Abstract
Accurate detection and quantification of HIV-1 group O viruses have been challenging for currently available HIV assays. We have developed a novel time-resolved fluorescence (TRF) europium nanoparticle immunoassay for HIV-1 group O detection using a conventional microplate enzyme-linked immunosorbent assay (ELISA) and a microchip platform. We screened several antibodies for optimal reactivity with several HIV-1 group O strains and identified antibodies that can detect all the strains of HIV-1 group O that were available for testing. The antibodies were used to develop a conventional ELISA format assay and an in-house developed europium nanoparticle-based assay for sensitivity. The method was evaluated on both microwell plate and microchip platforms. We identified two specific and sensitive antibodies among the six we screened. The antibodies, C65691 and ANT-152, were able to quantify 15 and detect all 17 group O viruses, respectively, as they were broadly cross-reactive with all HIV-1 group O strains and yielded better signals compared with other antibodies. We have developed a sensitive assay that reflects the actual viral load in group O samples by using an appropriate combination of p24 antibodies that enhance group O detection and a highly sensitive TRF-based europium nanoparticle for detection. The combination of ANT-152 and C65690M in the ratio 3:1 was able to give significantly higher signals in our europium-based assay compared with using any single antibody.
Introduction
The first case of HIV-1 group O was described from a Cameroonian patient 25 years ago and the recent data show that it is spreading epidemically in western central Africa. The prevalence of group O is very low at 1%–2% in Cameroon, and some European countries with strong colonial ties to central Africa have reported the highest prevalence out of Africa. The sequence diversity between HIV-1 group O and M strains may be up to 50% and 30% in the envelope and pol, respectively. This enormous diversity has hindered diagnosis, monitoring, and treatment of group O-infected patients. Due to the intrinsic presence of mutation in the C181 in group O, more than 60% of the ∼30,000 individuals who are infected with this virus are faced with the challenge of drug resistance to some currently used antiretroviral therapies, notably the non-nucleoside reverse transcriptase inhibitors (NNRTIs).1,2
The sequence divergence of HIV group strains makes it challenging to accurately detect and measure viral load. Inaccurate estimation of HIV-1 group O infection in the population is due to lack of adequate sensitivity of assays for group O detection. Accurate diagnosis of group O is necessary for maintaining safety of the blood supply, epidemiological survey, incidence estimation, and targeted intervention of HIV infection. We have developed a novel p24 time-resolved fluorescence (TRF) microplate europium nanoparticle immunoassay (ENIA) and microchip ENIA to detect HIV group O with higher degree of sensitivity.
The first HIV-1 group O viruses to be described were the ANT-70 and MVP-5180 strains, both isolated in persons from Cameroon. These isolates had only 50% homology with the other HIV-1 isolates in the env gene.3,4 Additional HIV-1 group O variants have been described and the majority of the strains have been obtained from Cameroon or from Cameroonians living in Europe. Some cases have been reported in other African countries such as Gabon, Nigeria, Equatorial Guinea, and Kenya, and in Benin, a dual HIV-1 group M and O infection has been described. At least more than one case has been reported in the United States.5,6
Analysis of group O viruses shows that these viruses are genetically very diverse.7 HIV-1 group O viruses may present a public health challenge because several of them escape detection by some conventional screening immunoassays and can produce indeterminate Western blot patterns.8–10 Different serological approaches have been used to diagnose HIV-1 group O infection. Some researchers use a competitive immunoblot or a negative result in HIV-1 competitive assays.11,12 Studies among HIV-infected individuals from different African countries, using an enzyme-linked immunosorbent assay (ELISA) based on the V3 peptide from ANT-70 with confirmation by a specific ANT-70 Western blot, indicated that HIV-1 group O infection is present in Cameroon and Gabon.13 Sequence data on a limited number of samples confirmed that this strategy can lead to the identification of HIV-1 viruses from group O.14 HIV-1 group O strains are highly divergent from the major group M, leading to their designation as outliers. These strains also display marked intragroup genetic diversity: they are divided into three clades (clades A, B, and C), but numerous divergent strains lie outside these clades.15 This genetic diversity has important implications for the diagnosis and monitoring of HIV-1 group O infection, including a risk of false negativity and viral load underestimation.16–19
In regions where HIV-1groups M and O cocirculate, HIV-O infection has to be diagnosed specifically before treatment initiation; otherwise, it can have harmful consequences on the patients as HIV-O strains are known to harbor natural resistance to NNRTIs, efavirenz and nevirapine.20–22 The specific diagnosis of HIV-O is therefore necessary to avoid rapid virological failure when using NNRTI in biotherapy for the patient and the use of NNRTIs in treatments for the prevention of mother-to-child transmission. The diagnosis is also important where follow-up is required for children born to infected mothers.23,24 Therapeutic management, especially for resistance analysis using specific tools, requires accurate diagnosis of HIV 1 group O. Finally, it has been shown that dual HIV-M/HIV-O infection/superinfection can occur in areas in which both groups circulate, including France,25–28 necessitating the precise monitoring of each strain to determine the course of infection in the absence of treatment and specific response to treatment.
The need for HIV/AIDS care and treatment is increasing in resource-poor settings, especially in Africa, which has the majority of people infected with HIV. Increased access to accurate and specific HIV testing is essential to achieve universal access to HIV prevention and treatment in resource-limited countries. The World Health Organization and the Joint United Nations Program on HIV/AIDS have recommended that prevention of new HIV transmissions should be one of the key elements in the global strategy to fight HIV/AIDS.29 Antiretroviral treatment (ART), combined with present prevention approaches, could have a major effect on severe generalized HIV/AIDS epidemics.30 However, efficient implementation of prevention and care strategies requires correct identification of uninfected and infected people.
