two additional subjects were reported to be SMA carriers and two others were reported as alpha-thalassemia carriers. One of the latter carriers who self-identified as African American (Donor No. 15) was also reported to be a Tay-Sachs disease carrier based on borderline HexA enzyme levels, but he did not have any disease-associated mutations in the HEXA gene.
FIG. 1.
Results of commercial carrier tests and systematic analytics. (A) Donors identified as carriers with each testing panel or methodology are indicated with colored boxes. (B) The proportion of total dysfunctional variants identified as DCV or HLGD by NGS and the subportions detected by the combined commercial carrier testing panels. DCV, defined clinical variants; HLGD, high likelihood of gene dysfunction; NGS, next-generation sequencing. Color images available online at www.liebertpub.com/gtmb
The third assessment was based on a proxy for the 23andMe Personal Genome Service test panel, which identified only a single donor, No. 21, as a carrier based on heterozygosity for the most prominent ACADM mutation associated with a metabolic condition known as MCAD deficiency.
The fourth carrier assessment was based on a proxy for the standard Counsyl Family Prep Screen 1.0. In addition to the MCAD deficiency carrier also identified by 23AndMe, the Counsyl proxy panel identified three other carriers. Only one, Donor No. 6, was also detected as a carrier by GSG based on heterozygosity for a Gaucher disease mutation in GBA. Another donor, No. 1, was identified with two mutations: a DHCR7 mutation associated with multiple congenital abnormalities, intellectual disability, and fetal or infant death (Sulem et al., 2015), and an IDUA mutation associated with a severe progressive multisystem disorder leading to death before the age of 10. The final Counsyl identified carrier, Donor No. 24, is heterozygous for a PYGM mutation associated with McArdle disease, a glycogen storage disorder that can manifest as muscle pain and kidney disease.
Both proxy panels target mutations in the BLM gene associated with Bloom syndrome but they failed to detect the GSG-predicted Bloom syndrome carrier. Neither proxy panel included the genes responsible for SMA or alpha-thalassemia and, as a result, they failed to identify an additional four of the carriers reported by GSG (Table 1 and Fig. 1A).
Systematic analysis demonstrates all donors are carriers
The fifth carrier assessment was based on a systematic analysis of 4330 NGS variants detected in our donor cohort. Within this set of variants, we identified 30 DCVs, an average of 1.1 per donor (Table 2). Twenty-five were not found on any of the carrier screening panels examined in this study (Fig. 1B and Table 2). We also confirmed the GSG-reported SMA carrier status of Donors Nos. 10 and 11 (carrier probabilities of 0.83 and 1.0, respectively). Our NGS data are also consistent with the alpha-thalassemia carrier status of donors as reported by GSG (Supplementary Fig. S1). In total, 23 of the 27 donors carry at least one DCV.
Table 2.
Defined Clinical Variants with an Uncontested Pathogenic Classification
