Abstract
Glycosylation of biomolecules is one of the most prevalent post- and co-translational modification in a human body, with more than half of all human proteins being glycosylated. Malignant transformation of cells influences glycosylation machinery resulting in subtle changes of the glycosylation pattern within the cell populations as a result of cancer. Thus, an altered terminal glycan motif on glycoproteins could provide a warning signal about disease development and progression and could be applied as a reliable biomarker in cancer diagnostics. Among all highly effective glycoprofiling tools, label-free electrochemical impedance spectroscopy (EIS)-based biosensors have emerged as especially suitable tool for point-of-care early-stage cancer detection. Herein, we highlight the current challenges in glycoprofiling of various cancer biomarkers by ultrasensitive impedimetric-based biosensors with low sample consumption, low cost fabrication and simple miniaturization. Additionally, this review provides a short introduction to the field of glycomics and lectinomics and gives a brief overview of glycan alterations in different types of cancer.
Keywords: glycosylation, lectins, electrochemical impedance spectroscopy, cancer biomarkers, biosensors
1. Introduction
Carbohydrates are an essential part of every living organism and are considered to be the most abundant organic molecules found in nature [1–3]. It is widely known that glycans modulate or mediate cell-cell or cell-biomolecule interactions, cell signalling, host-pathogen interactions, disease progression or metastasis [4–7]. Changes in the glycosylation, mediated by multiple enzymes, play critical roles in regulation of numerous biological processes. Oligosaccharides may covalently link to a protein backbone in order to stabilize, functionalize it and create highly specific sites for biorecognition. There are two main types of glycan attachments to glycoproteins: (i) N-linked glycosylation e.g. glycans are covalently bound to asparagine residues in a consensus sequence Asn-X-Ser/Thr (X can be any amino acid except for proline) via N-acetylglucosamine (N-GlcNAc); and (ii) O-linked glycosylation e.g. attachment of glycans to the hydroxyl groups of serine or threonine [8]. Glycosylation is quite a complex process catalysed by glycosyltransferases in the endoplasmic reticulum (ER)-Golgi apparatus [9,10].
The structural variety of glycans derives from the various ways in which monosaccharides can be linked together and from many available isomers of monosaccharides [9]. Covalent glycosidic bond can be designed in two possible positions at an anomeric carbon; i.e. via either an α- or a β- glycosidic linkage. A vast complexity of glycans can be illustrated by a theoretical number of all possible saccharides formed from 4 building blocks, when 4 different amino acids can form 24 different tetrapeptides, but four different hexoses may potentially generate 35,560 unique tetrasaccharides [1,11]. In addition, glycans can be enzymatically modified, which further increases the number of possible saccharidic units potentially present in biological systems [12].
In order to understand the function of glycans on a molecular level, advanced mass spectrometry (MS), liquid chromatography (LC), capillary electrophoresis (CE), microarray techniques, and biosensors have been employed [13,14]. Glycan arrays and arrays of glycan-binding biomolecules have become “a must have” component for highly robust and parallel glycoprofiling in glycomics and for glycoprofiling of various diseases [3,15–20]
2. Lectins
Lectins are ubiquitous natural proteins specifically recognizing and binding carbohydrate complexes. Structural studies indicated that the carbohydrate-binding activity of lectins was generated by a limited polypeptide segment designated as the carbohydrate recognition domain (CRD) [1,21]. Most lectins interact with terminal non-reducing carbohydrate residues of a glycoprotein and glycolipid component of the cell membrane. Variety of lectins are localized in different parts of the organism, depending on their functional role (e.g. intracellular lectins are involved in protein trafficking, membrane-bound lectins mediate host-pathogen interactions, etc.) [22]. The term lectin is evolved from the Latin word legere meaning to choose, pick or select [23]. Furthermore, these carbohydratebinding molecules are able to agglutinate cells (e.g. erythrocytes). It is believed that the earliest description of ability to agglutinate erythrocytes was by Peter Hermann Stillmark in 1888 [21]. He described the agglutination activity of toxin Ricinus communis in his doctoral thesis. However, the modern age of lectinomics began almost one hundred years later [1].
Lectins have been isolated from various sources, such as plants, bacteria, viruses, and animals [18,24]. Plant lectins are the biggest family of lectins; one of the best characterized types of plant lectin are the ones isolated from Legeminosae sp. The Leguminosae lectins are Ca2+- and Mn2+-dependent metalloproteins, such as concanavalin A (ConA), lentil lectin (LCA), soybean agglutinin (SBA), and others [24]. Some of the plant lectins (Ricinus communis agglutinin, and lectin from Abrus precatorius) exhibit toxic effect to animal cells [25]. Animal lectins belong to endogenous lectins and are further classified into a C-type (Ca2+-dependent) and S-type (sulphhydryl-dependent galectins usually occurring in a soluble form) [9,25]. Viral lectins are known as hemagglutinins and the influenza virus hemagglutinin was the first glycosyaminoglycan-binding protein isolated from lower organisms in 1950 [9]. The overview of lectins with carbohydrate specificity, source, and molecular weight is given in Table 1.
Table 1.
List of lectins with their common abbreviations, source, preferred carbohydrate specificity and molecular weight [17,31,36,38].
| Lectin from | Abbr. | S | Carbohydrate specificity | Mw |
|---|---|---|---|---|
| Aleuria aurantia | AAL | A | α-L-Fuc | 72 |
| Anguilla anguilla | AAA | F | α-L-Fuc | 50 |
| Aspergillus oryzae | AOL | MO | α-1,6Fuc | n/a |
| Concanavalin A | Con A | P | α-D-Man, α-D-Glc; branched N-linked hexa-saccharide | 104 |
| Datura stramonium | DSL/DSA | P | GlcNAcβ-1,4GlcNAc | 86 |
| Dolichos biflorus | DBA | P | GalNAc α or β-1,3Gal | 111 |
| Erythrina cristagalli | ECL/ECA | P | Galβ-1,4GlcNAc | 54 |
| Euonymus europaeus | EEL | P | Galα-3Gal | 140 |
| Galanthus nivalis | GNL | P | α-D-Man | 50 |
| Griffonia simplicifolia I | GSL-I | P | Galα -1,3Gal | 114 |
| Griffonia simplicifolia II | GSL-II | P | α or β-GlcNAc | 113 |
| Jacalin | Jacalin (AIL) | P | Galβ-1,3GalNAc | 66 |
| Lens culinaris/Lentil lectin | LCA | P | α-D-Man, α-D-Glc | 50 |
| Lotus tetragonolobus | LTA/LTL | P | Fucα-1,2Galβ | 107 |
| Maackia amurensis I | MAA/MAL | P | α-2,3Neu5Ac | 130 |
| Narcissus pseudonarcissus | NPA/NPL | P | α-D-Man | 59 |
| Phaseolus vulgaris | E-PHA | P | N-linked bi-antennary | 126 |
| Phaseolus vulgaris | L-PHA | P | Branched β-1,6GlcNAc; N-linked tri/tetra-antennary | 126 |
| Peanut agglutinin | PNA | P | Galβ-1,3GalNAc | 110 |
| Ricunus communis I | RCA-I | P | β-D-Gal | 120 |
| Ricinus communis II | RCA-II | P | Galβ-1,4GalNAc | 60 |
| Sambucus nigra | SNA-I | P | Neu5Acα-2,6Gal/ GalNAc | 140 |
| Soybean agglutinin | SBA | P | α or β-Gal; α or β-GalNAc | 120 |
| Ulex europaeus I | UEA-I | P | Fucα-1,2Gal | 63 |
| Ulex europaues II | UEA-II | P | GlcNAcβ-1,4GlcNAc | 63 |
| Vicia villosa | VVL/VVA | P | α-D-GalNAc | 144 |
| Wheat Germ | WGA | P | β-D-GlcNAc, Neu5Ac | 36 |
S: source, Mw: molecular weight in kDa, A: animal, F: fungi, MO: microorganism, P: plant, Fuc: fucose, Gal: galactose, GalNAc: N-acetylgalactosamine, Glc: glucose, GlcNAc: N-acetylglucosamine, Man: mannose, Neu5Ac: N-acetylneuraminic acid (sialic acid)
Lectins can also be classified, based on their carbohydrate binding specificity, into five main groups with a specificity to: L-fucose, galactose/N-acetylgalactose, sialic acids, mannose and/or glucose, and N-acetylglucosamine [22].
2.1. Lectin-carbohydrate interactions
Details about lectin-saccharide interactions and about structural basics of the carbohydrate specificity of lectins have been provided by several studies based on X-ray crystallography and other instrumental techniques [17,21,24,26]. Lectin-glycoconjugate biorecognition is specific, like in the case of antibody-antigen or enzyme-substrate binding [10,27]. These interactions are mediated by hydrogen bonds, van der Waals interactions, and hydrophobic binding [28]. For instance, polar parts of galactose are recognized by a lectin through hydrogenbond interactions, while less polar side of galactose interacts with a lectin via hydrophobic interactions (i.e. tryptophan residues in lectin) [10]. Many lectins contain two or more carbohydrate-binding sites. The binding-sites for monosaccharides are stabilized via numerous hydrogen bonds, most often by Asp, Asn, and Gly residues [28]. Moreover, based on thermodynamic studies it was concluded that the dominant forces stabilizing the complex would appear to be intermolecular hydrogen bonds and van der Waals interactions [29]. Electrostatic interactions are limited to specific monosaccharides such as various forms of sialic acid [28]. In general, lectins exhibit low affinity (KD = 10−3−10−4 mol L−1) while binding with carbohydrates [30]. On the other hand, antigen-antibody interactions typically exhibit KD in the subnanomolar range [30].
Lectins as biorecognition elements and valuable glycan affinity reagents have been broadly utilized in numerous applications, such cancer diagnostics [18,25,31], drug delivery [32], immunohistological studies [28], analysis of pathogenic bacteria [33], HIV research [34], etc. Plenty of analytical techniques have been employed to study lectin-carbohydrate binding profiles and subtle changes in the glycosylation pattern. The most high-throughput methods in glycomics are advanced mass spectrometry (MS) ones combined with liquid chromatography and electrophoresis [14]; and lectin/carbohydrate microarrays. Additionally, numerous laboratory techniques have been adapted to integrate lectins (lectin affinity chromatography, enzyme-linked lectin assay (ELLA), lectin blotting, agglutination methods, histo- and cyto-chemical methods, electrochemical impedance spectroscopy (EIS), quartz crystal microbalance (QCM), flow cytometry, surface plasmon resonance (SPR), etc.) for effective glycoprofiling of a diverse range of samples [17,35–37]. A distinct advantage of using lectin-based approaches compared to the instrumental techniques is a direct glycoprofiling of intact proteins, and even cells without a need to release glycans for subsequent analysis. Moreover, in some cases lectins can provide information about spatial distribution of glycans e.g. Sambucus nigra agglutinin (SNA) recognizes sialic acid linked to galactose via an α-2,6 linkage, while Maackia amurensis agglutinin recognizes sialic acid linked to galactose via an α-2,3 linkage. Considering that glycan alternating motifs have been frequently occurred on malignant cells and tissues, lectins have been copiously utilized in cancer-related research areas.
3. Cancer study
3.1. Glycosylation changes in cancer
Glycan alteration is a universal feature of malignant transformation and tumour progression. Due to changes associated with a biological function, cancer cells frequently show fundamentally different glycan structure than those observed on and within normal non-malignant cells [9,39,40]. This change in a carbohydrate content was first described by Meezan et al. in 1969 in a study focused on characterization of healthy and virus-transformed mouse fibroblasts [41].