The aim of this work was to design and evaluate a new more sensitive and specific assay in a simplified format for detection of group O. Most diagnostic tests were historically developed for detection of HIV-1 subtype B strains. These tests showed limitations in detecting HIV-1 group O8 and some group M variants, especially during the serological window period.31 Broadly, cross-reactive antigens or inclusion of HIV-1 group O-specific antigens increased detection of most HIV strains. The simultaneous detection of HIV antigens (p24) and anti-HIV antibodies by fourth-generation HIV assays has helped reduce the diagnostic window period.32,33 Despite these efforts, the performance of certain serological assays is still suboptimal for detection of emerging variants and group O, as illustrated by some studies.34–36 Many studies have shown that in African countries, especially Cameroon, the available tests are unable to detect all HIV-1 group O strains. Therefore, there is a continued need to validate and develop tests that are more sensitive for detection of all strains of HIV.29
Highly specific and sensitive nucleic acid tests are not well suited for HIV diagnosis in resource-limited regions because of their high cost, instrumentation complexity, demands for highly trained operators, and well-controlled environment. There exist many low-cost portable point-of-care (POC) assay kits for rapidly diagnosing HIV epidemics in remote settings and most of them are based on lateral flow assay (LFA)/immunochromatographic strip test. Unfortunately, LFA has mediocre analytical sensitivity and would be unable to reliably detect early HIV infections. Current LFA design limits its multiplex assay capability, thus affecting its ability to identify different HIV strains and HIV coinfections on a single platform. Lab-on-a-chip/microchip (LOC) technology, on the other hand, may provide a new avenue to a new generation of POC HIV test platforms. LOC technology borrows concepts from techniques of microelectromechanical system technology and applies it in chemical and biological fields. Besides advantages such as high reaction efficiency, low reagent consumption, and small footprint, this technology allows integration of multiple assay components to a single platform for complex assays or high-throughput parallel assay.37 We have developed in our laboratory a sensitive europium nanoparticle-based microtiter plate immunoassay capable of detecting target analytes at subpicogram per milliliter levels without the use of catalytic enzymes and signal amplification processes. The use of europium nanoparticless further permits the assay to be adapted to an ELISA format that is already widely used in testing laboratories since the antibody–antigen sandwich complex bound to europium nanoparticles coated with streptavidin (SA) can be directly measured with a fluorescence reader.
Materials and Methods
All monoclonal anti-HIV p24 antibodies were purchased from commercial vendors. The HIV-1 group O viruses were obtained from the NIH AIDS Reagent Program (BCF01, BCF02, BCF03, BCF06, BCF07, BCF11, BCF13, and MVP5180) and others were from strains cultured in our laboratory in peripheral blood mononuclear cells (MD1422, MD1312, MD5267, MD4354, MD47, Spain 152, Spain 153, and German O). Polyclonal anti-HIV p24 antibodies were purchased from Perkin Elmer. Europium nanoparticles, 107 nm in diameter, were obtained from Fisher. The preparation and characterization of Eu3+ nanoparticles coated with SA have been described previously and is the modified procedure described below.
Europium nanoparticle immunoassay
Although there are many commercially available ELISA format assays, the ENIA successfully avoids the use of enzymes and achieves high sensitivity, with significant improvements in detection of HIV-1 infection. Therefore, we evaluated other NPs to simplify the assay format and found that ENIA using Eu3+ NP is suitable for rapid and sensitive detection of HIV-1 p24. In the ENIA, Eu3+ NPs modified with SA are bound to the antigen–antibody sandwiched complex, followed by the binding of biotinylated anti-SA antibody and SA-coated europium chelates (Perkin Elmer). Because each Eu3+ NP contains ∼30,000 europium ions, the Eu3+ NPs can produce intense long-lifetime fluorescence lights that are identical to those in the dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) method and can be measured directly in the Victor Multilabel Counter (Perkin Elmer).38 The procedure to perform the ENIA involves dissolving the capture antibody in carbonate–bicarbonate buffer (pH 9.6, 100 mM) to a concentration of 2 μg/ml and adding 55 μl of this mixture into a Nunc MaxiSorp plate (Thermo Fisher Scientific). The plate was incubated at 4°C for 1 or 2 days, and the plate was washed five times with wash buffer (Perkin-Elmer) and added 250 μl of blocking buffer [Starting Block T20 phosphate-buffered saline (PBS) Blocking Buffer from Pierce]. Then, the plates were kept in blocking buffer for 30 min. In the next step, the blocking buffer was removed and different HIV group O strain samples were added. Then, the plate was incubated for one hour at 37°C with shaking. The HIV culture supernatant was diluted in blocking buffer containing 55 μl of 10% Triton X-100 per milliliter. Plates were washed five times with wash buffer and incubated with 100 μl of (1 × 108 particles/ml) SA-conjugated europium nanoparticle for half an hour at 37°C with shaking. The plate is washed and read with the TRF reader (Perkin Elmer Victor) with excitation at 340 nm and emission at 615 nm (decay time 0.4 ms, measurement window 0.4 ms). The assay included controls and a blank well, which does not contain HIV group O. The results are compared with the p24 standard ranging from a dynamic range of 1–500 pg/ml.
Preparation of SA-conjugated europium nanoparticle probes
Briefly, europium nanoparticles were first added to 10 mM phosphate buffer (pH 7.0) with 10 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and 10 mM sulfo-N-hydroxysuccinimide. The activation was allowed to proceed at room temperature for 30 min. After a buffer exchange with 10 mM carbonate buffer (pH 9.0), 0.5 mg/ml SA in the carbonate buffer was added to the activated europium nanoparticles. After a 2-h reaction at room temperature, the unreacted reagents and background buffer were removed and washed five times with 10 mM glycine buffer (pH 8.5). The product was stored at 4°C overnight before use. Buffer exchange and EuNP wash were performed using NanoSep® centrifugal ultrafiltration devices with a molecular weight cutoff of 300 kDa (Pall Life Sciences).