| Gene | Chr:position | Genotype | HGVSp/c | OMIM id | Donors | Panels |
|---|---|---|---|---|---|---|
| ABCA12 | 02:215813331 | C/T | p.D2365N | 607800.0008 | 17 | |
| ACADM | 01:076226846 | A/G | p.K333E | 607008.0001 | 21 | CSY, TAM |
| ADA | 20:043251694 | C/T | p.R211H | 608958.0004 | 4 | |
| ADA | 20:043280227 | C/T | p.D8N | 608958.0021 | 4, 5 | |
| ALDH7A1 | 05:125882068 | C/G | p.G505R | 14 | ||
| ALDOB | 09:104184173 | G/A | p.A338V | 9 | ||
| CBS | 21:044483184 | A/G | p.I278T | 613381.0004 | 14 | |
| CEP290 | 12:088477713 | T/A | p.K1575* | 610142.0007 | 4 | |
| CYP21A2 | 06:032007887 | G/T | p.V282L | 613815.0002 | 24, 7 | |
| CYP21A2 | 06:032008198 | C/T | p.Q318* | 613815.0020 | 3, 19 | |
| CYP21A2 | 06:032008783 | C/T | p.P454S | 613815.0010 | 26 | |
| DCLRE1C | 10:014977469 | C/T | p.G153R | 1, 13 | ||
| DHCR7 | 11:071146886 | C/G | c.964-1G>C | 602858.0001 | 1 | CSY |
| GBA | 01:155205634 | T/C | p.N409S | 606463.0003 | 6 | CSY, GSG |
| GJB2 | 13:020763612 | C/T | p.V37I | 121011.0023 | 17 | |
| GNPTAB | 12:102164255 | T/G | p.I348L | 27 | ||
| HBB | 11:005247914 | C/T | p.G70S | 141900.0050 | 18 | |
| IDUA | 04:000981646 | C/T | p.Q70* | 252800.0002 | 1 | CSY |
| MEFV | 16:003293403 | T/C | p.K695R | 608107.0010 | 19 | |
| MMACHC | 01:045973216 | T/+A | p.R91Kfs*14 | 609831.0001 | 23 | |
| MVK | 12:110034320 | G/A | p.V377I | 251170.0002 | 10 | |
| MYO7A | 11:076916644 | G/A | p.R1873Q | 1 | ||
| PKLR | 01:155261709 | G/A | p.R486W | 609712.0009 | 7 | |
| PYGM | 11:064527223 | G/A | p.R50* | 608455.0001 | 24 | CSY |
| SMPD1 | 11:006412867 | C/-T | p.S192Afs | 7 | ||
| TSEN54 | 17:073518081 | G/T | p.A307S | 608755.0001 | 1, 13 | |
| TYMP | 22:050967020 | C/T | p.R146H | 6 | ||
| WNT10A | 02:219755011 | T/A | p.F228I | 606268.0003 | 16, 18 | |
| ZNF469 | 16:088502971 | G/* | p.L3004_T3008 | 25 | ||
| ZNF469 | 16:088505063 | G/A | p.G3701S | 2 |
In the second stage of our systematic analysis, we identified 66 HLGD variants (2.44 average per donor, Table 3) that have not been classified by ClinVar (as of November 2, 2015). Only one of these non-ClinVar variants (in the Bloom syndrome gene BLM) was detected by any of the examined commercial screening panels.
Table 3.
Non-DCV High Likelihood of Gene Dysfunction Variants
| Gene | Chr:bpa | Geno | HGVSp/c | Donors | type | PP2 | CADD | PROVEAN |
|---|---|---|---|---|---|---|---|---|
| ALG9 | 11:111657228 | G/A | p.R413W | 6 | Missense | 1.00 | 16.3 | −7.13 |
| ATM | 11:108121543 | C/T | p.R451C | 10 | Missense | 1.00 | 34.0 | −2.17 |
| ATR | 03:142274739 | A/+T | p.I774Nfs*3 | 9 | Frameshift | |||
| BLM | 15:091290624 | T/C | p.M1T | 17 | Startloss | |||
| CD19 | 16:028943901 | A/C | p.Q108P | 6 | Missense | 1.00 | 21.6 | −4.98 |
| CFTR | 07:117174349 | G/A | p.R170H | 2 | Missense | 1.00 | 35.0 | −1.74 |
| CHRND | 02:233396160 | C/T | p.P307S | 8 | Missense | 1.00 | 31.0 | −7.86 |
| CHRNG | 02:233404829 | C/A | p.N61K | 1 | Missense | 1.00 | 22.8 | −4.42 |
| CLN3 | 16:028500658 | C/T | p.A59T | 7 | Missense | 1.00 | 35.0 | −3.71 |
| COL4A4 | 02:227915700 | C/T | p.G1048D | 12 | Missense | 1.00 | 26.3 | −6.63 |
| COL4A4 | 02:227915847 | C/T | p.G999E | 16, 7 | Missense | 1.00 | 24.2 | −7.08 |