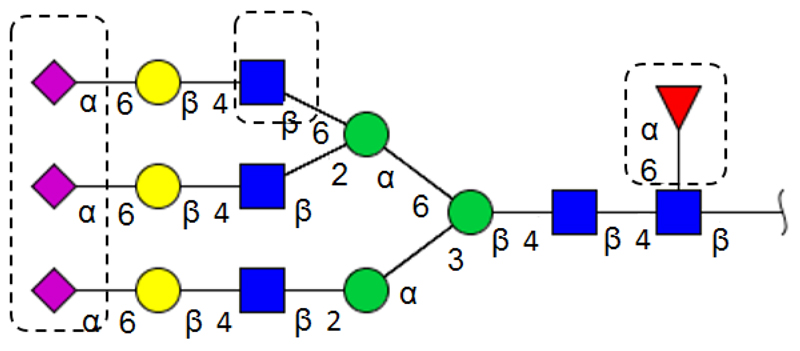
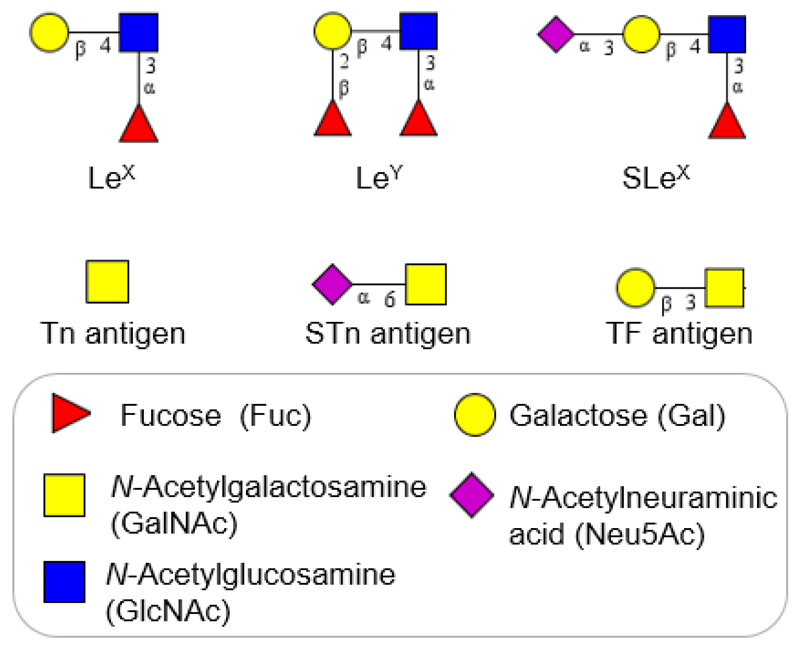
Aberrant glycosylation results from changes in expression levels of glycosyltransferases in the Golgi apparatus of malignant cells. One of the most frequent glycan alteration is an increase in the size and branching of N-linked glycans (Fig. 1) [39]. Increasing β-1,6 branching arises from upregulation of N-acetylglucosaminyltransferase V (GnT-V) [9,42, 43]. In a lectin histochemical study, β-1,6 branching has been correlated with the pathological stage in human breast and colon neoplasia [44]. While using the Phaseolus vulgaris leukoagglutinating lectin (L-PHA) it was observed that increasing branching is associated with metastasis resulted in decreased survival time in patients having colorectal cancer [45]. Furthermore, β-1,6 branching offers additional sites for attachment of terminal sialic acid residues. Another cancer-related change is the upregulation of cell surface expression of specific monosaccharides (N-acetylneuraminic acid, fucose) [31]. A general increased activity of sialyltransferases and fucosyltransferases leads to augmentation in the sialic acid and fucose content in malignant cells [46]. For instance, fucosylated glycoproteins are elevated in ovarian, prostate and colorectal cancer [47–49]. Global sialylation is often expressed as an increase in α-2,6-linked sialic acids attached to the outer Galβ-1,4-GlcNAc units on N-glycans or to the inner GalNAc-α1-O-Ser/Thr units on O-glycans [9]. Several alterations in overexpression of specific carbohydrate antigens, such as Lewis carbohydrate antigens (LeX, LeY), sialyl-LewisX (SLeX) and polysialic acid, have been also reported in carcinomas (Fig. 2) [9,50]. In a clinicopathological and immunohistochemical study, an increased expression of SLeX correlating with survival of colorectal carcinoma patients has been reported [51].
Figure 1.
The most common N-glycan alteration observed in tumourigenesis (sialylation, increased β-1,6 branching and core fucosylation). Structure was drawn using a program GlycoWorkbench.
Figure 2.
The major tumour-associated glycan structures observed in various types of cancer (Lewis antigens and O-glycans). Carbohydrate structures were created in GlycoWorkbench.
3.2. Cancer biomarkers
A biomarker is defined by the National Institute of Health as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathological processes, or pharmacological responses to a therapeutic intervention” [52]. Ideally, a biomarker should be able to confirm specific disease cases, detect the early stages of a disease and should be easily detected in the patient’s blood, urine or tissues. Biomarkers can be used as a tool to distinguish various stages of the disease, as an indicator of disease prognosis and as an index of the intensity of disease or other physiological state in the organism [52,53]. As a result of changes in the protein expression corresponding with the risk of a disease, biomarkers can be specific molecules or genes, gene products, enzymes or hormones [54]. Development of novel biomarkers is crucial in medical/clinical practice in order to increase specificity and selectivity of disease prognosis.
The early detection of cancer plays the essential role in prognosis and patient survival and ultimately may affect the quality of patient’s life and the efficacy of used treatments. It is widely agreed that many of common biomarkers used nowadays in cancer diagnosis still lack either sensitivity or specificity or both (CA125, CA15-3, prostate specific antigen (PSA), carcinoembryonic antigen (CEA), etc.) [40]. These cancer biomarkers are glycosylated proteins that turn out to have aberrant changes in carbohydrate contents during carcinogenesis (Table 2). In order to find tumours at an early stage, before they spread and become incurable, advances in the fields of glycomics and proteomics can help [55]. In the future, glycan alterations may be used as a reliable biomarker and even indicate a progression of the disease.
Table 2.
List of cancer biomarkers with their aberrant glycosylation.
| Cancer | Biomarker | Type of biomarker | Glycan modification | Ref. |
|---|---|---|---|---|
| Ovarian | α1-acid glycoprotein | Glycoprotein | Sialylation/SLex | [56] |
| α1-antichymotrypsin | Glycoprotein | Sialylation/Slex | [56] | |
| α1-antitrypsin (AAT) | Glycoprotein | ↑α-2,6Neu5Ac; core fucosylation (α-1,6) | [57] | |
| CA125 (MUC16) | Glycoprotein (mucin) | Truncated Tn (O-linked), sTn, | [58] | |
| core fucosylation | [59] | |||
| Haptoglobin | Glycoprotein | Sialylation/Slex α-1,3Fuc, branching | [56,57] | |
| IgG | Glycoprotein | ↓galactosylation and sialylation | [56] | |
| Breast | CA15-3 (MUC1) | Glycoprotein (mucin) | Truncated Tn, sTn | [60] |
| Colorectal | β-haptoglobin | Glycoprotein | ↑fucosylation | [61] |
| CA19-9 | Glycolipid | High mannan structures | [62] | |
| Carcinoembryonic antigen (CEA) | Glycoprotein | ↑Lex, Ley; high mannan structures | [62,63] | |
| ↓core fucosylation | [64] | |||
| ↑branching, ↑Neu5Ac | [63] | |||
| Complement C3, kininogen-I | Protein | ↑Neu5Ac, Fuc | [65] | |
| Pancreatic | α1-β-glycoprotein | Glycoprotein | ↑Neu5Ac | [66] |
| Antithrombin-III | Glycoprotein | ↑sialylation and fucosylation | [67] | |
| β-haptoglobin | Glycoprotein | ↑fucosylation | [68] | |
| Kininogen-I | Protein | ↑sialylation and fucosylation | [67] | |
| Prostate | β-haptoglobin | Glycoprotein | ↑fucosylation and branching | [69] |
| Prostate specific antigen (PSA) | Glycoprotein | ↑α-2,3 Neu5Ac | [70] | |
| ↑α-1,2Fuc, β-GalNAc | [47] | |||
| Thyroid | Thyroglobulin (Tg) | Glycoprotein | Terminal galactosylation, | [71] |
| Asialylation | [72] | |||
| Liver | α1-antitrypsin (AAT) | Glycoprotein | Fucosylation | [73] |
| α-fetoprotein (AFP) | Glycoprotein | Fucosylation | [73] | |
| Transferrin | Glycoprotein | Fucosylation | [73] | |
| Lung | β-haptoglobin | Glycoprotein | SLex, ↑fucosylation | [69,74] |
Interestingly, among many various cancer biomarkers that have been reported in scientific publications, only nine cancer biomarkers have been approved by the US FDA for clinical use [75]. Importantly, all of the following biomarkers are glycosylated: AFP (liver cancer), CA125 and HE4 (ovarian cancer); thyroglobulin (thyroid cancer); PSA (prostate cancer); CEA (colorectal cancer); HER2/NEU and CA15-3/CA27-29 (breast cancer) [75,76].
3.2.1. Colorectal cancer
Colorectal cancer (CRC) is the third most commonly occurring cancer among men and women worldwide. Nowadays, flexible sigmoidoscopy, colonoscopy, double-contrast barium enema, and blood testing for CEA (also called CEACAM5) level are broadly used for screening [77]. Unfortunately, some of these techniques are invasive, painful and uncomfortable for patients. The normal range for CEA value in an adult non-smoker is < 2.5 ng mL-1 and for smoker < 5.0 ng mL-1; a rising CEA level is associated with occurrence of the cancer or metastasis. However, it has been previously reported that CEA lacks sensitivity and specificity in the range of 2.5–5.0 ng mL-1 [76]. CEA is an oncofetal, a 180-kDa glycoprotein, normally present in the membranes of mucosal cells, on the luminal surface of the adult colon, and overexpressed in adenocarcinomas, especially in colorectal cancer [78]. Due to a low specificity of tumour associated CEA, high levels of CEA expression have been observed in epithelial tumour in the lung, breast, thyroid and ovaries [78]. CEA has 28 N-linked glycosylation sites and during carcinogenesis exhibit abnormal glycosylation [61,76]. It has been noticed that CEA in CRC patients contained high level of the blood group antigens, Lewis X (LeX) and Lewis Y (LeY) [63,79]. Additionally, an increased mannose expression and branched N-glycans were observed, as well [63]. In another study [64], 347 individuals were analysed, including CRC and colorectal adenoma patients, and healthy individuals. Results indicated that the level of total core fucose residues present on proteins in the CRC patients was significantly decreased compared to the healthy individuals [64].
Glycoprofiling of β-haptoglobin is another biomarker for early detection of colorectal carcinoma. β-Haptoglobin is an acute phase protein secreted into plasma which binds free haemoglobin to prevent haemoglobin-induced oxidative injury in the vascular system [80]. Human β-haptoglobin consists of four N-glycosylation sites and one O-glycosylation site [61]. Generally, the level of serum β-haptoglobin is enhanced in several carcinomas, but its abnormal carbohydrate structure is different in various types of cancer [61,81]. From a colon cancer viewpoint, increased fucosylation of β-haptoglobin in patients with CRC compared to the healthy individuals was observed [61].
3.2.2. Ovarian cancer
Ovarian cancer is the most lethal cancer among gynaecologic malignancies affecting women in the Western world and has the highest mortality rate of all gynaecological cancer types [82–84]. Due to several factors, including morphological heterogeneity, the anatomical position of the ovaries within the abdominal cavity, and the fact that approximately 70% of ovarian cancer cases are detected in the advanced stage III or stage IV, ovarian cancer is associated with a poor prognosis [76,85]. Only about 19% of ovarian cancer cases are detected while still confined to the ovary and about 7% are diagnosed with pelvic spread. Unfortunately, the majority of ovarian cancer cases (68%) are diagnosed when the cancer spreads over the abdomen and extra-abdominal part [86]. Most patients with distant metastasis are usually treated with maximal cytoreductive surgery [87] followed by chemotherapy [86]. Early detection raises the 5-year survival rate up to 90% while a detection at the late stage provides only 10–20% survival rate [59]. Therefore, it is urgently needed to distinguish benign cases from the malignant ones..