Microchip ENIA of HIV samples
The detailed fabrication protocol and design of testing microchips are described in the previously published article.39 The microchip was fabricated using polydimethylsiloxane (PDMS) silicone. There are 12 independent microreactors on a single device to accept multiple capturing biomolecule coatings for parallel or multiplex antigen/antibody assays. In a typical microchip ENIA experiment, the capture antibody was diluted in a PBS (pH 7.2) solution to a concentration of 10 μg/ml and loaded to a microchip. The device was incubated at 4°C for at least 24 h and then blocked with phosphate buffered saline Tween 20 obtained from Thermo Scientific Product #37539 (PBST) solution for 30 min at 37°C before assay. In step 1 of HIV assays, sample solutions were loaded to the microchip and the device was incubated at 37°C for 15 min. In step 2, a biotinylated secondary antibody solution was filled in the device and the on-chip reaction was allowed to proceed at 37°C for 30 min. In the final step, Eu SA solution (5 × 108 particles/ml) was loaded to the microchips and the biotin-SA coupling reactions were incubated again at 37°C for 15 min. The devices were subjected to a final wash and then placed on a modified plate for time-resolved fluorescence signal reading. Microchip fluid manipulation was achieved using a manual micropump assembled using a rubber pipette filler (Thermo Fisher Scientific) and a section of Tygon tubing (0.02′′ ID ×0.060′′′′ OD; Cole-Plamer).
Results
Comparison of antibodies for sensitivity in p24 detection
We screened several different HIV 1group O strains with different commercially available HIV 1 p24 antibodies. We used the following HIV1 group O viruses, which were in our laboratory and cultured to get high titer (MD1422, MD1312, MD5267, MD4354, MD47, Spain 152, Spain 153, and German O). We also obtained BCF01, BCF02, BCF03, BCF06, BCF07, BCF11, BCF13, and MVP5180 from NIH AIDS reagent repository. We purchased HIV 1 p24 antibodies, C65690M, C65941M, and C65489M, from Meridian Life Science, Inc.; ANT-152 from ProSpec-Tany TechnoGene Ltd.; NB500-473 from Novus Biologicals; and 012-A from Virogen. These HIV 1 p24 antibodies were coated and screened against the HIV 1 group O viruses listed above using in-house developed europium nanoparticle assay and a commercial Perkin Elmer ELISA kit. When comparing the results, we subtract the negative well and use only results that are above the 1.5 signal-to-background ratio.
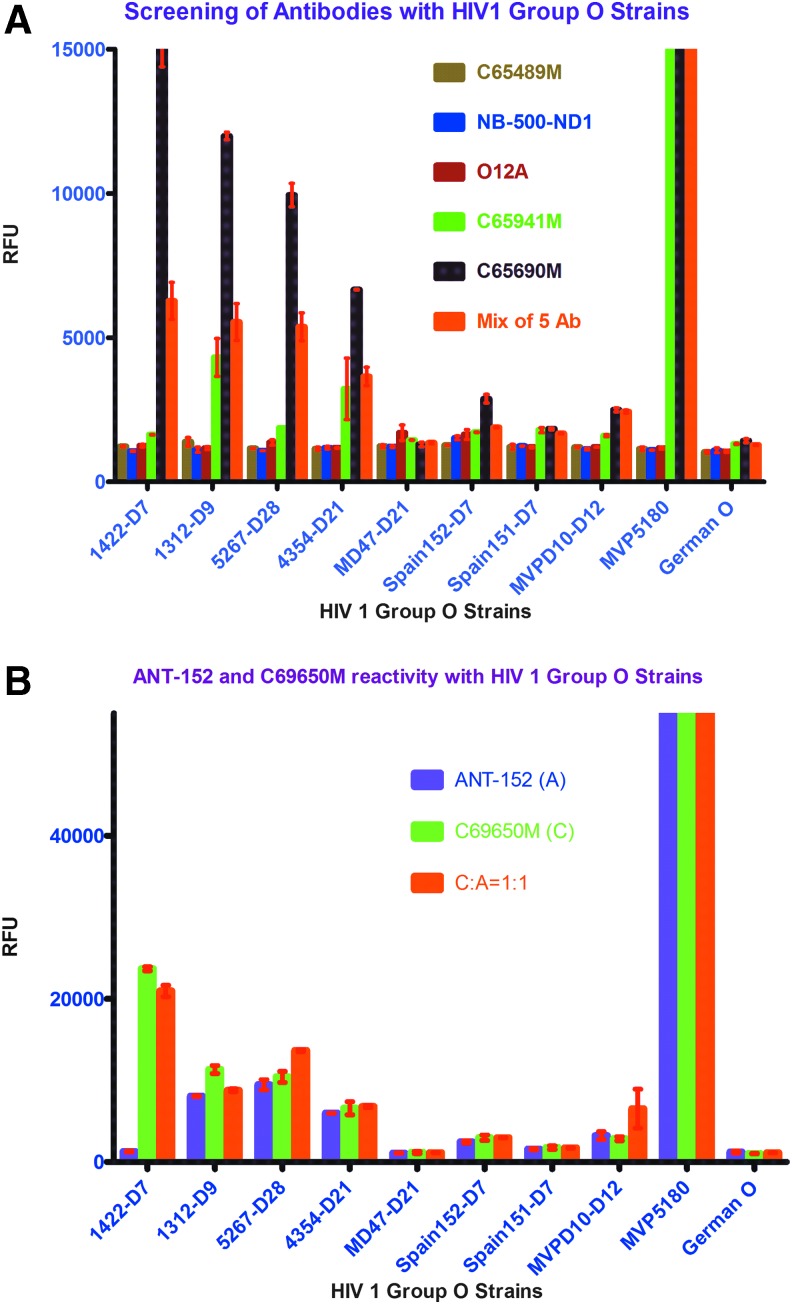
We identified two antibodies, C65690M and ANT-152, which were highly sensitive for detecting HIV 1 group O viruses. The combination of the five antibodies (Fig. 1A) and the combination of C65690M and ANT-152 antibodies in a 1:1 ratio (Fig. 1B) that showed high sensitivity in our laboratory were used in our in-house europium nanoparticle assay.