| COL4A4 | 02:227946893 | C/G | p.G545A | 2, 4 | Missense | 1.00 | 25.5 | −5.21 |
| COL6A2 | 21:047545926 | G/A | p.G733R | 12 | Missense | 1.00 | 32.0 | −7.23 |
| COL6A3 | 02:238285919 | G/A | p.R856C | 20 | Missense | 1.00 | 24.6 | −6.33 |
| COL7A1 | 03:048609571 | G/A | p.R2338* | 5 | Nonsense | |||
| DBT | 01:100684204 | C/T | p.R178H | 8 | Missense | 1.00 | 23.1 | −4.57 |
| DGUOK | 02:074184313 | A/G | p.Y218C | 4 | Missense | 1.00 | 21.5 | −7.92 |
| DMP1 | 04:088584363 | A/C | p.D478A | 2 | Missense | 0.99 | 6.8 | −4.94 |
| DPYD | 01:098205946 | A/G | c.321 + 2T>G | 5 | Splice site | |||
| DPYD | 01:098165030 | T/C | p.Y186C | 2 | Missense | 1.00 | 26.9 | −5.73 |
| ERCC4 | 16:014029033 | G/A | p.R415Q | 1 | Missense | 1.00 | 35.0 | −2.82 |
| ERCC5 | 13:103527849 | G/G | p.G1053* | 3 | Nonsense | |||
| ERCC5 | 13:103527930 | G/G | p.G1080* | 3 | Nonsense | |||
| ERCC6 | 10:050686482 | C/A | p.R735L | 11 | Missense | 1.00 | 36.0 | −6.21 |
| FAH | 15:080473480 | G/A | p.G387R | 11 | Missense | 1.00 | 34.0 | −5.54 |
| FRAS1 | 04:079387464 | A/A | p.K2378* | 12, 3 | Nonsense | |||
| FRAS1 | 04:079343055 | C/T | p.R1527W | 13 | Missense | 1.00 | 29.4 | −4.48 |
| FREM2 | 13:039454452 | C/T | p.T3013M | 6 | Missense | 1.00 | 34.0 | −3.4 |
| FUCA1 | 01:024194769 | G/-C | p.A3 | 2 | Frameshift | |||
| HADH | 04:108948869 | G/A | p.R221H | 2 | Missense | 1.00 | 34.0 | −4.92 |
| HGSNAT | 08:043054553 | T/T | p.Y583* | 15 | Nonsense | |||
| IKBKAP | 09:111653606 | C/T | p.G1013S | 3 | Missense | 1.00 | 31.0 | −2.66 |
| IKBKAP | 09:111656228 | T/A | p.K952I | 5 | Missense | 1.00 | 31.0 | −5.8 |
| IL1RN | 02:113885264 | A/G | c.74-2A>G | 5 | Splice site | |||
| ITGA6 | 02:173368930 | G/+A | p.Q1079Tfs*10 | 5 | Frameshift | |||
| JAK3 | 19:017951097 | T/C | p.Y399C | 2 | Missense | 1.00 | 26.2 | −7.06 |
| LAMA3 | 18:021424991 | C/A | p.P1208T | 8, 18 | Missense | 1.00 | 11.0 | −6.9 |
| LAMA3 | 18:021479356 | C/T | p.R1981W | 15 | Missense | 1.00 | 10.2 | −5.94 |
| LIG4 | 13:108861878 | C/T | p.R580Q | 22 | Missense | 1.00 | 34.0 | −3.8 |
| MAN2B1 | 19:012763007 | G/A | p.P669L | 11 | Missense | 0.99 | 15.7 | −5.43 |
| MUT | 06:049409599 | G/A | p.R588C | 3 | Missense | 1.00 | 35.0 | −2.64 |
| NDUFS4 | 05:052899281 | G/A | c.99-1G>A | 4 | Splice site | |||
| NEB | 02:152374912 | T/A | p.K5873* | 10 | Nonsense | |||
| NEB | 02:152471050 | G/A | p.R3781W | 9 | Missense | 1.00 | 33.0 | −3.31 |
| NPHS2 | 01:179528881 | A/T | p.L156* | 9 | Nonsense | |||
| PAH | 12:103246697 | A/-G | p.A246Vfs*95 | 23 | Frameshift | |||
| PCCB | 03:136046072 | C/T | p.P425L | 21 | Missense | 1.00 | 35.0 | −9.43 |
| PEX5 | 12:007354852 | C/T | p.R235W | 19 | Missense | 1.00 | 36.0 | −3.49 |
| PKHD1 | 06:051523922 | C/A | p.D3668Y | 3 | Missense | 1.00 | 32.0 | −4.81 |
| PRX | 19:040903208 | G/A | p.P351S | 15 | Missense | 1.00 | 23.2 | −6.73 |
| PRX | 19:040903267 | G/A | p.P331L | 12 | Missense | 1.00 | 25.8 | −7.72 |
| RAB3GAP1 | 02:135893167 | C/T | p.R530C | 10 | Missense | 1.00 | 22.7 | −5.56 |
| RNASEH2B | 13:051544085 | G/-A | p.Q253. | 1 | Frameshift | |||
| SLC22A5 | 05:131705708 | G/A | p.G15E | 20 | Missense | 1.00 | 22.1 | −6.77 |