To date, CA125 (also called MUC16) is a routinely used serum marker for detection, disease progression, and for evaluation of a response to treatment of ovarian cancer [82,88]. Despite of broadly used measurement of the CA125 level, analysis of this tumour-associated glycoprotein lacks the sensitivity and specificity and has a limited screening capability [89]. The main limitation comes from the fact that an elevated CA125 level has been found in benign cases, such as endometriosis, pregnancy, ovulatory cycles, and liver diseases [59]. Advances in MS, electrophoretic methods, hydrophilic interaction liquid chromatography (HILIC), and high performance liquid chromatography (HPLC) allowed to explore differences in glycosylation status of CA125 between the serum from patients with ovarian cancer and the healthy controls. It was found that there is an increase in core-fucosylated bi-antennary monosialylated glycans, as well as a decrease in non-fucosylated glycans in cancer patients compared to the control group [59]. In a follow up study using a microarray platform of analysis, specific aberrant O-glycoforms present on CA125 were observed. Glycoprofiling of CA125 showed a surface expression of sialyl-Thomsen-Friedenreich structures (sTn antigen, Neu5Acα-2,6-GalNAcα-O-Ser/Thr) and Thomsen-nouvelle antigen (Tn, GalNAcα-O-Ser/Thr) in patients with primary ovarian cancer (Fig. 2). This technique was able to distinguish benign cases from epithelial ovarian cancer with a specificity of 61.1% at 90% sensitivity [58].
HE4, a novel biomarker for efficient early stage detection of ovarian cancer in premenopausal patients [90], is slightly more specific compared to CA125 [91]. Several studies demonstrated that a combined clinical analysis of HE4 and CA125 could improve the sensitivity and the specificity of disease detection [91–93]. Recently, a study on ovarian cancer has reported aberrant changes in glycosylation status of β-haptoglobin, α1-acid-glyco-protein, α1-antichymotrypsin which contained elevated levels of sialyl Lewis X (SLeX) antigen. In this study it was also noticed that heavy chain of immunoglobulin (IgG) clearly showed core fucosylated agalactosylated biantennary glycan structures in patients with ovarian cancer, which was not present in the samples from the healthy control [56].
3.2.3. Prostate cancer
Prostate cancer (PCa) has been ranked globally as the second leading cause of death among men [83]. At present, prostate specific antigen (PSA) is a premier tumour biomarker available for prostate cancer diagnosis. Nevertheless, PSA is not considered as being a sufficiently specific biomarker for PCa detection in the diagnostic grey zone of 4–10 ng mL1, and does not provide a clear difference between benign and malignant cases [94]. Due to inherent limitations of PSA testing, the United States Preventive Services Task Force (USPSTF) against PCa screening has pointed out to an urgent need for novel diagnostic tool with a higher specificity and sensitivity [95]. The new auspicious molecular biomarkers, such as precursor forms of PSA (proPSAs), prostate health index (phi), prostate cancer antigen (PCA3), TMPRSS2- ERG gene, etc. are reflecting the growing efforts for improvement in clinical management of PCa [96,97]. Promising results of the new biomarkers generation show a possible supplementation or replacement of the PSA blood screening over time [98].
PSA (also known as hK3) is a secreted glycoprotein (serine-protease) with a single glycosylation site at Asn-45, containing approximately 8% of N-glycans [47]. Glycans attached to the PSA surface have been characterized as sialylated complex biantennary carbohydrates, mostly core fucosylated [99]. Circulating PSA includes formation of many molecular forms of PSA in human body: free PSA (fPSA), complexed PSA (cPSA) form with plasma proteins, especially serine protease inhibitor α1-antichymotrypsin, and inactive PSA (iPSA) [100].
Peracaula et al. presented N-glycan characterization of a normal PSA from seminal fluid and PCa cells using a sequencing analysis and mass spectrometry. The PSA from prostate cancer cells contained higher fucose amount, particularly α-1,2-Fuc-linked to galactose. Moreover, GalNAc was increased to 65% in the cancer samples, whereas in the control samples the PSA contained only 25% of GalNAc on carbohydrate structures [47]. Interestingly, another study based on elucidation of the structure of PSA purified from human seminal fluid revealed differential binding of free serum PSA to Maackia amurensis lectin. M. amurensis lectin recognizing α-2,3-linked sialic acid was increasingly bound to the prostate cancer samples compared to the benign prostate hypertrophy patients [70]. Kuno et al. published a novel practical system for glycan analysis with the platform based on antibody-assisted lectin profiling. In this study, a single antibody assisted in: (i) immunoprecipitation; (ii) Western blotting; and (iii) glycan profiling by antibody-lectin microarray. Surprisingly, a drastic decrease in α-2,6 sialylation and an increase in terminal α-2,3Neu5Ac on PSA from the PCa cell lines were observed [101].
Since it is known that β-haptoglobin level is significantly enhanced in various types of cancer, the analysis of this acute phase protein is widely studied [61, 68]. Fujimura et al. analysed the glycosylation status of β-haptoglobin in serum of PCa patients, benign prostate cases, and normal subjects. They noticed enhanced branching as well as antenna fucosylation at N-glycans in the PCa patients [69].
3.3. Current methods for glycoprofiling in cancer research
At present, there has been a rapid increase in number of techniques which can be applied to glycoprofiling of biomarkers in various types of cancer. Plentiful studies have employed advanced mass spectrometry (MS), liquid chromatography (LC), lectin or/and antibody-based methods, microarrays techniques, electrochemical investigation, capillary electrophoresis (CE), microfluidic platforms, etc. In order to obtain a detailed, clear, and atomistic structure of glycans, MS based on matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI) has been widely used. Samples are subsequently separated by mass/charge ratio using time of flight (TOF), Fourier transform ion-cyclotron resonance (FT-ICR) or quadrupole-based approaches, and finally analysed. Importantly, the purity of samples is an important requirement for the MS analysis, thus pretreatment methods are needed. The majority of the MS methods utilize enzymatic or chemical release of glycans from a protein backbone with a subsequent glycan modification (labeling, permethylation, separation, etc.), followed by MS analysis [85]. MS has recently emerged as an important tool in identification of novel glycan biomarkers due to fast and efficient analysis, high sensitivity and small sample volume. Nevertheless, structural complexity, heterogeneity, and vast variation of potential carbohydrate structures require manual interpretation and time-consuming data evaluation [40].
Another promising strategy for investigation of the glycan structures are: lectin-based platforms (lectin affinity chromatography, lectin microarrays, enzyme-linked lectin assays (ELLA), lectin histochemical staining, lectin blotting). The obvious benefit of application of lectin arrays is high-throughput detection of a minute amount of sample without a need for glycan removal from a glycoconjugate prior to analysis [17,18,40]. This approach is a convenient biorecognition tool for distinguishing between the normal and tumourigenic glycosylation patterns. Additionally, these lectin-based techniques have been used for biomarker detection and for visualization of cell-surface carbohydrates [13]. Unfortunately, lectin-based formats are associated with relatively low sensitivity of analysis and require application of fluorescent labels [40]. Thus, other techniques, such as surface plasmon resonance (SPR) platforms, flow cytometry, and microfluidic systems, are increasingly employed for investigation of binding affinities, kinetic parameters, biospecific interactions, and real-time quantitative measurements in the field of glycomics [102,103].
Recently, nanotechnology has been increasingly utilized to develop reliable, rapid and sensitive tools for biomarker detection. Due to the fact that nanomaterials exhibit unique optical, chemical, mechanical, and physical properties, it is not surprising that nanomaterials have been applied in biomarker analysis [104,105]. One of the most extensively studied nanomaterials are gold nanoparticles (GNPs). Since GNPs have special optical properties, can absorb and scatter light from the visible to near-infrared region, they are widely used as stable molecular imaging agents [104,106]. For instance, nontoxic nanoparticles were applied for in vivo tumour targeting and detection [105].
Furthermore, quantum dots (QDs), semiconducting, light-emitting nanocrystals, are widely exploited for multiplexed molecular diagnosis [105], for in vivo imaging [107], and for drug delivery [104,108]. QDs have several exceptional properties, such as resistance against photobleaching, simultaneous excitation of multiple fluorescence dyes, and nanometer scale size [104,107,108]. Among nanomaterials, nanoparticles like carbon nanotubes (CNTs) [109,110], graphene [111–113], and nanowires have been used most frequently [13,114,115]. CNTs modulate redox interfacial properties, allow label-free detection and could be easily functionalize with many functional groups [104]. Besides, CNTs modified with multifunctional dendrimers can be applied as platforms in biomedical sensing, diagnosis, and for therapeutic purposes [13]. Park et al. designed a biosensor able to detect CEA biomarker using single-walled CNT field effect transistors (SWCNT-FETs) [116].
4. Biosensor technology
Within the last few decades biosensing has become a rapidly developing field involving knowledge from the fields of chemistry, biochemistry, biomedicine, biotechnology, and material sciences [117, 118]. Biosensors with high sensitivity and specificity, of nanoscale size and high speed, simple handling, and using a small volume of sample are rapidly developed in cancer research [119]. With advanced properties of nanomaterials and processing power of micro/opto-electronics, biosensors represent suitable analytical tools which can detect low concentrations of analytes [120–124].
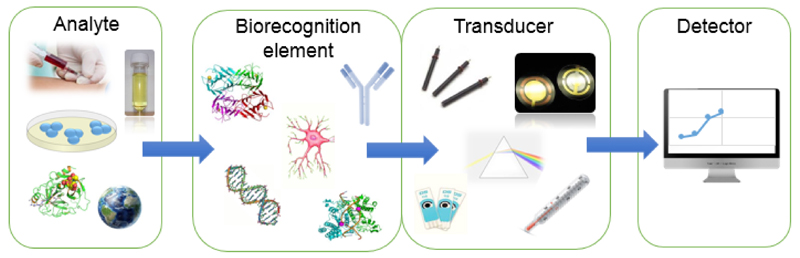
The basic feature of a biosensor is the interaction of an analyte with a biorecognition element, which is in direct contact with a physico-chemical transducer. The detector transduces a physico-chemical signal into an electric output signal, proportional to the analyte concentration (Fig. 3). Biological molecules, including enzymes, antibodies/antigens, oligonucleotides, receptors, etc. [125] translate the information from the biochemical domain into a chemical or physical output signal [126]. Biosensors are mostly classified into several groups according to their transduction: (i) electrochemical (amperometric, impedimetric, potentiometric), (ii) optical, (iii) massdetecting (piezoelectric, acoustic) and (iv) enthalpic [127]. Additionally, biosensors may be divided into two main groups based on the type of a biorecognition element. The first type are catalytic biosensors (enzymes, various types of cells and tissues) and the second group are affinity biosensors applying antibodies, receptors, lectins, and nucleic acids/aptamers as the biorecognition elements [120].
Figure 3.
A scheme of the biosensor with an analyte, a biorecognition element, a transducer, and a detector.
Electrochemical platforms in biosensing constitute a low cost, fast, and sensitive approach to the investigation of interactions between different biomolecules, which is applied to development of molecular electronic devices and in the field of biotechnology, medicine, and pharmacy [13,119,128]. The common electrochemical techniques include cyclic voltammetry (CV), potentiometry, amperometry, and electrochemical impedance spectroscopy (EIS). Additionally, differential pulse voltammetry (DPV) and square-wave voltammetry have also been applied quite frequently [13]. In recent decades, nanomaterials have been increasingly applied in many fields (molecular electronics, (bio)sensors, optical communications, quantum dot-based devices, biomedical applications, photoelectrochemical cells, etc.) [129]. Nanomaterials exhibit unique properties, such as reactive surfaces, coercive force in magnetic materials, nanoscale size, and high strength [130]. Many research groups have utilized various nanomaterial-based strategies to improve the detection sensitivity (e.g. graphene, nanoparticles, CNTs and others) [131].