FIG. 1.
(A) Screening of different commercially available monoclonal HIV p24 antibodies with HIV 1 group O strains on ENIA platform. (B) Comparision of monoclonal HIV p24antibodies, ANT-152 with C65690M, on ENIA platform. ENIA, europium nanoparticle immunoassay. Color images available online at www.liebertpub.com/aid
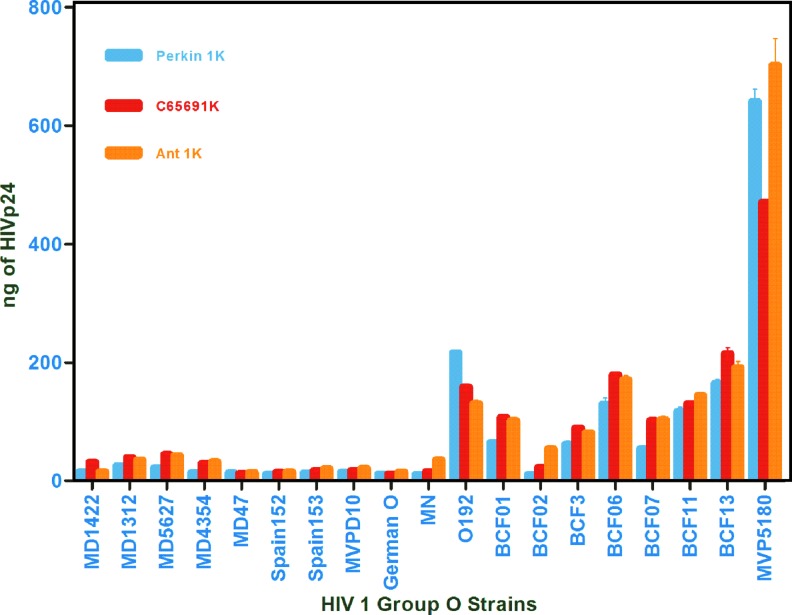
We compared the two antibodies, C65690m and ANT-152, using our in-house europium nanoparticle laboratory assay with a commercial Perkin Elmer ELISA kit to verify quantification of HIV 1 group O. (Fig. 2; Table 1)
FIG. 2.
Comparision of HIV 1 group O quantification by Perkin Elmer Kit and ENIA for ANT-152 with C65690M antibodies. Color images available online at www.liebertpub.com/aid
Table 1.
Quantification of HIV 1 Group O Using Perkin Elmer and Europium Nanoparticle Immunoassays
| HIV 1 group O strains | Perkin Elmer | C65690M | ANT 152 |
|---|---|---|---|
| MD1422 | 15.66667 | 31.65098 | 15.53191 |
| MD1312 | 25.78788 | 39.57647 | 35.50355 |
| MD5267 | 21.95152 | 44.79608 | 42.53191 |
| MD4354 | 14.09091 | 29.15686 | 32.51064 |
| MD47 | 13.83636 | 12.18039 | 14.22695 |
| Spain152 | 11.4 | 14.84314 | 15.53191 |
| Spain153 | 13.50303 | 17.06275 | 20.7305 |
| MVPD10 | 14.55758 | 17.34902 | 21.06383 |
| German O | 11.90909 | 11.87451 | 15.01418 |
| O192 | 216.3455 | 158.4902 | 130.5674 |
| BCF01 | 64.68485 | 107.1059 | 102.1489 |
| BCF02 | 10.89091 | 22.65882 | 54.36879 |
| BCF3 | 61.64242 | 89.64314 | 81.58156 |
| BCF06 | 129.8727 | 179.3098 | 170.5319 |
| BCF07 | 54.2 | 102.1137 | 103.3901 |
| BCF11 | 117.3515 | 130.8627 | 144.0426 |
| BCF13 | 164.6848 | 214.8588 | 191.4823 |
| MVP5180 | 640.5697 | 470.5882 | 701.7305 |
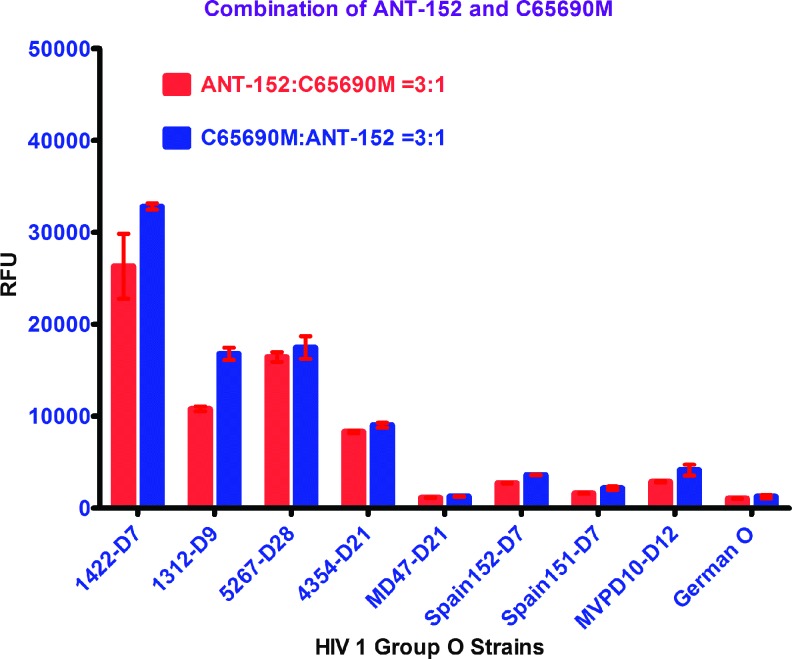
As noted in Figure 2 and Table 1 using the HIV 1 p24 monoclonal antibody C65690M from Meridian Life Science, Inc., coated on the plate, our in-house europium nanoparticle assay was able to quantify 14 of 18 HIV1 group O viruses better than the commercial Perkin Elmer kit. Similarly, using the HIV 1 p24 monoclonal antibody ANT-152 from ProSpec-Tany TechnoGene Ltd., coated on the plate, our in-house europium nanoparticle assay was able to quantify 16 of 18 HIV1 group O viruses more efficiently than the commercial Perkin Elmer kit. In summary, monoclonal antibodies, C65690M from Meridian and ANT-152 from ProSpec-Tany TechnoGene Ltd., were able to quantify HIV 1 group O viruses using our laboratory-developed in-house europium nanoparticle assay more accurately than the commercial Perkin Elmer kit. To enhance efficiency of quantification and detection of HIV 1 group O viruses for all the strains, compared with the commercial Perkin Elmer kit, we coated both monoclonal antibodies, C65690M and ANT-152, in 1:1, 1:3, and 3:1 ratios to find out the best combination of these antibodies that would improve detection of group O. The combination of these two antibodies in equal proportion gave similar results for all the HIV 1 group O strains, whereas a 3:1 ratio of C65690M and ANT-152 yielded enhanced sensitivity for group O detection as shown in in Figures 2 and 3.