| SLC26A2 | 05:149360744 | G/A | p.G530S | 13 | Missense | 1.00 | 27.6 | −5.9 |
| SMPD1 | 11:006415539 | C/T | p.P533L | 20 | Missense | 1.00 | 11.6 | −7.18 |
| STXBP2 | 19:007710082 | G/C | c.1247-1G>C | 8 | Splice site | |||
| TBCE | 01:235599092 | T/G | p.L257* | 7 | Nonsense | |||
| TYK2 | 19:010469975 | A/C | p.I684S | 14 | Missense | 1.00 | 28.8 | −5.13 |
| UNC13D | 17:073832671 | C/T | p.R427Q | 8 | Missense | 1.00 | 35.0 | −3.2 |
| VLDLR | 09:002648286 | G/A | p.R634H | 16 | Missense | 1.00 | 35.0 | −4.93 |
| ZNF469 | 16:088497233 | G/T | p.E1091* | 11 | Nonsense |
Additional details on each variant reported in this study can be accessed at http://research.genepeeks.com
All 27 donors carry one or more variants classified as DCV or HLGD that are likely to be associated with severe recessive pediatric disease. The 96 predicted dysfunctional NGS variants detected in the donor cohort are distributed among 81 genes (Supplementary Table S1). Only 6% of these variants were detected by the screening panels examined in this study (Fig. 1B).
Discussion
Carrier screening is a universal component of donor eligibility protocols performed by U.S. sperm banks. Surprising to most prospective clients, however, actual carrier screening practices are highly variable across donor banks (Sims et al., 2010). Different banks screen their donor applicants using different commercial carrier tests, which have each been developed based on a different set of largely self-imposed scientific, social, and economic constraints. Some screens emphasize the depth of individual gene analysis over the scope of covered disease genes. Some companies choose to restrict their analysis to clinically characterized variants; others do not. Some screens focus their disease coverage on conditions prevalent in Ashkenazi Jewish and northern European populations, a historical artifact from the origins of human genetics research and testing. As in the case of 23andMe, the design of some mutation panels has been highly influenced by the testing company's choice of business model and corresponding regulatory pathway (Servick, 2015; Levenson, 2016).
A donor bank's selection of screening provider, therefore, can drive very different outcomes in the determination of donor eligibility—and ultimately the exposure of recipient offspring to disease risk. Our analysis of a cohort of prospective donors demonstrates how large these differences can be. At the lower bound, as per the study's design, none of the screened donor applicants tested positive on the Integrated Genetics CFplus screen, although one did carry a CF-associated variant. In subsequent screenings, GSG reported six of these donors to be carriers of disease-causing variants based on its GeneVu panel, and proxies for tests sold by 23andMe and Counsyl identified one and four carriers, respectively. Because each panel is designed to examine different variants and distinct diseases, no single donor was uniformly identified as carrier positive by all screens. This striking inconsistency challenges the validity of carrier screening as a categorical diagnostic device.