4.2. Electrochemical impedance spectroscopy (EIS)
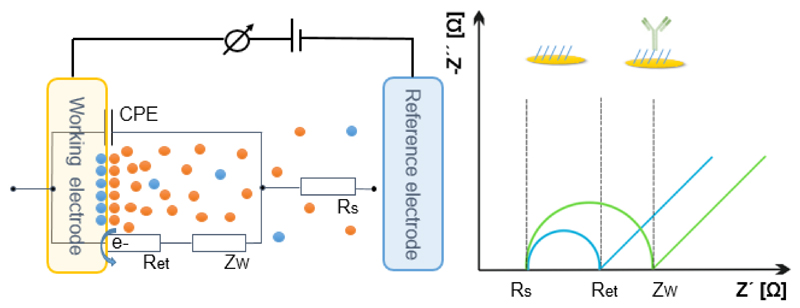
EIS is a powerful technique working in a label-free mode for probing interfacial interactions on surface-modified electrodes [125]. This method was pioneered by Sluyters-Rehbach et al. in 1969 [132]. Basically, a small sinusoidal potential (2–10 mV) is applied to an electrochemical cell, and the response of the resulting current is measured [13]. Consequently, impedance is calculated as the ratio between the system voltage U(jω) and the current signal I(jω), where and ω and f are the excitation frequencies expressed in units rad s-1 and Hertz (Hz), respectively [125]. A common way to represent a complex impedance is to use a sum of the real (resistive) component of the impedance ZRe(ω) and the imaginary (capacitive) part ZIm(ω). In analogy to Ohm’s law, the electrochemical impedance is given by:
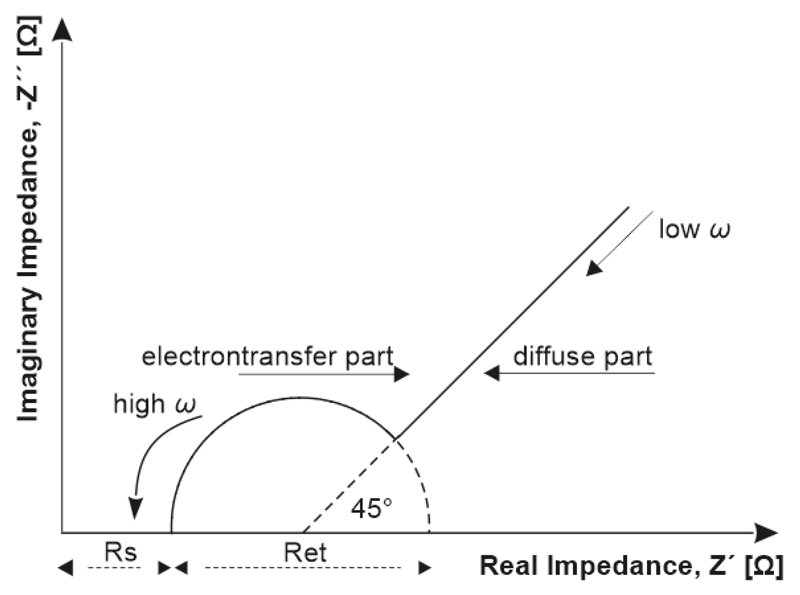
The impedance data are typically represented in a Nyquist plot showing dependence of the real impedance on the imaginary impedance (Fig. 4). A Nyquist diagram provides a visual insight into the dynamics of an electrochemical system with a semicircle and a linear part. The semicircle part, at higher frequencies, matches up with an electron transfer-limited process whilst the linear part, at lower frequencies, expresses the diffusion-limited process [13]. If a very fast electron-transfer process is present, the Nyquist impedance plot could include only a linear region. On the other hand, in case of a very slow electron-transfer process, a large semicircle part without a straight line region is obtained [125]. A typical equivalent circuit (according to the Randles and Ershler model [133]) can provide the resistance of the electrolyte solution, Rs, the double layer capacitance CPE, the Warburg coefficient Zw, and the electron transfer resistance, Ret (Fig. 5).
Figure 4.
A typical Nyquist plot made from EIS investigation to obtain key EIS characteristics.
Figure 5.
A schematic equivalent circuit with electrochemical processes occurring on the working electrode surface (on left); Immunosensor based on EIS with an increase in Ret after successful immobilization of antibody (on right).
Using a redox couple, typically a mixture of ferricyanide and ferrocyanide, the change in the charge transfer resistance Ret is obtained. Generally, the charge transfer resistance is inversely proportional to the rate of electron transfer. The double layer capacitance (CPE) and the charge transfer resistance (Ret) describedielectric and isolation features of electrode-electrolyte interface. However, the electrolyte resistance (Rs) and the Warburg impedance (Zw) characterize the properties of an electrolytic solution and diffusion limitation for redox probe to reach the electrode surface and do not affect electron transfer at the electrode surface. The detection in the broad frequencies range (10−4−106 Hz) makes the EIS strategy useful for diffusion analysis and for providing kinetics characteristics [92,118]. Generally, at low frequencies (f < 1 mHz) the impedance is determined by the DC-conductivity of the electrolyte solution and at higher frequencies (f > 100 kHz), inductance of the electrochemical cell and connecting wires dominate the system [125].
Label-free monitoring is an indisputable advantage of the EIS analysis. It was observed that the labeling process might affect the bioaffinity between the probes and their target, resulting in a false positive/negative results [37,134,135]. Furthermore, the EIS detection provides very low limits of detection (pM-aM range) [135]. Due to its high sensitivity and label-free characteristics, EIS is gaining in popularity in lectin-carbohydrate, protein-glycan, and antibody-antigen studies. Additionally, EIS has been tremendously utilized in study of corrosion, electrodeposition, batteries, and fuel cells [136]. EIS methods are employed in development and examination of DNA-sensors as well. DNA-sensor devices are applied in gene analysis, tissue matching, analysis of genetic disorders, and in the field of forensic [125]. The remaining challenges in the DNA-based biosensors are the modification of surfaces with different DNA-composites and specific detection of single-base mismatches in DNA sequences [125].
4.3. Immobilization techniques in biosensor design
Immobilization of biomolecules plays a critical role in the fabrication of the biosensors and can greatly influence their performance. Biomolecules can be attached to the biosensor surface using several techniques, such as covalent immobilization, physical adsorption, bioaffinity binding, entrapment in gel, and crosslinking by a multifunctional reagent [137]. Physical adsorption of biomolecules is a reversible, simple, and fast process exploiting non-covalent interactions (hydrophobic forces, ionic binding, hydrogen bonding, and van der Waals interactions) [138,139]. Besides, adsorbed biomaterials are highly susceptible to the environmental changes (pH, ionic strength, temperature). Despite of the simplicity and the short processing time the drawbacks of the immobilization method are: unfavorable orientation, decreased functionality/stability of biomolecules, and weak binding [139].
Covalent immobilization involves direct covalent binding between biomolecules and a biosensor surface [139]. Usual working surfaces for covalent immobilization are chosen from a relatively small group of materials: metals (gold, silver, platinum), natural hydroxylic polymers, different carbonaceous surfaces (graphene, glassy carbon), glass, and silica [137]. Covalent immobilization may utilize chemical modification of the surface to create reactive functional groups, which react with the biorecognition molecules. Nowadays, amine coupling and thiol coupling are the most common immobilization methods in biosensing [140,141]. Light-assisted immobilization, utilizing photolabile agents forming covalent bonds upon UV light activation, and click chemistry, involving cycloaddition of an azide and an alkyne, are other examples of covalent immobilization techniques [139,142].
4.4. Self-assembled monolayers (SAMs)
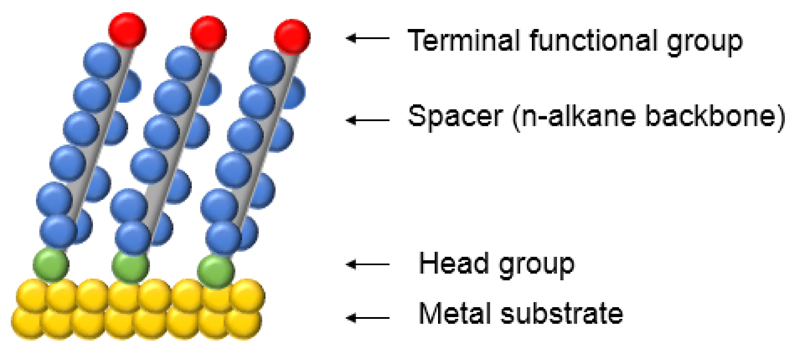
Self-assembled monolayers (SAMs) provide a well-studied, convenient and simple system allowing for the density and thickness control at the transducer surface [139]. SAMs are organic assemblies spontaneously formed by strong adsorption onto a solid surface (gold, silver, platinum, etc.) [143]. Modification of the gold surface using alkanethiols with different terminal functional groups (-SH, -NH2 or -COOH) is currently the best characterized model in the SAMs strategy (Fig. 6) [139,144]. Gold is broadly used in electrochemistry as a standard substrate for SAMs due to several factors: (i) gold is a reasonably inert metal resisting oxidation, (ii) gold has fewer defects, as a substrate for SAMs fabrication, than silver or copper, (iii) gold is easily available as a thin film and as a colloid, (iv) gold substrate can be simply fabricated with different patterns by chemical and lithographic tools, or their combination [128,143,145].
Figure 6.
A schematic diagram of SAMs on metal surface.
A highly packed and ordered SAM can be prepared on surfaces using gold-thiol bonds [144]. Moreover, the stability of the sulphur bond with gold makes the thiol-gold chemistry convenient for further immobilization reactions. It was observed that the chain length affects the organization of the monolayers. Hence, the addition of shorter alkyl derivatives may decrease the density of the coverage, shows a better electron transfer from the soluble redox probes and improves the electrochemical response of proteins and other substances [139]. Due to the high affinity of thiols for the noble metals, alkanethiols are useful and highly tunable chemicals for biosensor surface modifications [143].
Additionally, SAM formation is highly sensitive to the pre-treatment of the metal surface. Several techniques of metal surface cleaning have been previously reported [128,146]. Such protocols may include electrochemical cleaning (reductive desorption of previously bound adsorbates, electrochemical polishing and gold oxide stripping), mechanical polishing of the electrode, chemical treatment with hot piranha solution (a mixture of concentrated H2SO4 and concentrated H2O2 in a 3:1 ratio), thermal methods, etc. [128,147]. Furthermore, in a comparative study of nine gold cleaning methods, a solution of KOH + H2O2 combined with the potassium hydroxide potential sweep method have been found to deliver the cleanest gold surface [148].
The initial adsorption of alkanethiols on gold surface takes only from milliseconds to minutes, but a slow reorganization phase which follows takes several hours to maximize the coverage density and minimize the defects in the SAM [143]. The most commonly used procedure for preparation of SAMs on gold, silver, and other materials is immersion of thiols for 12–18 h at room temperature [143]. The alkyl chains tilt at an angle of 20–30° from the surface normal [143,144]. Details about the mass coverage and organization of SAM have been provided using several techniques, such as atomic force microscopy (AFM), X-ray photoelectron spectroscopy (XPS), electrochemistry, scanning tunneling microscopy (STM), contact angle goniometry, advanced mass spectroscopy, etc. [139,143,144].
4.5. Amine coupling
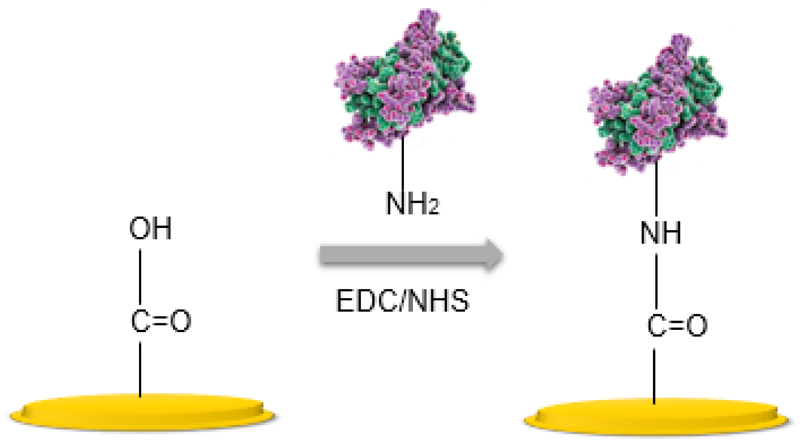
Surface modification with SAMs allows for simple regeneration of the active surface and controls the organization of the attached probes. However, in order to successfully bind biomolecules, another step is required; i.e. activation of the terminal functional groups. One of the most widely used approaches to the biomolecule attachment is covalent binding via activation agents, such as 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) or N,N’-dicyclohexylcarbodiimide (DCC) and N-hydroxysulfosuccinimide (NHS) [127,139]. With amine coupling, carboxyl groups on the surface of the functionalized material are first activated to give a reactive succinimide esters, then the ester spontaneously reacts with a primary amine of the biomolecules (Fig. 7). EDC/NHS chemistry is a pH dependent strategy and in case of protein immobilization the protein solution should have a pH below the pI of the protein in order to maximize electrostatic forces between protein and the negatively charged carboxyl groups [142].
Figure 7.