FIG. 3.
Comparision of HIV 1 group O signals on ENIA with 3:1 and 1:3 ratios of ANT-152 with C65690M antibodies. Color images available online at www.liebertpub.com/aid
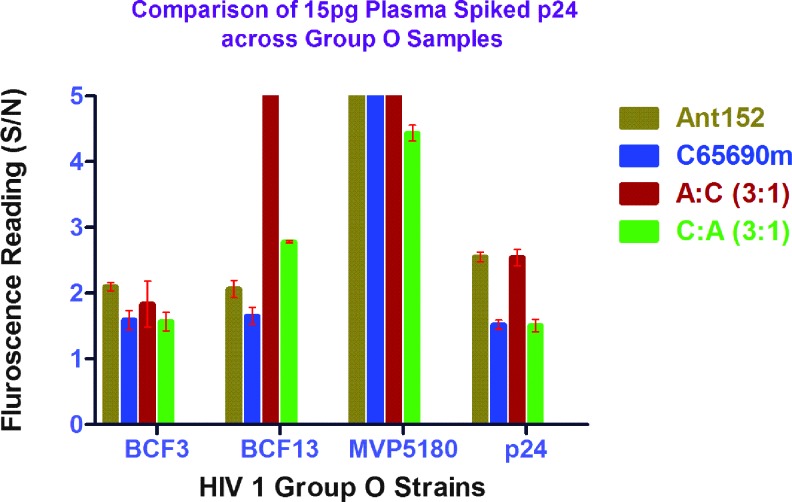
Determination of detection limit for ENIA in spiked plasma samples
Using the signal-to-background ratio of above 1.5 as the detection threshold, we found the detection limit of the antigen to be at 5 pg/ml. The detection limit was more or less similar with both antibodies, ANT-152 and C65690M, as seen in Figure 4. Since clinical samples for group O are hard to obtain, we tested a limited number HIV 1 group O viruses spiked in plasma to mimic clinical sample conditions. We had previously tested negative samples in our laboratory using these two antibodies with the europium nanoparticle assay and found no false-positive or false-negative results. We prepared p24 at a concentration of 25 pg/ml by spiking it in pooled plasma (George Medical, ID: PNP3469) and in PBST, both containing Triton X-100, and the results are shown in Figure 5. We compared the conserved HIV 1 group O P24 domains of the BH10 and ABT325, which showed 76% identity match. HIV 1 p24 detection assays are thus not geared toward accurate detection of p24 antigen concentration of group O owing to sequence diversity. However, in this study, we identified two antibodies, C65690M and ANT-152, which are able to better quantify using our in-house ENIA platform.
FIG. 4.
Detection of 5 pg of p24 in group O strains with different antibodies. Color images available online at www.liebertpub.com/aid
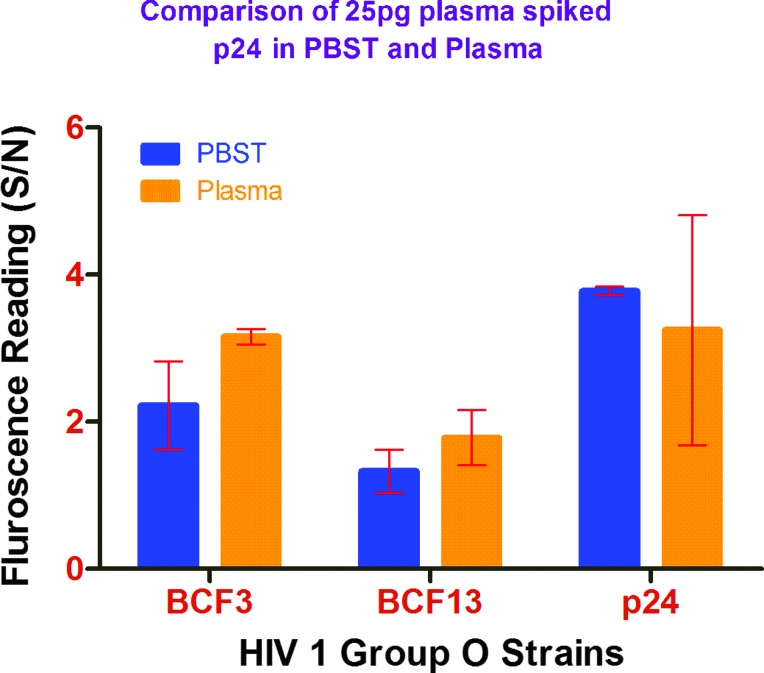
FIG. 5.