The problem of inconsonance, however, may be less harmful than the low sensitivity of all of these screens in identifying carrier-positive subjects. We observed at least one highly dysfunctional recessive gene variant in each prospective donor, a finding that is consistent with expectations from large population NGS results (Cooper et al., 2013; Henn et al., 2015). Two-thirds of these dysfunctional variants were not contained in the ClinVar database (as of November 10, 2015). This is a consequence of both the population biases embedded in clinical research (Lim et al., 2015), and the fact that so-called novel recessive disease mutations are usually transmitted silently from one generation of heterozygotes to the next without clinical detection until it is too late and a child is affected by disease. A variety of experimental approaches have led researchers to the same conclusion: every human being is a carrier of multiple disease-causing mutations, most of which are rare and not clinically defined (Bell et al., 2011; Xue et al., 2012; Simons et al., 2014; Tabor et al., 2014; Henn et al., 2015; Schrodi et al., 2015). The tools of modern molecular biology enable both observation and characterization of these clinically unobserved variants, an advance that has particular utility in preconception risk assessment.
Perhaps a more fundamental flaw in the carrier screening paradigm is illustrated by our detection of the clinically characterized disease-contributing variants HBB:p.G70S and CFTR:p.R170H, which were not reported by any of the carrier screening panels (Cooper et al., 2013). Each of these amino acid substitutions only partially reduces gene function, allowing homozygotes to remain asymptomatic (Rahbar et al., 1984; Paradisi et al., 2010; LaRusch et al., 2014). Based on the industry's working definition of a carrier, the application of a carrier-positive label to individuals harboring these partial-function variants would be considered incorrect. And yet each of these variants unambiguously contributes to disease when combined with a more severely dysfunctional allele carried by a reproductive partner: compound heterozygosity of CFTR:p.R170H with CFTR:p.F508del causes atypical CF (Alibakhshi et al., 2008; Baker et al., 2011); compound heterozygosity of HBB:p.G70S with the sickle cell mutation or a loss-of-function HBB allele results in beta0-thalassemia intermedia (Yang et al., 1989; Paradisi et al., 2010; Vinciguerra et al., 2014). Offspring of donor sperm recipients who carry these more damaging variants would be highly exposed to disease risk despite a donor's designation as carrier negative for mutations in the relevant genes.
The same ambiguity in carrier status applies to two donors who were, in fact, identified by GSG as carriers of an alpha-thalassemia allele (referred to as α+) that retains ∼50% functionality. As is the case with HBB:p.G70S and CFTR:p.R170H, homozygosity for an α+ allele does not cause disease; indeed, it is actually the norm in some Asian populations, although infrequent in northern Europeans (Yenchitsomanus et al., 1985; Harteveld and Higgs, 2010; Purohit et al., 2014). Thus, α+ carrier designation and donor disqualification result in higher rates of unnecessary stigmatization and counseling for minority donors in the United States.
As these findings illustrate, the true complexity of human reproduction is not reflected by the standard of care. To reject donor applicants who are carrier positive for a subset of clinically characterized variants found primarily in individuals with northern European ancestry—and without any corresponding analysis of recipient DNA—is an outdated protocol that violates the current tenets of recessive disease genetics. These include the universality of positive carrier status for every human being; the pervasiveness of novel highly damaging mutations; the widespread occurrence of partially damaging variants that are disease-causing in particular genotypes but defy simple classification as pathogenic or benign; and wide population variance in disease risk and mutation frequency. The persistence of this anachronistic standard harms both donors and recipients. Donors who test positive are led to believe incorrectly that their status is atypical and risk of disease transmission is materially higher than the general population. In contrast, a negative result followed by inclusion in a sperm bank program can elicit a false belief that a donor is “mutation-free” (Callum et al., 2010).
