Abstract
Two functional variants in the inosine triphosphatase (ITPA) gene causing inosine triphos-phatase (ITPase) deficiency protect against ribavirin (RBV)-induced hemolytic anemia and the need for RBV dose reduction in patients with genotype 1 hepatitis C virus (HCV). No data are available for genotype 2/3 HCV. We evaluated the association between the casual ITPA variants and on-treatment anemia in a well-characterized cohort of genotype 2/3 patients treated with variable-duration pegylated interferon alfa-2b (PEG-IFN-α2b) and RBV. Two hundred thirty-eight Caucasian patients were included in this retrospective study [185 (78%) with genotype 2 and 53 (22%) with genotype 3]. Patients were treated with PEG-IFN-α2b plus weight-based RBV (1000/1200 mg) for 12 (n = 109) or 24 weeks (n = 129). The ITPA polymorphisms rs1127354 and rs7270101 were genotyped, and an ITPase deficiency variable was defined that combined both ITPA variants according to their effect on ITPase activity. The primary endpoint was hemoglobin (Hb) reduction in week 4. We also considered Hb reduction over the course of therapy, the need for RBV dose modification, and the rate of sustained virological response (SVR). The ITPA variants were strongly and independently associated with protection from week 4 anemia (P = 10−6 for rs1127354 and P = 10−7 for rs7270101). Combining the variants into the ITPase deficiency variable increased the strength of association (P = 10−11). ITPase deficiency protected against anemia throughout treatment. ITPase deficiency was associated with a delayed time to an Hb level < 10 g/dL (hazard ratio = 0.25, 95% confidence interval = 0.08–0.84, P = 0.025) but not with the rate of RBV dose modification (required per protocol at Hb < 9.5 g/dL). There was no association between the ITPA variants and SVR.
Conclusion
Two ITPA variants were strongly associated with protection against treatment-related anemia in patients with genotype 2/3 HCV, but they did not decrease the need for RBV dose reduction or increase the rate of SVR.
Chronic infection with hepatitis C virus (HCV) affects up to 170 million individuals worldwide and is a leading cause of end-stage liver disease.1 HCV is curable, but the therapy may be difficult to tolerate and is associated with significant morbidity. Ribavirin (RBV)-induced hemolytic anemia is common, with dose modification being required in 9% to 22% of patients enrolled in the phase 3 registration trials of pegylated interferon (PEG-IFN) and RBV therapy.2,3 RBV dose reduction has the potential to limit treatment efficacy.4
We recently identified strong and independent associations between two functional variants in the inosine triphosphatase (ITPA) gene on chromosome 20 that caused an inosine triphosphatase (ITPase) deficiency5–8 and protection from early RBV-induced hemolytic ane-mia in genotype 1 HCV–infected patients enrolled in the Individualized Dosing Efficacy Versus Flat Dosing to Assess Optimal Pegylated Interferon Therapy trial.9,10 In a follow-up study of genotype 1 HCV–infected patients from the Viral Resistance to Antiviral Therapy of Chronic Hepatitis C study, the ITPA variants protected against anemia throughout 48 weeks of therapy and were associated with a lesser frequency of RBV dose reduction.11 ITPA variants were not significantly associated with virological treatment outcome in either study.
The impact of these genetic variants on anemia in patients with genotype 2/3 HCV, who are typically treated with a shorter course of therapy than those with genotype 1 HCV, has not yet been investigated. We have, therefore, performed a retrospective analysis of a well-characterized cohort of European patients with genotype 2 or 3 HCV infection who were treated with PEG-IFN-α2b and weight-based RBV for 12 or 24 weeks according to the week 4 virological response.12 We investigated the association between the functional ITPA variants and anemia in week 4, week 12, and/or week 24 of therapy. By assessing the relationship with the need for RBV dose reduction as well as treatment outcomes, we considered whether and how ITPA genotyping might affect clinical decision making.