A scheme of covalent attachment of the biomolecules via EDC/NHS surface chemistry.
4.6. Antibody assisted lectin glycoprofiling
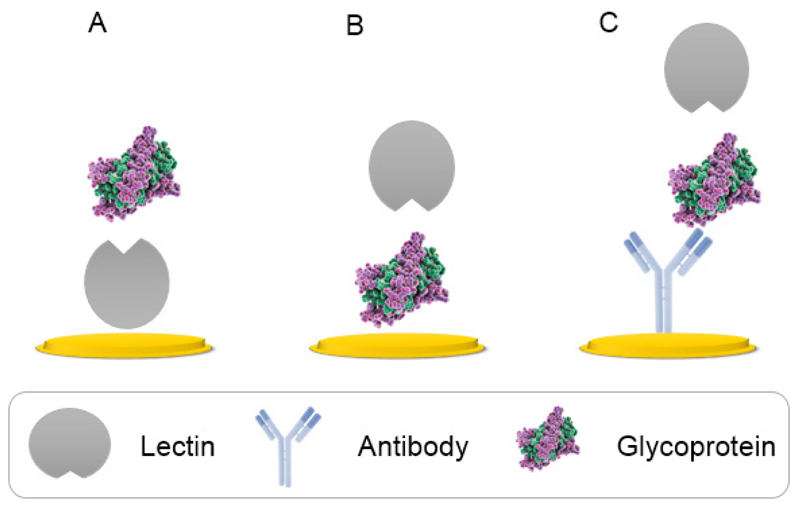
To date, practically three different lectin-based formats have been employed for studying glycosylation patterns in bioanalysis (Fig. 8). The direct lectin-based detection utilizing an immobilization of lectin onto a solid surface followed by an incubation with glycoprotein (Fig. 8A), an inverse format with adsorption of glycoproteins first with a subsequent incubation with lectin (Fig. 8B) and a sandwich configuration based on the capture of antibody (Ab) recognizing a glycoprotein on the surface, followed by an addition of a glycoprotein of interest and finally the incubation with lectin for the complete biorecognition to take place (Fig. 8C) [37].
Figure 8.
Configuration of lectin-based analysis with applied direct (A), reverse (B) or sandwich immobilization protocol (C).
Antibody-lectin based arrays combine the advantages and unique features of lectins as biorecognition elements with immunoreactions exhibiting high sensitivity towards analytes. Thus, the glycoprofiling of a particular biomarker can be highly specific even in quite complex samples. Such valuable strategy is known as the antibody-lectin sandwich array (ALSA), pioneered by Chen et al. in 2007 [149]. This strategy allows for the measurement of the glycosylation patterns on specific glycoproteins, which is required for the early-stage cancer diagnostics. Hence, ALSA is a well suited, sensitive and comprehensive approach for detection of glycan alteration on a biomarker surface [150].
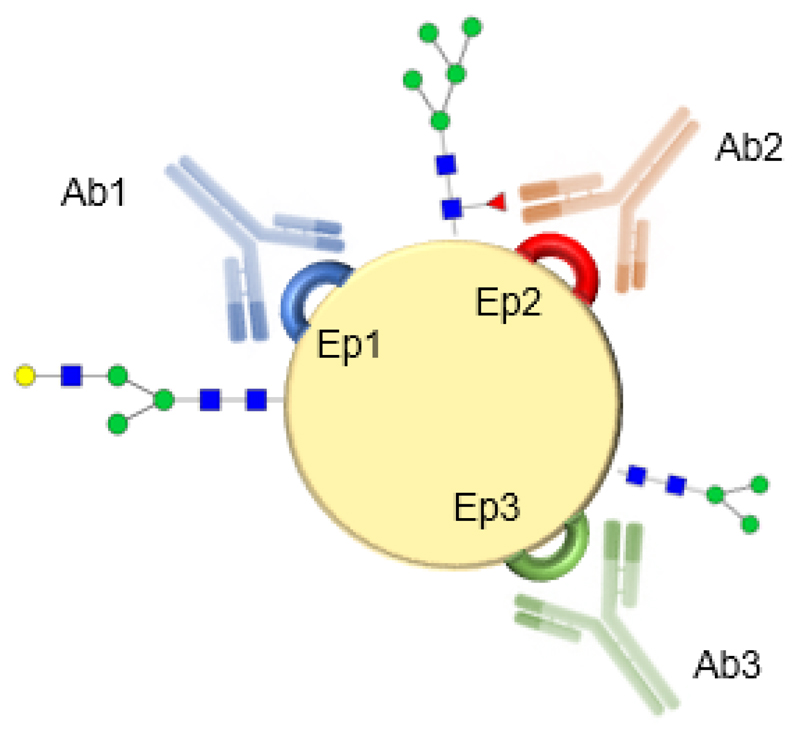
In the antibody related platforms the affinity constant and cross-reactivity of antibodies should be considered [151]. There are two types of antibodies, polyclonal and monoclonal. Monoclonal antibodies are highly specific to one epitope on an antigen and require quite a sophisticated production technology. Moreover, the production of monoclonal antibodies is more expensive and prolonged in comparison to polyclonal ones. On the other hand, polyclonal antibodies recognize multiple epitopes on any antigen [151]. Polyclonal antibodies for subsequent glycoprofiling of antigen have to be selected in a way that glycan can be still available for lectin binding after antibody is bound to the antigen (Fig. 9).
Figure 9.
An effective glycoprofiling of an antigen (biomarker) based on multiple epitopes targeting. Polyclonal antibodies will be selected in way to do not interfere with glycoprofiling of a particular glycan moiety by a lectin i.e. Ab3 will be selected for attachment of a biomarker for subsequent glycoprofiling of glycan close to epitope Ep1 by lectin.
The ALSA platform has naturally a few limitations, such as the lack of information about the precise character of the glycan structures and diversity of glycans at various attachment sites [150]. The ALSA can be directly built up on the already developed immunoassay formats with numerous advantageous characteristics [131], especially when combined with the electrochemical detection methods [151]. Thus, the ALSA approach with the support of advanced mass spectrometry could provide a reliable, highly effective, and detailed analysis of glycoproteins.
4.7. Recent impedimetric-based strategies in glycan analysis
Biosensors based on the electrochemical transduction mechanism have recently emerged as an efficient tool in glycan analysis. Here we provide the current trends in the electrochemical label-free detection of glycans and discuss discuss various approaches in biosensor construction (Table 3).
Table 3.
Summary of selected parameters of impedimetric biosensors.
| Target | Linear range | Limit of detection | Ref. |
|---|---|---|---|
| Asf/ Fet | 13 fM - 15 pM | 13 fM | [152] |
| AFP | 1-100 ng mL-1 | 0.1 ng mL-1 | [153] |
| SG from patients infected by DF | 10-80 dilution fold | 80 dilution fold | [154] |
| Fet/ pancreatic cancer cells (BXPC-3) | n/a | 20 fM | [155] |
| SG from patients with RA | aM - nM | 1 aM | [156] |
| Human liver cancer cells Bel-7404 | 103-106 cells mL-1 | 234 cells mL-1 | [134] |
| PSA | 152 fM - 3.65 pM | 50 fM | [160] |
| Cell line K562 | n/a | 106 cells mL-1 | [161] |
Asf: asialofetuin, Fet: fetuin, AFP: α-fetoprotein, SG: serum glycoproteins, DF: dengue fever, RA: rheumatoid arthritis PSA: prostate specific antigen.
For the first time, a label-free EIS in conjunction with lectins, was developed by La Belle and co-workers for detection of a glycan-lectin interaction [152]. A galactosebinding lectin (PNA) and sialic acid-binding lectin (SNA) were covalently attached to the layered Cu/Ni/Au printed circuit board electrodes with a subsequent incubation of the lectin-modified electrode with artificial and natural glycoproteins. The artificial and natural glycoconjugates consisted of: (i) gold nanoparticles encapsulated with TF-antigens; (ii) the glycoprotein asialofetuin (Asf) containing both LacNAc (Galβ-1,4GlcNAc) and TF-antigen; and (iii) fetuin (Fet) glycoconjugate, the sialylated glycoform of Asf. The EIS measurements carried out in the presence of the redox couple ferrocyanide/ferricyanide demonstrated that TF-GNP and glycoprotein Asf were rapidly and reliably detected down to 1 pg mL-1 (13 fM) concentration on the PNA-modified electrodes, while the SNA electrodes yielded no response. Fet glycoprotein was detected on the SNA-modified electrodes with a limit of detection down to 10 pg mL-1 (150 fM) [152].
A label-free EIS investigation was utilized for a sensitive determination and evaluation of α-fetoprotein (AFP), a reliable biomarker for hepatocellular carcinoma [153]. The EIS biosensor was designed by adsorbing carboxyl-functionalized single-wall carbon nanotubes (SWNTs) onto a screen-printed carbon electrode with the WGA lectin being immobilized as a biomolecular recognition element. In order to block the surface active sites on the SWNT-modified electrode, bovine serum albumin (BSA) was applied for 30 min. Upon binding of AFP to a WGA-modified electrode, Ret response was increased with a linear proportion to the logarithm of the AFP concentration in the range from 1 ng mL-1 to 100 ng mL-1, with a limit of detection of 0.1 ng mL-1. In this study, the electrochemical measurements were carried out with the ferrocyanide/ferricyanide redox probe [153].
Oliveira et al. used a electrochemical biosensor for examination of serum glycoproteins from patients infected by dengue fever and from healthy individuals [154]. With an emphasis on improvement of the sensitivity of glycans detection, they modified the gold electrodes using the sol-gel method with integration of GNPs and a polymer. Furthermore, the electrodes were treated with lectin Con A and blocking agent BSA. The results showed a large Ret increase after binding of glycoproteins from the infected patients, and on the other hand a smaller increase in Ret obtained from binding of glycoproteins from the healthy (control) individuals [154].
An ultrasensitive diagnostic platform called “NanoMonitor” has been developed by Nagaraj´s group [155]. The surface of a silicon chip with an array of gold electrodes was modified with a nanoporous alumina membrane on the top of each electrode. Using lectins SNA and MAA subtle glycosylation alterations of Fet and human pancreatic cancer cell line (BXPC-3) were identified. The data resulted from the NanoMonitor platform were correlated very well with a conventional laboratory technique ELLA. Due to approximately five orders of magnitude lower limit of detection of the biosensor (20 fM) compared to ELLA, a very short assay time (15 min) and a small sample consumption (10 μl), the NanoMonitor device has a great potential in clinical applications [155].
Recently, our group constructed an impedimetric biosensor for the glycoprofiling of human serum [156]. Three different lectins (SNA, RCA, Con A) were covalently immobilized on the activated mixed SAMs layer (11-mercaptoundecanoic acid mixed with a betaine terminated thiol to avoid nonspecific interactions). The glycobiosensor was capable of detecting extremely low concentrations of glycoproteins, especially in a sandwich configuration (down to 1 aM). The EIS measurements revealed the distinct glycan pattern in patients with rheumatoid arthritis and in healthy controls. Furthermore, a non-specific interaction of proteins for the Con A modified electrode was only 6.1% [156]. Reproducibility of assays by our EIS biosensors, expressed as an average relative standard deviation (RSD) of analysis in a diluted serum sample, is around 28%, which is a value similar to the other devices based on the screen printed electrodes having an average RSD of 19% [157] or 27% [158]. It is worth noting that this RSD is not the RSD of the assay itself, but rather it expresses the reproducibility of the biosensor preparation involving numerous steps. In a recent study it was shown that reproducibility of an assay by the EIS biosensor can be quite high with an average RSD of 4.1% or 7.8% for two different analytes for numerous electrodes present on the same chip (i.e. having an array of electrodes on the chip) and that the chip to chip reproducibility of an analysis for the same two analytes was 7.0% or 11.2% [159]. When a proper modification of the lectin EIS biosensor will be carried out using efficient blocking agents (i.e. either based on proteins or other molecules having betaine or glycol moieties), the device should work properly with complex samples such as human serum. Even though the lectin EIS-based biosensors seem to be promising in analysis of a wide range of analytes and samples, so far it is very difficult to say whether the EIS-based lectin biosensors could be applied in a routine clinical assays in a future.