Despite tremendous advances in variant identification, understanding, and analysis, the vast majority of disease-causing mutation combinations remain undetected in the carrier screening process. Most important, the exposure of recipient offspring to recessive disease risk is not reduced sufficiently by this protocol (Lim et al., 2014). To achieve the ultimate screening goal of protecting future children from highly heritable disease, industry screening standards must be modernized to reflect the biological reality that both donors and recipients carry a wide range of known and novel disease-causing variants that cause a spectrum of damage to the underlying gene. The inputs for this more clinically effective approach are now available and affordable. Systematic, high-resolution analysis of recessive disease risk associated with particular donor/recipient pairings has been developed, but industry practice must be modified to incorporate its use.
Supplementary Material
Acknowledgments
The authors thank Andrew Remis and Juliana Cooper for data support and Jeffrey Field for his insightful comments regarding the development of this project. Finally, the authors are grateful to the cohort of donors who contributed their genomes to this analysis. GenePeeks, Inc. funded this project in its entirety.
Author Disclosure Statement
A.J.S., J.L.L., M.J.S., R.M.L., C.B., B.S., A.M., L.M.S. are employed by, and have equity interest in, GenePeeks, Inc. GenePeeks, Inc. is a fully owned subsidiary of the Lifeprint Group, which also owns Manhattan Cryobank. A.M. and L.M.S. are directors of the Lifeprint Group.
References
- Adzhubei IA, Schmidt S, Peshkin L, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibakhshi R, Kianishirazi R, Cassiman J-J, et al. (2008) Zamani M, Cuppens H. Analysis of the CFTR gene in Iranian cystic fibrosis patients: Identification of eight novel mutations. J Cyst Fibros 7:102–109 [DOI] [PubMed] [Google Scholar]
- Baker MW, Groose M, Hoffman G, et al. (2011) Optimal DNA tier for the IRT/DNA algorithm determined by CFTR mutation results over 14 years of newborn screening. J Cyst Fibros 10:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Dinwiddie DL, Miller NA, et al. (2011) Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 3:65ra4–65ra4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callum P, Iger J, Ray M, et al. (2010) Outcome and experience of implementing spinal muscular atrophy carrier screening on sperm donors. Fertil Steril 94:1912–1914 [DOI] [PubMed] [Google Scholar]
- Centola GM. (2010) Sperm banking, donation, and transport in the age of assisted reproduction: Federal and state regulation. In: Carrell DT, Peterson CM. (eds) Reproductive Endocrinology and Infertility. Springer Science and Business Media, LLC, New York, NY [Google Scholar]
- Choi Y, Sims GE, Murphy S, et al. (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Polychronakos C, et al. (2013) Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet 132:1077–1130[Internet]. Available at: http://link.springer.com/article/10.1007/s00439-013-1331-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JG, Feldman G, Goldberg J, et al. (2015) Expanded carrier screening in reproductive medicine—Points to consider. Obstet Gynecol 125:653–662 [DOI] [PubMed] [Google Scholar]
- Harteveld CL, Higgs DR. (2010) α-Thalassaemia. Orphanet J Rare Dis 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Bustamante CD, et al. (2015) Estimating the mutation load in human genomes. Nat Rev Genet 16:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina (2016) Sequencing panel for 4813 genes with known associated clinical phenotypes. Available at www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/technote_trusight_one_panel.pdf (accessed January15, 2016)
- Isley L, Callum P. (2013) Genetic evaluation procedures at sperm banks in the United States. Fertil Steril 99:1587–1587 [DOI] [PubMed] [Google Scholar]