Comparison of detection of 25 pg of group O strains in phosphate buffered saline Tween 20 obtained from Thermo Scientific Product #37539 versus plasma. Color images available online at www.liebertpub.com/aid
Evaluation of microchip detection
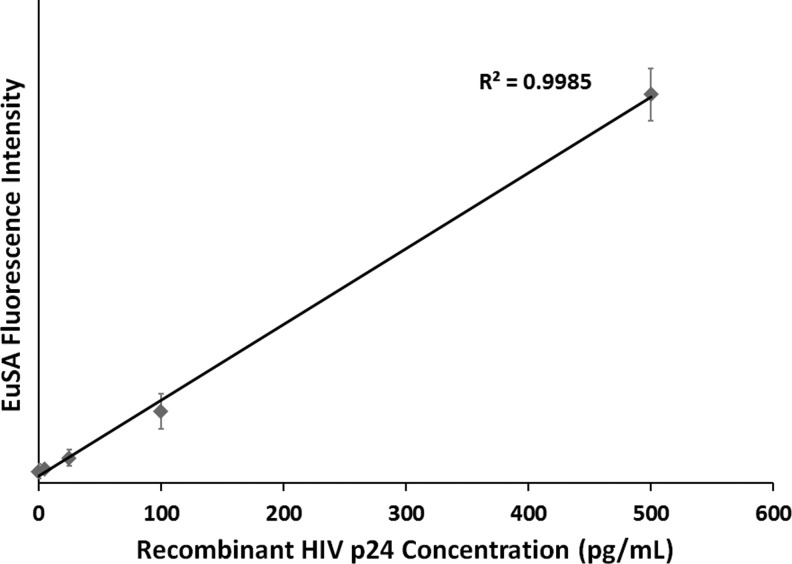
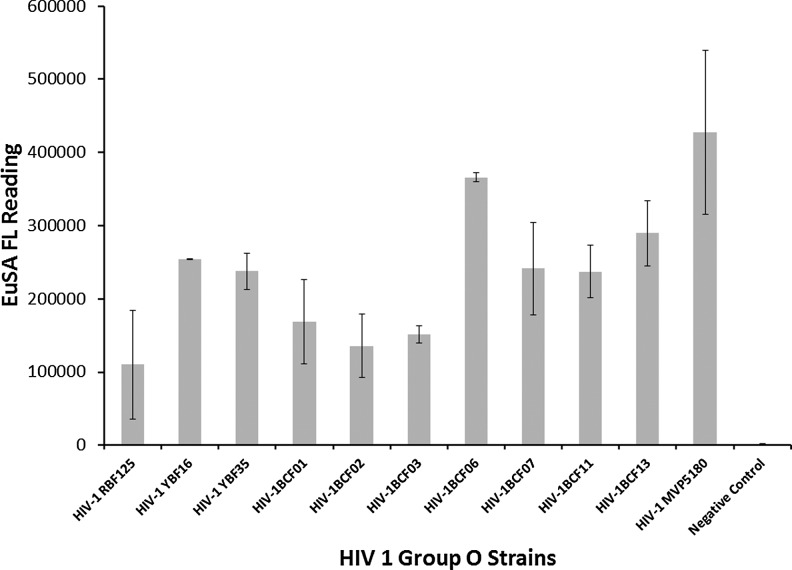
Performances of PDMS microchips coated with antibody C65690M were first evaluated in HIV antigen assay using recombinant HIV-1 p24 (Perkin-Elmer) as a standard. We can confidently detect 5 pg/ml of p24 antigen with a signal-to-background (S/B) ratio above 1.5 and with a minimum limit of detection: 3.3 pg/ml, as shown in Figure 6. The linear dynamic range was found to extend from 0 to 500 pg/ml. Following the initial test with C65690M, we continued evaluating performances of microchips coated with antibody, ANT-152 (Fig. 7), and found that similar sensitivity and linear dynamic range were achieved using the ANT-152-coated device. We did not identify significant performance difference in the two antibodies on the microchip platform. We investigated 11 HIV-1 group O virus strains (diluted 100-folds in PBST containing Triton X-100) on ANT-152-coated microchips using ENIA as the detection assay and found that all strains could be readily detected at S/B ratios higher than 1.5 (Fig. 7). This result confirms that the PDMS microchip platform can also be employed in the detection of HIV-1 group O virus. Compared with benchtop ENIA, the microchip ENIA platform offers reasonably similar high sensitivity, low reagent consumption and waste generation, shorter assay time, and ability to perform on-site rapid tests when combined with portable readers.
FIG. 6.
HIV 1 p24 sensitivity and dynamic range curve for microchip ENIA.
FIG. 7.
HIV 1 group O p24 detection on microchip platform using ENIA.
Discussion
In this article, we describe a novel highly sensitive TRF-based ENIA for the detection and quantification of HIV-1 group antigen in plasma and culture supernatant. Our ENIA showed high sensitivity compared with commercially available ELISA formats due to enhancement of sensitivity by a combination of the unique properties of time-resolved fluorescent europium nanoparticles and the use of the monoclonal antibodies, C65691 and ANT-152, which are capable of capturing HIV-1 p24 antigen efficiently compared with other antibodies tested. An advantage of the ENIA is its ultrasensitive detection capability in the absence of enzymatic reactions. In this study, we use novel labeling technology to improve detection; the frequently used labeling technologies in immunoassays include enzyme activity and chemicals (chemiluminescence and fluorescence). A major advantage of the ENIA over enzyme-based colorimetric ELISA is its high sensitivity and low background. In the past decade, lanthanide chelates have been successfully used in immunoassays, such as DELFIA technology. The ENIA is a highly sensitive assay with a stable signal, high signal-to-noise ratio, no enzymatic step, flexible platform, and short incubation time. Polystyrene NP-containing lanthanide chelates were reported to significantly improve detection sensitivity of prostate-specific antigen.40,41 The improved detection sensitivity is attributable to the high content of Eu3+ ions in a single NP and, in particular, the unique characteristic of the time-resolved lanthanide chelates without self-quenching, even at high millimolar concentrations. It has been shown that the Eu3+ NPs, especially SA-coated NPs, may be more useful than Eu3+ chelates in developing assays with high amplification ratios and extremely high detection sensitivity.