Patients and Methods
Patient and Control Populations
The primary study was an Italian, multicenter, randomized controlled trial.12 In the original study, 70 patients were randomized to the standard-duration treatment (24 weeks), and 213 were randomized to a variable-duration treatment: patients achieving rapid virological response (RVR) were treated for 12 weeks, whereas those not achieving RVR were treated for 24 weeks. RVR was defined as the finding of undetectable serum/plasma HCV RNA with a sensitive qualitative assay (lower limit of detection < 50 IU/mL) in week 4. Inclusion and exclusion criteria have been reported previously.12 Patients received PEG-IFN-α2b (PEG-Intron, Schering Plough, Kenilworth, NJ; 1.0 μg/kg/week) and RBV (Rebetol, Schering Plough; 1000/1200 mg daily) according to body weight ± 75 kg. PEG-IFN and RBV dose modification followed the standard criteria and procedures.12 Specifically, if hemoglobin (Hb) levels were lower than 9.5 g/dL, the dose of RBV was lowered to 800 mg/day, and RBV was discontinued if the Hb level fell to <8.0 g/dL. Per the study protocol, erythropoietin was not allowed. Patients provided informed consent for the collection and storage of peripheral blood mononuclear cells for DNA testing for research purposes consistent with the current study. The study was approved by a central ethics committee and was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines.
ITPA Genotyping
The two functional variants in the ITPA gene (encoding ITPase) on chromosome 20 are rs1127354 (a missense variant in exon 2, P32T) and rs7270101 [a splice-altering single-nucleotide polymorphism (SNP) in intron 2]. Patients were genotyped at the polymorphic sites with the ABI TaqMan allelic discrimination kit and the ABI7900HT sequence detection system (Applied Biosystems, Carlsbad, CA), as previously described.10 The possible genotypes for each biallelic polymorphism are as follows: C/C, A/C, and A/A (A is the minor allele) for rs1127354 and A/A, A/C, and C/C (C is the minor allele) for rs7270101. This cohort was previously tested for the interleukin-28B (IL-28B) polymorphism rs12979860.13
Definition of a Composite ITPase Deficiency Variable According to the Genotype at rs1127354 and rs7270101
The ITPA polymorphisms rs1127354 and rs7270101 have previously been well characterized and validated as functional variants in studies of patients with ITPase deficiency.5–8 The predicted severity of ITPase deficiency was defined according to these functional studies (Table 1).5–8,10
Table 1.
Predicted Levels of ITPase Deficiency According to the ITPA Haplotype
| rs1127354 | rs7270101 | Prediction
|
|
|---|---|---|---|
| ITPase Activity (%) | ITPase Deficiency | ||
| Wild type (C/C) | Wild type (A/A) | 100 | – |
| Wild type (C/C) | Heterozygosity (A/C) | 60 | + |
| Heterozygosity (C/A) | Wild type (A/A) | 30 | ++ |
| Wild type (C/C) | Homozygosity (C/C) | 30 | ++ |
| Heterozygosity (C/A) | Heterozygosity (A/C) | 10 | +++ |
| Homozygosity (A/A) | Wild type (A/A) | <5 | +++ |
The levels of ITPase deficiency predicted according to the ITPA haplotype and in vitro studies5–8,10 are presented. ITPase activity was defined as the accumulated presence of minor alleles at the respective polymorphic sites [the minor alleles for both SNPs never occurred together on the same chromosome (3)].10
Definition of Clinical Endpoints
The primary genetic association analyses considered the following: (1) the absolute Hb reduction from the baseline to week 4 (continuous trait) and (2) Hb reductions >3 g/dL in week 4 (categorical trait). We also considered the relationship between the ITPA variants and (1) Hb reductions over the course of 12 to 24 weeks of therapy, which were defined both qualitatively and quantitatively (reduction >3 g/dL); (2) the need for RBV dose reduction; and (3) the rate of sustained virological response (SVR).