- Kingsmore S. (2012) Comprehensive carrier screening and molecular diagnostic testing for recessive childhood diseases. PLoS Curr 4:e4f9877ab8ffa9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, et al. (2014) Technical reports. Nat Genet 46:310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landaburu I, Gonzalvo MC, Clavero A, et al. (2013) Genetic testing of sperm donors for cystic fibrosis and spinal muscular atrophy: evaluation of clinical utility. Eur J Obstet Gynecol Reprod Biol 170:183–187 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, et al. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JL, Silver AJ, Chan D, et al. (2015) Validation of a high resolution NGS method for detecting spinal muscular atrophy carriers among phase 3 participants in the 1000 Genomes Project. BMC Med Genet 16:919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRusch J, Jung J, General IJ, et al. (2014) Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS Genet 10:e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarin GA, Haque IS, Nazareth S, et al. (2013) An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet Med 15:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson D. (2016) 23andMe markets carrier screening service directly to consumers. Am J Med Genet A 170:293–294 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RM, Silver AJ, Borroto C, et al. (2014) Autosomal recessive disease risk in offspring of qualified sperm bank donors and clients: proof of principle for a novel analysis based on virtual progeny. Fertil Steril 102:e305–e306 [Google Scholar]
- Lim RM, Silver AJ, Silver MJ, et al. (2016) Targeted mutation screening panels expose systematic population bias in detection of cystic fibrosis risk. Genet Med 18:174–179 [DOI] [PubMed] [Google Scholar]
- Nazareth SB, Lazarin GA, Goldberg JD. (2015) Changing trends in carrier screening for genetic disease in the United States. Prenat Diagn 35:931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradisi I, González N, Hernández A, et al. (2010) Hemoglobin S/hemoglobin City of Hope compound heterozygote with a SubSaharan genetic background and severe bone marrow hypoplasia. Invest Clin 51:403–414 [PubMed] [Google Scholar]
- Perreault-Micale C, Davie J, Breton B, et al. (2015) A rigorous approach for selection of optimal variant sets for carrier screening with demonstration of clinical utility. Mol Genet Genomic Med 3:363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit P, Dehury S, Patel S, et al. (2014) Prevalence of deletional alpha thalassemia and sickle gene in a tribal dominated malaria endemic area of eastern India. ISRN Hematol 2014:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbar S, Asmerom Y, Blume KG. (1984) A silent hemoglobin variant detected by HPLC: hemoglobin City of Hope beta 69 (E13) Gly—Ser. Hemoglobin 8:333–342 [DOI] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, et al. (2015) ClinGen—the Clinical Genome Resource. N Engl J Med 372:2235–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servick K. (2015) Can 23AndMe have it all? Science 349:1472–1477 [DOI] [PubMed] [Google Scholar]
- Schrodi SJ, DeBarber A, He M, Ye Z, Peissig P, Wormer JJ, et al. (2015) Prevalence estimation for monogenic autosomal recessive diseases using population‐based genetic data. Hum Genet 134:659–669 [DOI] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, et al. (2014) The deleterious mutation load is insensitive to recent population history ng.2896 (1). Nat Genet 46:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims CA, Callum P, Ray M, et al. (2010) Genetic testing of sperm donors: survey of current practices. Fertil Steril 94:126–129 [DOI] [PubMed] [Google Scholar]
- Sulem P, Helgason H, Oddson A, et al. (2015) Identification of a large set of rare complete human knockouts. Nat Genet 47:448–452 [DOI] [PubMed] [Google Scholar]
- Tabor HK, Auer PL, Jamal SM, et al. (2014) Pathogenic variants for Mendelian and complex traits in rxomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet 95:183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 11:11..10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M, Passarello C, Leto F, et al. (2014) Co-inheritance of the rare β hemoglobin variants Hb Yaounde, Hb Görwihl and Hb City of Hope with other alterations in globin genes: impact in genetic counseling. Eur J Haematol 94:322–329 [DOI] [PubMed] [Google Scholar]
- Xue Y, Chen Y, Ayub Q, et al. (2012) Deleterious- and disease-allele prevalence in healthy individuals: Insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 91:1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KG, Kutlar F, George E, et al. (1989) Molecular characterization of β-globin gene mutations in Malay patients with Hb E-β-thalassaemia and thalassaemia major. Br J Haematol 72:73–80 [DOI] [PubMed] [Google Scholar]
- Yenchitsomanus PT, Summers KM, Bhatia KK, et al. (1985) Extremely high frequencies of alpha-globin gene deletion in Madang and on Kar Kar Island, Papua New Guinea. Am J Hum Genet 37:778–784 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.