A novel label-free electrochemical impedance spectroscopy biosensor exploiting the interactions between glycans and lectin in order to analyse the carbohydrate expression on cancer cells was developed [134]. Firstly, the pre-treated electrodes were incubated with activated Con A with a subsequent immobilization of human hepatocellular carcinoma cells (Bel-7404) and normal liver cells (L02). This biosensor allowed to detect cancer cells with a detection limit of 234 cells mL-1 and was able to distinguish between the cancer cells and the normal liver cells [134].
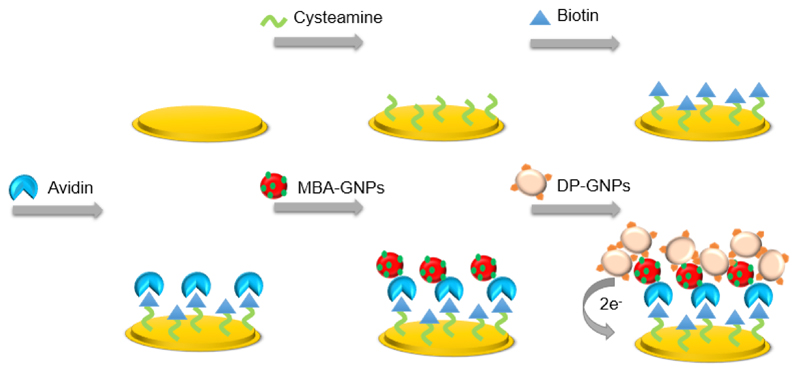
A sub-pM detection limit has been reached by a sandwich-type electrochemical biosensor based on a dual-amplification of 4-mercaptophenylboronic acid-capped gold nanoparticles (MBA-GNPs) and dopaminegold nanoparticles (DA-GNPs) [160]. At first, the cysteamine SAMs were formed on a gold disk electrode, followed by covering with a biotin-avidin containing film (Fig. 10). The capture of PSA was carried out by immobilization of thiolated single strand DNA1 (ssDNA1) and a subsequent application of a given concentration of PSA. ssDNA or RNA aptamers could serve as biorecognition elements able to strongly bind to various proteins, even whole cells [160]. Due to a simple and controllable chemical modification and long-term stability, aptamers have distinct advantages over antibodies.
Figure 10.
A scheme of the sandwich-type biotin-avidin detection based on dual amplification of MBA-GNPs and DA-GNPs. Reprinted from Biosensors and Bioelectronics, Copyright (2013), with permission from Elsevier [160].
In the last report presented here, a microfluidic platform for analysis of the multi-glycan expressions on a cell line K562 was used [161]. In this study, indium tin oxide electrodes were modified with a poly(diallyldimethyl-ammonium chloride) (PDDA) aqueous solution and a GNPs solution. In this step, the negatively charged GNPs were easily adsorbed on the PDDA-modified electrode surface with a positive charge. In the following step, the electrodes were incubated with Con A, PNA or WGA and BSA for 30 min. Diverse expression of carbohydrates on the cell surface was confirmed by the EIS measurement as a decreased binding ability in the order WGA < Con A < PNA. [161].
5. Conclusions
As discussed in this article, cancer development and progression is associated with altered glycosylation. Changes in glycosylation machinery observed in various cancers were briefly described. One of the most common pathological alterations in glycosylation is an increase in the size and β-1,6 branching of N-linked carbohydrates with an increased activity of sialyltransferases and fucosyltransferases.
Since the construction of lectin biosensors is a subject of a particular interest, EIS as a reliable, highly robust and sensitive detection method represent highly applicable tool in glycan patterning for early cancer diagnosis. Moreover, the EIS platform offers a label-free mode of operation while providing low-cost analysis and small sample consumption.
Additionally, a combination of ultrasensitive electrochemical detection with advanced mass spectrometric techniques can identify novel prospective cancer biomarkers. Such a combined effort can provide an elegant way for glycoanalysis of cancer biomarkers with tremendous potential for highly effective cancer monitoring.
Acknowledgement
The financial support from the Slovak research and development agency APVV 0282-11 and VEGA 2/0162/14 is acknowledged. The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement no 311532 and this work has received funding from the European Union’s Seventh Framework Program for research, technological development and demonstration under grant agreement no 317420.This publication was made possible by NPRP grant # 6-381-1-078 from the Qatar National Research Fund (a member of Qatar Foundation). The statements made herein are solely the responsibility of the authors.
Contributor Information
Dominika Pihíková, Department of Glycobiotechnology, Institute of Chemistry, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 38 Bratislava, Slovakia.
Peter Kasák, Center for Advanced Materials, Qatar University, P.O.Box 2713 Doha, Qatar.
Jan Tkac, Department of Glycobiotechnology, Institute of Chemistry, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 38 Bratislava, Slovakia.
References
- 1.Ghazarian H, Idoni B, Oppenheimer SB. A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2011;113:236–247. doi: 10.1016/j.acthis.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirabayashi J, Yamada M, Kuno A, Tateno H. Lectin microarrays: concept, principle and applications. Chem Soc Rev. 2013;42:4443–4458. doi: 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]
- 4.Tong L, Baskaran G, Jones MB, Rhee JK, Yarema KJ. Glycosylation changes as markers for the diagnosis and treatment of human disease. Biotechnol Gen Eng Rev. 2003;20:199–244. doi: 10.1080/02648725.2003.10648044. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Boons G-J. Carbohydrate recognition: Biological problems, methods and applications. John Wiley & Sons, Inc; 2011. [Google Scholar]
- 6.Tkac J, Bertok T, Nahalka J, Gemeiner P. Perspectives in glycomics and lectin engineering. Methods in Molecular Biology. 2014;1200:421–445. doi: 10.1007/978-1-4939-1292-6_37. [DOI] [PubMed] [Google Scholar]
- 7.Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:37. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 8.Choi E, Hill MM. Targeted high-throughput glycoproteomics for glyco-biomarker discovery. Integrative Proteomics InTech. 2012 [Google Scholar]
- 9.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al. Essential of glycobiology. 2nd. Cold Spring Harbor Laboratory Press (NY); 2009. [PubMed] [Google Scholar]
- 10.Nelson DL, Cox MM. Chapter 7 in Lehninger principles of biochemistry. 4. W.H. Freeman & Company; 2004. Carbohydrates and glycobiology. [Google Scholar]
- 11.Sharon N, Lis H. Carbohydrates in cell recognition. Sci Am. 1993;268:82–89. doi: 10.1038/scientificamerican0193-82. [DOI] [PubMed] [Google Scholar]
- 12.Cummings RD, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21:1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuel NF, Mu B, Zhang J, Hinckley A, Strano MS. Nanoengineered glycan sensors enabling native glycoprofiling for medicinal applications: towards profiling glycoproteins without labeling or liberation steps. Chem Soc Rev. 2012;41:5744–5779. doi: 10.1039/c2cs35142k. [DOI] [PubMed] [Google Scholar]
- 14.Alley WR, Mann BF, Novotny MV. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem Rev. 2013;113:2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur CM, Cummings RD, Stowell SR. Using glycan microarrays to understand immunity. Curr Opin Chem Biol. 2014;18:55–61. doi: 10.1016/j.cbpa.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissner A, Anish C, Seeberger PH. Glycan arrays as tools for infectious disease research. Curr Opin Chem Biol. 2014;18:38–45. doi: 10.1016/j.cbpa.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Gemeiner P, Mislovicova D, Tkac J, Svitel J, Patoprsty V, Hrabarova E, et al. Lectinomics II. A highway to biomedical/ clinical diagnostics. Biotechnol Adv. 2009;27:1–15. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Katrlik J, Svitel J, Gemeiner P, Kozar T, Tkac J. Glycan and lectin microarrays for glycomics and medicinal applications. Med Res Rev. 2010;30:394–418. doi: 10.1002/med.20195. [DOI] [PubMed] [Google Scholar]
- 19.Palma AS, Feizi T, Childs RA, Chai W, Liu Y. The neoglycolipid (NGL)-based oligosaccharide microarray system poised to decipher the meta-glycome. Curr Opin Chem Biol. 2014;18:87–94. doi: 10.1016/j.cbpa.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Gildersleeve JC, Blixt O, Shin I. Carbohydrate microarrays. Chem Soc Rev. 2013;42:4310–4326. doi: 10.1039/c2cs35401b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson CL. Lectins: Analytical technologies. Oxford: Elsevier; 2007. [Google Scholar]
- 23.Boyd WC. The protein of the immune reactions. The Proteins. 1954;2:756–844. [Google Scholar]
- 24.Lis H, Sharon N. Lectin-carbohydrate interactions. Curr Opin Struct Biol. 1991;1:741–749. [Google Scholar]
- 25.Mody R, Joshi S, Chaney W. Use of lectins as diagnostic and therapeutic tools for cancer. J Pharmacol Toxicol Methods. 1995;33:1–10. doi: 10.1016/1056-8719(94)00052-6. [DOI] [PubMed] [Google Scholar]
- 26.Arnaud J, Audfray A, Imberty A. Binding sugars: from natural lectins to synthetic receptors and engineered neolectins. Chem Soc Rev. 2013;42:4798–4813. doi: 10.1039/c2cs35435g. [DOI] [PubMed] [Google Scholar]
- 27.Minko T. Drug targeting to the colon with lectins and neoglycoconjugates. Adv Drug Deliv Rev. 2004;56:491–509. doi: 10.1016/j.addr.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Varrot A, Blanchard B, Imberty A. Lectin binding and its structural basic. In: Wang B, Boons G-J, editors. Carbohydrate Recognition: Biological Problems, Methods and Applications. Wiley; 2011. [Google Scholar]
- 29.Goldstein IJ, Poretz RD. The Lectins: Properties, Functions, and Applications in Biology and Medicine. Orlando: Academic Press Inc; 1986. Isolation, physicochemical characterization, and carbohydrate-binding specificity; pp. 33–243. [Google Scholar]
- 30.Ieth C, Lütteke T, Frank M. Bioinformatics for glycobioogy and glycomics: an introduction. Wiley-Blackwell; 2009. [Google Scholar]
- 31.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, et al. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki N, Kojima S, Bovin NV, Andre S, Gabius S, Gabius HJ. Endogenous lectins as targets for drug delivery. Adv Drug Deliv Rev. 2000;43:225–244. doi: 10.1016/s0169-409x(00)00071-5. [DOI] [PubMed] [Google Scholar]
- 33.Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protocols. 2006;1:543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 34.Hirabayashi J. Glycome ,fingerprints’ provide definitive clues to HIV origins. Nat Chem Biol. 2009;5:198–199. doi: 10.1038/nchembio0409-198. [DOI] [PubMed] [Google Scholar]
- 35.Mislovičová D, Gemeiner P, Kozarova A, Kožár T. Lectinomics I. Relevance of exogenous plant lectins in biomedical diagnostics. Biologia. 2009;64:1–19. [Google Scholar]
- 36.Bertók T, Šefčovicová J, Gemeiner P, Tkáč J. Lektinomika: Nástroj pre klinickú diagnostiku. Chem Listy. 2012;106:10–26. [Google Scholar]
- 37.Bertók T, Katrlík J, Gemeiner P, Tkac J. Electrochemical lectin based biosensors as a label-free tool in glycomics. Microchim Acta. 2012;180:1–13. doi: 10.1007/s00604-012-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.2012 https://www.vectorlabs.com/ Vector Laboratories.