The excellent analytical sensitivity of this assay at 0.5 pg/ml makes it suitable for use in the diagnosis of HIV1 group O infection, which may go undetected in other assays38 or in discordant samples due to the high sensitivity of the C65691 and ANT-152. Discordant results of HIV antibody assays, p24 antigen assays, and nucleic acid-based assays might be suggestive of either false-positive or false-negative reactive samples. They strongly complicate the early diagnosis of acute HIV-1 group O infection and are often instrumental in the decision to stop ART soon thereafter. The failure to detect HIV antibodies because of HIV genetic diversity has been regularly reported, even when using highly sensitive assays.42 Direct detection by p24 antigen assays is particularly useful for detection of window period cases and in cases of indeterminate antibody response. Therefore, sensitive assays that accurately identify HIV-1 group O strains are important for public health prevention efforts particularly in Africa where group O and other highly divergent non B subtype variants are prevalent. In this study, we also demonstrate that this assay can be adapted to a microchip microfluidic platform wherein a 4.5-fold reduction in sample/reagent consumption and a 2-fold reduction in assay time were achieved with the microchip compared with conventional microtiter plate assay. These features of a microchip format make it suitable for POR use.
Acknowledgments
This work was supported by the NHLBI Nano IAA- A-HL-12-001 and a CBER Intramural grant MOD SCI 2014. The authors are thankful to the NIH AIDS Reagent Program for providing HIV 1 group O strains.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Food and Drug Administration, U.S. Department of Health and Human Services.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Villabona-Arenas CJ, Domyeum J, Mouacha F, Butel C, Delaporte E, Peeters M, et al. : HIV-1 group O infection in Cameroon from 2006 to 2013: Prevalence, genetic diversity, evolution and public health challenges. Infect Genet Evol 2015;36:210–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush S, Tebit DM: HIV-1 group O origin, evolution, pathogenesis, and treatment: Unraveling the complexity of an outlier 25 years later. AIDS Rev 2015;17:147–158 [PubMed] [Google Scholar]
- 3.Vanden Haesevelde M, Decourt JL, De Leys RJ, Vanderborght B, van der Groen G, van Heuverswijn H, et al. : Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol 1994;68:1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurtler LG, Hauser PH, Eberle J, von Brunn A, Knapp S, Zekeng L, et al. : A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol 1994;68:1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, et al. : Geographical distribution of HIV-1 group O viruses in Africa. AIDS 1997;11:493–498 [DOI] [PubMed] [Google Scholar]
- 6.Sullivan PS, Do AN, Ellenberger D, Pau CP, Paul S, Robbins K, et al. : Human immunodeficiency virus (HIV) subtype surveillance of African-born persons at risk for group O and group N HIV infections in the United States. J Infect Dis 2000;181:463–469 [DOI] [PubMed] [Google Scholar]
- 7.Loussert-Ajaka I, Chaix ML, Korber B, Letourneur F, Gomas E, Allen E, et al. : Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol 1995;69:5640–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loussert-Ajaka I, Ly TD, Chaix ML, Ingrand D, Saragosti S, Courouce AM, et al. : HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet 1994;343:1393–1394 [DOI] [PubMed] [Google Scholar]
- 9.Schable C, Zekeng L, Pau CP, Hu D, Kaptue L, Gurtler L, et al. : Sensitivity of United States HIV antibody tests for detection of HIV-1 group O infections. Lancet 1994;344:1333–1334 [DOI] [PubMed] [Google Scholar]
- 10.Simon F, Ly TD, Baillou-Beaufils A, Fauveau V, De Saint-Martin J, Loussert-Ajaka I, et al. : Sensitivity of screening kits for anti-HIV-1 subtype O antibodies. AIDS 1994;8:1628–1629 [DOI] [PubMed] [Google Scholar]
- 11.Zekeng L, Gurtler L, Afane Ze E, Sam-Abbenyi A, Mbouni-Essomba G, Mpoudi-Ngolle E, et al. : Prevalence of HIV-1 subtype O infection in Cameroon: Preliminary results. AIDS 1994;8:1626–1628 [DOI] [PubMed] [Google Scholar]
- 12.Gurtler L: Difficulties and strategies of HIV diagnosis. Lancet 1996;348:176–179 [DOI] [PubMed] [Google Scholar]
- 13.Nkengasong JN, Peeters M, vanden Haesevelde M, Musi SS, Willems B, Ndumbe PM, et al. : Antigenic evidence of the presence of the aberrant HIV-1ant70 virus in Cameroon and Gabon. AIDS 1993;7:1536–1538 [PubMed] [Google Scholar]
- 14.Janssens W, Nkengasong JN, Heyndrickx L, Fransen K, Ndumbe PM, Delaporte E, et al. : Further evidence of the presence of genetically aberrant HIV-1 strains in Cameroon and Gabon. AIDS 1994;8:1012–1013 [DOI] [PubMed] [Google Scholar]
- 15.Roques P, Robertson DL, Souquiere S, Damond F, Ayouba A, Farfara I, et al. : Phylogenetic analysis of 49 newly derived HIV-1 group O strains: High viral diversity but no group M-like subtype structure. Virology 2002;302:259–273 [DOI] [PubMed] [Google Scholar]
- 16.Leoz M, Depatureaux A, Vessiere A, Roquebert B, Damond F, Rousset D, et al. : Integrase polymorphism and HIV-1 group O diversity. AIDS 2008;22:1239–1243 [DOI] [PubMed] [Google Scholar]
- 17.Depatureaux A, Charpentier C, Leoz M, Unal G, Damond F, Kfutwah A, et al. : Impact of HIV-1 group O genetic diversity on genotypic resistance interpretation by algorithms designed for HIV-1 group M. J Acquir Immune Defic Syndr 2011;56:139–145 [DOI] [PubMed] [Google Scholar]