- 39.Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 40.Svarovsky SA, Joshi L. Cancer glycan biomarkers and their detection - past, present and future. Anal Methods. 2014;6:3918–3936. [Google Scholar]
- 41.Meezan E, Wu HC, Black PH, Robbins PW. Comparative studies on the carbohydrate-containing membrane components of normal and virus-transformed mouse fibroblasts. II. Separation of glycoproteins and glycopeptides by Sephadex chromatography. Biochemistry. 1969;8:2518–2524. doi: 10.1021/bi00834a039. [DOI] [PubMed] [Google Scholar]
- 42.Kim EH, Misek DE. Glycoproteomics-based identification of cancer biomarkers. Int J Proteom. 2011;2011:601937. doi: 10.1155/2011/601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YJ, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconjugate J. 1997;14:569–576. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes B, Sagman U, Auger M, Demetrio M, Dennis JW. Beta 1-6 branched oligosaccharides as a marker of tumor progression in human breast and colon neoplasia. Cancer Res. 1991;51:718–723. [PubMed] [Google Scholar]
- 45.Seelentag WK, Li WP, Schmitz SF, Metzger U, Aeberhard P, Heitz PU, et al. Prognostic value of beta1,6-branched oligosaccharides in human colorectal carcinoma. Cancer Res. 1998;58:5559–5564. [PubMed] [Google Scholar]
- 46.Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9:1307–1311. doi: 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- 47.Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, et al. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13:457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 48.Thompson S, Dargan E, Turner GA. Increased fucosylation and other carbohydrate changes in haptoglobin in ovarian cancer. Cancer Lett. 1992;66:43–48. doi: 10.1016/0304-3835(92)90278-4. [DOI] [PubMed] [Google Scholar]
- 49.Misonou Y, Shida K, Korekane H, Seki Y, Noura S, Ohue M, et al. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: Elucidation of aberrant glycosylation and its mechanistic causes in colorectal cancer cells. J Proteome Res. 2009;8:2990–3005. doi: 10.1021/pr900092r. [DOI] [PubMed] [Google Scholar]
- 50.Aubert M, Panicot L, Crotte C, Gibier P, Lombardo D, Sadoulet MO, et al. Restoration of alpha(1,2) fucosyltransferase activity decreases adhesive and metastatic properties of human pancreatic cancer cells. Cancer Res. 2000;60:1449–1456. [PubMed] [Google Scholar]
- 51.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, et al. Increased expression of sialyl Lewisx antigen correlates with poor survival in patients with colorectal carcinoma: clinicopathological and immunohistochemical study. Cancer Res. 1993;53:3632–3637. [PubMed] [Google Scholar]
- 52.Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharm Therap. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 53.Majkić-Singh N. What is a biomarker? From its discovery to clinical application. J. Med. Biochem. 2011:30. [Google Scholar]
- 54.Li J, Li S, Yang CF. Electrochemical biosensors for cancer biomarker detection. Electroanal. 2012;24:2213–2229. [Google Scholar]
- 55.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, et al. The case for early detection, Nat. Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 56.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 57.Turner GA, Goodarzi MT, Thompson S. Glycosylation of alpha-1-proteinase inhibitor and haptoglobin in ovarian cancer: evidence for two different mechanisms. Glycoconjugate J. 1995;12:211–218. doi: 10.1007/BF00731322. [DOI] [PubMed] [Google Scholar]
- 58.Chen K, Gentry-Maharaj A, Burnell M, Steentoft C, MarcosSilva L, Mandel U, et al. Microarray glycoprofiling of CA125 improves differential diagnosis of ovarian cancer. J Proteome Res. 2013;12:1408–1418. doi: 10.1021/pr3010474. [DOI] [PubMed] [Google Scholar]
- 59.Saldova R, Struwe W, Wynne K, Elia G, Duffy M, Rudd P. Exploring the glycosylation of serum CA125. Int J Mol Sci. 2013;14:15636–15654. doi: 10.3390/ijms140815636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cazet A, Julien S, Bobowski M, Burchell J, Delannoy P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010;12:204. doi: 10.1186/bcr2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SY, Yoon SJ, Jeong YT, Kim JM, Kim JY, Bernert B, et al. N-glycosylation status of beta-haptoglobin in sera of patients with colon cancer, chronic inflammatory diseases and normal subjects. Int J Cancer. 2010;126:142–155. doi: 10.1002/ijc.24685. [DOI] [PubMed] [Google Scholar]
- 62.Vercoutter-Edouart AS, Slomianny MC, Dekeyzer-Beseme O, Haeuw JF, Michalski JC. Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics. 2008;8:3236–3256. doi: 10.1002/pmic.200800151. [DOI] [PubMed] [Google Scholar]
- 63.Saeland E, Belo AI, Mongera S, van Die I, Meijer GA, van Kooyk Y. Differential glycosylation of MUC1 and CEACAM5 between normal mucosa and tumour tissue of colon cancer patients. Int J Cancer. 2012;131:117–128. doi: 10.1002/ijc.26354. [DOI] [PubMed] [Google Scholar]
- 64.Zhao YP, Ruan CP, Wang H, Hu ZQ, Fang M, Gu X, et al. Identification and assessment of new biomarkers for colorectal cancer with serum N-glycan profiling. Cancer. 2012;118:639–650. doi: 10.1002/cncr.26342. [DOI] [PubMed] [Google Scholar]
- 65.Qiu Y, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7:1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li C, Simeone DM, Brenner DE, Anderson MA, Shedden KA, Ruffin MT, et al. Pancreatic cancer serum detection using a lectin/glyco-antibody array method. J Proteome Res. 2009;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6:1864–1874. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- 68.Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: detailed analyses of oligosaccharide structures. Proteomics. 2008;8:3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 69.Fujimura T, Shinohara Y, Tissot B, Pang PC, Kurogochi M, Saito S, et al. Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int J Cancer. 2008;122:39–49. doi: 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- 70.Ohyama C, Hosono M, Nitta K, Oh-eda M, Yoshikawa K, Habuchi T, et al. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14:671–679. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- 71.Takeya A, Hosomi O, Nishijima H, Ohe Y, Sugahara K, Sagi M, et al. Presence of beta-linked GalNAc residues on N-glycans of human thyroglobulin. Life Sci. 2007;80:538–545. doi: 10.1016/j.lfs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto K, Tsuji T, Tarutani O, Osawa T. Structural changes of carbohydrate chains of human thyroglobulin accompanying malignant transformations of thyroid glands. Eur J Biochem. 1984;143:133–144. doi: 10.1111/j.1432-1033.1984.tb08352.x. [DOI] [PubMed] [Google Scholar]
- 73.Naitoh A, Aoyagi Y, Asakura H. Highly enhanced fucosylation of serum glycoproteins in patients with hepatocellular carcinoma. J Gastroenter Hepatol. 1999;14:436–445. doi: 10.1046/j.1440-1746.1999.01882.x. [DOI] [PubMed] [Google Scholar]
- 74.Hoagland LFt, Campa MJ, Gottlin EB, Herndon JE, 2nd, Patz EF., Jr Haptoglobin and posttranslational glycan-modified derivatives as serum biomarkers for the diagnosis of nonsmall cell lung cancer. Cancer. 2007;110:2260–2268. doi: 10.1002/cncr.23049. [DOI] [PubMed] [Google Scholar]
- 75.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 76.Badr HA, Alsadek DM, Darwish AA, Elsayed AI, Bekmanov BO, Khussainova EM, et al. Lectin approaches for glycoproteomics in FDA-approved cancer biomarkers. Expert Rev Proteomics. 2014;11:227–236. doi: 10.1586/14789450.2014.897611. [DOI] [PubMed] [Google Scholar]
- 77.Balog CI, Stavenhagen K, Fung WL, Koeleman CA, McDonnell LA, Verhoeven A, et al. N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol Cell Proteomics. 2012;11:571–585. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmidt MM, Thurber GM, Wittrup KD. Kinetics of anti- carcinoembryonic antigen antibody internalization: effects of affinity, bivalency, and stability. Cancer Immunol Immunother. 2008;57:1879–1890. doi: 10.1007/s00262-008-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahn YH, Ji ES, Shin PM, Kim KH, Kim YS, Ko JH, et al. A multiplex lectin-channel monitoring method for human serum glycoproteins by quantitative mass spectrometry. Analyst. 2012;137:691–703. doi: 10.1039/c1an15775b. [DOI] [PubMed] [Google Scholar]
- 80.Cheng TM, Lee TC, Tseng SH, Chu HL, Pan JP, Chang CC. Human haptoglobin phenotypes and concentration determination by nanogold-enhanced electrochemical impedance spectroscopy. Nanotechnology. 2011;22:245105. doi: 10.1088/0957-4484/22/24/245105. [DOI] [PubMed] [Google Scholar]
- 81.Thompson S, Turner GA. Elevated levels of abnormally-fucosylated haptoglobins in cancer sera. Br J Cancer. 1987;56:605–610. doi: 10.1038/bjc.1987.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B, Cai FF, Zhong XY. An overview of biomarkers for the ovarian cancer diagnosis. Eur J Obstetrics Gynec Reprod. Biol. 2011;158:119–123. doi: 10.1016/j.ejogrb.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 83.World Health Organization I.A.f.R.o.C. Globocan. 2012 Available from http://globocan.iarc.fr/Default.aspx.
- 84.World Health Organization I.A.f.R.o.C. European Cancer Observatory. 2012 Available from http://eu-cancer.iarc.fr/
- 85.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 86.Bhoola S, Hoskins WJ. Diagnosis and management of epithelial ovarian cancer. Obstetrics Gynecol. 2006;107:1399–1410. doi: 10.1097/01.AOG.0000220516.34053.48. [DOI] [PubMed] [Google Scholar]
- 87.Eltabbakh GH, Mount SL, Beatty B, Simmons-Arnold L, Cooper K, Morgan A. Factors associated with cytoreducibility among women with ovarian carcinoma. Gynecol Oncol. 2004;95:377–383. doi: 10.1016/j.ygyno.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 88.Biskup K, Braicu EI, Sehouli J, Fotopoulou C, Tauber R, Berger M, et al. Serum glycome profiling: A biomarker for diagnosis of ovarian cancer. J Proteome Res. 2013;12:4056–4063. doi: 10.1021/pr400405x. [DOI] [PubMed] [Google Scholar]
- 89.Park CW, Jo Y, Jo EJ. Enhancement of ovarian tumor classification by improved reproducibility in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of serum glycans. Anal Biochem. 2013;443:58–65. doi: 10.1016/j.ab.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 90.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112:40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghasemi N, Ghobadzadeh S, Zahraei M, Mohammadpour H, Bahrami S, Ganje MB, et al. HE4 combined with CA125: favorable screening tool for ovarian cancer. Med Oncol. 2014;31:808. doi: 10.1007/s12032-013-0808-0. [DOI] [PubMed] [Google Scholar]
- 92.La Belle JT, Fairchild A, Demirok UK, Verma A. Method for fabrication and verification of conjugated nanoparticle-antibody tuning elements for multiplexed electrochemical biosensors. Methods. 2013;61:39–51. doi: 10.1016/j.ymeth.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 93.Yurkovetsky Z, Skates S, Lomakin A, Nolen B, Pulsipher T, Modugno F, et al. Development of a multimarker assay for early detection of ovarian cancer. J Clin Oncol. 2010;28:2159–2166. doi: 10.1200/JCO.2008.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vermassen T, Speeckaert MM, Lumen N, Rottey S, Delanghe JR. Glycosylation of prostate specific antigen and its potential diagnostic applications. Clin Chim Acta. 2012;413:1500–1505. doi: 10.1016/j.cca.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 95.Cary KC, Cooperberg MR. Biomarkers in prostate cancer surveillance and screening: past, present, and future. Ther Adv Urol. 2013;5:318–329. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hori S, Blanchet JS, McLoughlin J. From prostate-specific antigen (PSA) to precursor PSA (proPSA) isoforms: a review of the emerging role of proPSAs in the detection and management of early prostate cancer. BJU Int. 2013;112:717–728. doi: 10.1111/j.1464-410X.2012.11329.x. [DOI] [PubMed] [Google Scholar]
- 97.Velonas VM, Woo HH, Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Mol Sci. 2013;14:11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crawford ED, Ventii K, Shore ND. New biomarkers in prostate cancer. Oncology. 2014;28:135–142. [PubMed] [Google Scholar]
- 99.Okada T, Sato Y, Kobayashi N, Sumida K, Satomura S, Matsuura S, et al. Structural characteristics of the N-glycans of two isoforms of prostate-specific antigens purified from human seminal fluid. Biochim Biophys Acta. 2001;1525:149–160. doi: 10.1016/s0304-4165(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 100.Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O’Kennedy RJ. Aberrant PSA glycosylation--a sweet predictor of prostate cancer. Nat Rev Urol. 2013;10:99–107. doi: 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- 101.Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, et al. Focused differential glycan analysis with the platform antibody-assisted lectin profiling for glycan-related biomarker verification. Mol Cell Proteomics : MCP. 2009;8:99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Duverger E, Lamerant-Fayel N, Frison N, Monsigny M. Carbohydrate-lectin interactions assayed by SPR. Methods Mol Biol. 2010;627:157–178. doi: 10.1007/978-1-60761-670-2_10. [DOI] [PubMed] [Google Scholar]
- 103.Safina G, Duran Iu B, Alasel M, Danielsson B. Surface plasmon resonance for real-time study of lectin-carbohydrate interactions for the differentiation and identification of glycoproteins. Talanta. 2011;84:1284–1290. doi: 10.1016/j.talanta.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 104.Choi YE, Kwak JW, Park JW. Nanotechnology for early cancer detection. Sensors. 2010;10:428–455. doi: 10.3390/s100100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 106.Liu X, Dai Q, Austin L, Coutts J, Knowles G, Zou J, et al. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J Am Chem Soc. 2008;130:2780–2782. doi: 10.1021/ja711298b. [DOI] [PubMed] [Google Scholar]
- 107.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 108.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. In vivo molecular and cellular imaging with quantum dots. Curr Opin Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 109.Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 110.Iijima S, Ichihashi T. Single-shell carbon nanotubes of 1-nm diameter. Nature. 1993;363:603–605. [Google Scholar]
- 111.Geim AK, Novoselov KS. The rise of graphene. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 112.Geim AK. Graphene: status and prospects. Science. 2009;324:1530–1534. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- 113.Novoselov KS, Geim AK, Morozov S, Jiang D, Zhang Y, Dubonos S, et al. Electric field effect in atomically thin carbon films. Science. 2004;306:666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- 114.Dreyer DR, Park S, Bielawski CW, Ruoff RS. The chemistry of graphene oxide. Chem Soc Rev. 2010;39:228–240. doi: 10.1039/b917103g. [DOI] [PubMed] [Google Scholar]
- 115.Reichardt NC, Martín-Lomas M, Penadés S. Glyconanotechnology, Chem Soc Rev. 2013;42:4358–4376. doi: 10.1039/c2cs35427f. [DOI] [PubMed] [Google Scholar]
- 116.Park DW, Kim YH, Kim BS, So HM, Won K, Lee JO, et al. Detection of tumor markers using single-walled carbon nanotube field effect transistors. J Nanosci Nanotechnol. 2006;6:3499–3502. [PubMed] [Google Scholar]
- 117.Kerman K, Saito M, Tamiya E, Yamamura S, Takamura Y. Nanomaterial-based electrochemical biosensors for medical applications. Trends Anal Chem. 2008;27:585–592. [Google Scholar]
- 118.Perumal V, Hashum U. Advances in biosensors: Principle, architecture and applications. J Appl Biomed. 2014;12:1–15. [Google Scholar]
- 119.Lee SY, Hwang SY. Electrical and electrochemical immunosensor for cancer study. Biosensors and Cancer: Science Publishers. 2012:125–145. [Google Scholar]
- 120.Yun YH, Eteshola E, Bhattacharya A, Dong Z, Shim JS, Conforti L, et al. Tiny medicine: Nanomaterial-based biosensors. Sensors. 2009;9:9275–9299. doi: 10.3390/s91109275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu JJ, Zhao WW, Song S, Fan C, Chen HY. Functional nanoprobes for ultrasensitive detection of biomolecules: an update. Chem Soc Rev. 2014;43:1601–1611. doi: 10.1039/c3cs60277j. [DOI] [PubMed] [Google Scholar]
- 122.Wang Y, Qu K, Tang L, Li Z, Moore E, Zeng X, et al. Nanomaterials in carbohydrate biosensors. Trends Anal Chem. 2014;58:54–70. [Google Scholar]
- 123.Mu B, Zhang J, McNicholas TP, Reuel NF, Kruss S, Strano MS. Recent advances in molecular recognition based on nanoengineered platforms. Acc Chem Res. 2014;47:979–988. doi: 10.1021/ar400162w. [DOI] [PubMed] [Google Scholar]
- 124.Kluková L’, Bertók T, Kasák P, Tkac J. Nanoscale controlled architecture for development of ultrasensitive lectin biosensors applicable in glycomics. Anal Methods. 2014;6:4922–4931. doi: 10.1039/C4AY00495G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Katz E, Willner I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: Routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanal. 2003;15:913–947. [Google Scholar]
- 126.Thévenot DR, Toth K, Durst RA, Wilson GS. Electrochemical biosensors: recommended definitions and classification. Biosens Bioelectron. 2001;16:121–131. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 127.Bertok T, Šefčovičová J, Gemeiner P, Tkac J. Vývoj a súčasné trendy pri príprave nanoštrukturovaných biosenzorov. Chem Listy. 2012;106:174–181. [Google Scholar]
- 128.Tkac J, Davis JJ. An optimised electrode pre-treatment for SAM formation on polycrystalline gold. J Electroanal Chem. 2008;621:117–120. [Google Scholar]
- 129.Cao G, >Wang Y. Nanostructures and nanomaterials: Synthesis, properties, and applications. 2. World Scientific Publishing; 2011. [Google Scholar]
- 130.Alvarez TV. Highly sensitive nanomaterial based electrochemical biosensor. Arizona State University: 2009. [Google Scholar]
- 131.Zhou Y, Xu Z, Wang M, Meng X, Yin H. Electrochemical immunoassay platform for high sensitivity detection of indole-3-acetic acid. Electrochim Acta. 2013;96:66–73. [Google Scholar]
- 132.Sluyters-Rehbach M, Sluyters JH. On the impedance of galvanic cells XXVIII. The frequency-dependence of the electrode admittance for systems with first-order homogeneous chemical reactions and reactant adsorption occurring simultaneously. J Electroanal Chem Interf Electrochem. 1969;23:457–474. [Google Scholar]
- 133.Ershler B. Investigation of electrode reactions by the method of charging-curves and with the aid of alternating currents. Discuss Faraday Soc. 1947;1:269–277. [Google Scholar]
- 134.Hu Y, Zuo P, Ye BC. Label-free electrochemical impedance spectroscopy biosensor for direct detection of cancer cells based on the interaction between carbohydrate and lectin. Biosens Bioelectron. 2013;43:79–83. doi: 10.1016/j.bios.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 135.Bertok T, Sediva A, Katrlik J, Gemeiner P, Mikula M, Nosko M, et al. Label-free detection of glycoproteins by the lectin biosensor down to attomolar level using gold nanoparticles. Talanta. 2013;108:11–18. doi: 10.1016/j.talanta.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Suni II. Impedance methods for electrochemical sensors using nanomaterials. Trends Anal Chem. 2008;27:604–611. [Google Scholar]
- 137.Scouten WH, Luong JHT, Stephen Brown R. Enzyme or protein immobilization techniques for applications in biosensor design. Trends Biotechnol. 1995;13:178–185. [Google Scholar]
- 138.Li S, Singh J, Li H, Banerjee IA. Biosensor Nanomaterials. Wiley-VCH Verlag GmbH&Co.; 2011. [Google Scholar]
- 139.Putzbach W, Ronkainen N. Immobilization techniques in the fabrication of nanomaterial-based electrochemical biosensors: A review, Sensors. 2013;13:4811–4840. doi: 10.3390/s130404811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davis JJ, Tkac J, Humphreys R, Buxton AT, Lee TA, Ko Ferrigno P. Peptide aptamers in label-free protein detection: 2. Chemical optimization and detection of distinct protein isoforms. Anal Chem. 2009;81:3314–3320. doi: 10.1021/ac802513n. [DOI] [PubMed] [Google Scholar]
- 141.Davis JJ, Tkac J, Laurenson S, Ferrigno PK. Peptide aptamers in label-free protein detection: 1. Characterization of the immobilized scaffold. Anal Chem. 2007;79:1089–1096. doi: 10.1021/ac061863z. [DOI] [PubMed] [Google Scholar]
- 142.Ericsson E. Biosensor surface chemistry for oriented protein immobilization and biochip patterning. Linköping University: 2013. [Google Scholar]
- 143.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 2005;105:1103–1170. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 144.Gooding JJ, Mearns F, Yang W, Liu J. Self-assembled monolayers into the 21st century: Recent advances and applications. Electroanal. 2003;15:81–96. [Google Scholar]
- 145.Hushegyi A, Tkac J. Are glycan biosensors an alternative to glycan microarrays? Anal Methods. 2014;6:6610–6620. doi: 10.1039/C4AY00692E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ron H, Matlis S, Rubinstein I. Self-assembled monolayers on oxidized metals. 2. gold surface oxidative pretreatment, monolayer properties, and depression formation. Langmuir. 1998;14:1116–1121. [Google Scholar]
- 147.Bertok T, Gemeiner P, Mikula M, Gemeiner P, Tkac J. Ultrasensitive impedimetric lectin based biosensor for glycoproteins containing sialic acid. Microchim Acta. 2013;180:151–159. doi: 10.1007/-s00604-012-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fischer LM, Tenje M, Heiskanen AR, Masuda N, Castillo J, Bentien A, et al. Gold cleaning methods for electrochemical detection applications. Microelectronic Eng. 2009;86:1282–1285. [Google Scholar]
- 149.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 150.Haab BB. Antibody-lectin sandwich arrays for biomarker and glycobiology studies. Expert Rev Proteomics. 2010;7:9–11. doi: 10.1586/epr.09.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen H, Jiang C, Yu C, Zhang S, Liu B, Kong J. Protein chips and nanomaterials for application in tumor marker immunoassays. Biosens Bioelectron. 2009;24:3399–3411. doi: 10.1016/j.bios.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 152.La Belle JT, Gerlach JQ, Svarovsky S, Joshi L. Label-free impedimetric detection of glycan-lectin interactions. Anal Chem. 2007;79:6959–6964. doi: 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]
- 153.Yang H, Li Z, Wei X, Huang R, Qi H, Gao Q, et al. Detection and discrimination of alpha-fetoprotein with a label-free electrochemical impedance spectroscopy biosensor array based on lectin functionalized carbon nanotubes. Talanta. 2013;111:62–68. doi: 10.1016/j.talanta.2013.01.060. [DOI] [PubMed] [Google Scholar]
- 154.Oliveira MDL, Correia MTS, Diniz FB. A novel approach to classify serum glycoproteins from patients infected by dengue using electrochemical impedance spectroscopy analysis. Synth Met. 2009;159:2162–2164. [Google Scholar]
- 155.Nagaraj VJ, Aithal S, Eaton S, Bothara M, Wiktor P, Prasad S. NanoMonitor: a miniature electronic biosensor for glycan biomarker detection. Nanomedicine. 2010;5:369–378. doi: 10.2217/nnm.10.11. [DOI] [PubMed] [Google Scholar]
- 156.Bertok T, Klukova L, Sediva A, Kasak P, Semak V, Micusik M, et al. Ultrasensitive impedimetric lectin biosensors with efficient antifouling properties applied in glycoprofiling of human serum samples. Anal Chem. 2013;85:7324–7332. doi: 10.1021/ac401281t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Silva MLS, Gutiérrez E, Rodríguez JA, Gomes C, David L. Construction and validation of a Sambucus nigra biosensor for cancer-associated STn antigen. Biosens Bioelectron. 2014;57:254–261. doi: 10.1016/j.bios.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 158.Kongsuphol P, Ng HH, Pursey JP, Arya SK, Wong CC, Stulz E, et al. EIS-based biosensor for ultra-sensitive detection of TNF-α from non-diluted human serum. Biosens Bioelectron. 2014;61:274–279. doi: 10.1016/j.bios.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 159.Luo X, Xu Q, James T, Davis JJ. Redox and label-free array detection of protein markers in human serum. Anal Chem. 2014;86:5553–5558. doi: 10.1021/ac5010037. [DOI] [PubMed] [Google Scholar]
- 160.Xia N, Deng D, Zhang L, Yuan B, Jing M, Du J, et al. Sandwichtype electrochemical biosensor for glycoproteins detection based on dual-amplification of boronic acid-gold nanoparticles and dopamine-gold nanoparticles. Biosens Bioelectron. 2013;43:155–159. doi: 10.1016/j.bios.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 161.Cao JT, Hao XY, Zhu YD, Sun K, Zhu JJ. Microfluidic platform for the evaluation of multi-glycan expressions on living cells using electrochemical impedance spectroscopy and optical microscope. Anal Chem. 2012;84:6775–6782. doi: 10.1021/ac3013048. [DOI] [PubMed] [Google Scholar]