- 18.Gueudin M, Plantier JC, Lemee V, Schmitt MP, Chartier L, Bourlet T, et al. : Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J Acquir Immune Defic Syndr 2007;44:500–505 [DOI] [PubMed] [Google Scholar]
- 19.Plantier JC, Djemai M, Lemee V, Reggiani A, Leoz M, Burc L, et al. : Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J Clin Microbiol 2009;47:2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, et al. : Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: In vitro phenotypic and genotypic analyses. J Virol 1997;71:8893–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinones-Mateu ME, Soriano V, Domingo E, Menendez-Arias L: Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology 1997;236:364–373 [DOI] [PubMed] [Google Scholar]
- 22.Tuaillon E, Gueudin M, Lemee V, Gueit I, Roques P, Corrigan GE, et al. : Phenotypic susceptibility to nonnucleoside inhibitors of virion-associated reverse transcriptase from different HIV types and groups. J Acquir Immune Defic Syndr 2004;37:1543–1549 [DOI] [PubMed] [Google Scholar]
- 23.Chaix-Baudier ML, Chappey C, Burgard M, Letourneur F, Igual J, Saragosti S, et al. : First case of mother-to-infant HIV type 1 group O transmission and evolution of C2V3 sequences in the infected child. French HIV Pediatric Cohort Study Group. AIDS Res Hum Retroviruses 1998;14:15–23 [DOI] [PubMed] [Google Scholar]
- 24.Gueudin M, Lemee V, Ferre V, Beby-Defaux A, Pathe JP, Guist'hau O, et al. : Virologic diagnosis and follow-up of children born to mothers infected by HIV-1 group O. J Acquir Immune Defic Syndr 2004;36:639–641 [DOI] [PubMed] [Google Scholar]
- 25.Brand D, Beby-Defaux A, Mace M, Brunet S, Moreau A, Godet C, et al. : First identification of HIV-1 groups M and O dual infections in Europe. AIDS 2004;18:2425–2428 [PubMed] [Google Scholar]
- 26.Plantier JC, Lemee V, Dorval I, Gueudin M, Braun J, Hutin P, et al. : HIV-1 group M superinfection in an HIV-1 group O-infected patient. AIDS 2004;18:2444–2446 [PubMed] [Google Scholar]
- 27.Vergne L, Bourgeois A, Mpoudi-Ngole E, Mougnutou R, Mbuagbaw J, Liegeois F, et al. : Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 2003;310:254–266 [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi J, Bodelle P, Vallari AS, Coffey R, McArthur CP, Schochetman G, et al. : HIV infections in northwestern Cameroon: Identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res Hum Retroviruses 2004;20:944–957 [DOI] [PubMed] [Google Scholar]
- 29.Aghokeng AF, Mpoudi-Ngole E, Dimodi H, Atem-Tambe A, Tongo M, Butel C, et al. : Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PloS One 2009;4:e7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granich RM, Gilks CF, Dye C, De Cock KM, Williams BG: Universal voluntary HIV testing with immediate antiretroviral therapy as a strategy for elimination of HIV transmission: A mathematical model. Lancet 2009;373:48–57 [DOI] [PubMed] [Google Scholar]
- 31.Apetrei C, Loussert-Ajaka I, Descamps D, Damond F, Saragosti S, Brun-Vezinet F, et al. : Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS 1996;10:F57–F60 [DOI] [PubMed] [Google Scholar]
- 32.Gurtler L, Muhlbacher A, Michl U, Hofmann H, Paggi GG, Bossi V, et al. : Reduction of the diagnostic window with a new combined p24 antigen and human immunodeficiency virus antibody screening assay. J Virol Methods 1998;75:27–38 [DOI] [PubMed] [Google Scholar]
- 33.Weber B, Fall EH, Berger A, Doerr HW: Reduction of diagnostic window by new fourth-generation human immunodeficiency virus screening assays. J Clin Microbiol 1998;36:2235–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcoran C, Hardie D: HIV-1 subtype C seronegative AIDS. Clin Infect Dis 2008;46:322–323 [DOI] [PubMed] [Google Scholar]
- 35.Gautheret-Dejean A, Mesmin-Poho S, Birguel J, Lemee V, Huraux JM, Plantier JC: Unequal detection of HIV type 1 group O infection by simple rapid tests. Clin Infect Dis 2008;46:1936–1937 [DOI] [PubMed] [Google Scholar]
- 36.Novitsky V, Gaolathe T, Woldegabriel E, Makhema J, Essex M: A seronegative case of HIV-1 subtype C infection in Botswana. Clin Infect Dis 2007;45:e68–e71 [DOI] [PubMed] [Google Scholar]
- 37.Reyes DR, Iossifidis D, Auroux PA, Manz A: Micro total analysis systems. 1. Introduction, theory, and technology. Anal Chem 2002;74:2623–2636 [DOI] [PubMed] [Google Scholar]
- 38.Tang S, Hewlett I: Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis 2010;201 Suppl 1:S59–S64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Du B, Zhang P, Haleyurgirisetty M, Zhao J, Ragupathy V, et al. : Development of a microchip Europium nanoparticle immunoassay for sensitive point-of-care HIV detection. Biosens Bioelectron 2014;61C:177–183 [DOI] [PubMed] [Google Scholar]
- 40.Harma H, Soukka T, Lonnberg S, Paukkunen J, Tarkkinen P, Lovgren T. Zeptomole detection sensitivity of prostate-specific antigen in a rapid microtitre plate assay using time-resolved fluorescence. Luminescence 2000;15:351–355 [DOI] [PubMed] [Google Scholar]
- 41.Soukka T, Harma H, Paukkunen J, Lovgren T: Utilization of kinetically enhanced monovalent binding affinity by immunoassays based on multivalent nanoparticle-antibody bioconjugates. Anal Chem 2001;73:2254–2260 [DOI] [PubMed] [Google Scholar]
- 42.Henquell C, Jacomet C, Antoniotti O, Chaib A, Regagnon C, Brunet S, et al. : Difficulties in diagnosing group o human immunodeficiency virus type 1 acute primary infection. J Clin Microbiol 2008;46:2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]