Statistical Analysis
For descriptive statistics, continuous variables were summarized as medians and 25th and 75th percentiles. Categorical variables were described as frequencies and percentages. Comparisons between groups for demographic, clinical, and virological data were performed with a Wilcoxon test for skewed continuous data or the chi-square test/Fisher’s exact test for categorical data. Significance was defined at P < 0.05. Associations between the individual polymorphisms/ITPase deficiency variable and the anemia phenotypes were tested with single-marker genotype trend tests of association in linear/logistic regression models. Multivariate regression models with backward selection were used to identify independent predictors of anemia. A significance level of 0.05 was used for removal from the model. The covariates included the age, sex, weight (kg), METAVIR fibrosis stage according to pretreatment liver biopsy, baseline Hb level, baseline serum creatinine level, and RBV dose (mg/kg). The relationship between the ITPase deficiency variable and the need for RBV dose reduction was tested by survival analysis with the log-rank test of equality across strata. All statistical analyses were performed with SAS version 9.2 (SAS Institute, Cary, NC).
Results
Patients
The cohort has been well described previously.12,13 Two hundred thirty-eight patients who were successfully genotyped for the ITPA causal variants and for whom baseline and week 4 Hb levels were available were included in this analysis. All patients were Caucasian; 185 patients (78%) were infected with genotype 2 HCV, and 53 (22%) were infected with genotype 3 HCV. Baseline demographic, biochemical, and virological characteristics of the study cohort are reported in Table 2. One hundred nine patients were treated for 12 weeks, and 129 were treated for 24 weeks; the baseline features did not differ significantly between the two groups (data not shown).
Table 2.
Characteristics of the Patients
| Characteristic | Value |
|---|---|
| n | 238 |
| Age, years | 52 (41–62) |
| Male sex, n (%) | 142 (59) |
| Body mass index, kg/m2 | 26 (24–28) |
| Genotype 2, n (%) | 185 (78) |
| Genotype 3, n (%) | 53 (22) |
| HCV RNA, log10 IU/mL | 5.7 (5.3–6.0) |
| Alanine aminotransferase, IU/mL | 59 (31–117) |
| Baseline Hb, g/dL | 14.0 (13.0–15.0) |
| Baseline creatinine, μmol/L | 0.87 (0.74–1.0) |
| METAVIR F3-F4, n (%) | 48 (20) |
| RBV dose, mg/kg | 15 (13.5–16.7) |
| Need for RBV dose reduction, n (%) | |
| 12-week treatment | 11/109 (10)* |
| 24-week treatment | 24/129 (19) |
Data are presented as medians and 25th and 75th percentiles unless otherwise specified.
P = 0.06.
Population Distribution of the ITPA Gene Variants
The two polymorphisms, rs1127354 and rs7270101, were genotyped in the study population (Tables 3 and 4). The minor allele (A) frequency was 0.05 for rs1127354 and 0.11 for rs7270101. The two genotypes were then used to define the ITPase deficiency variable: the majority of patients were predicted to have wild-type (WT) ITPase activity, but a significant minority of patients were predicted to have reduced ITPase activity (29%; Tables 3 and 4).
Table 3.
Frequency of ITPA Genotypes: rs1127354 and rs7270101 Genotype Prevalence Within the Study Population
| n (238) | % | |
|---|---|---|
| rs1127354 | ||
| CC | 215 | 90 |
| CA | 20 | 8 |
| AA | 3 | 1 |
| Minor C allele frequency | 0.05 | – |
| rs7270101 | ||
| AA | 188 | 79 |
| AC | 46 | 19 |
| CC | 4 | 2 |
| Minor A allele frequency | 0.11 | – |
Table 4.
Frequency of ITPA Genotypes: Population Distribution of the Predicted Levels of ITPase Deficiency
| Predicted ITPase Deficiency | n (238) | % |
|---|---|---|
| − | 168 | 71 |
| + | 43 | 18 |
| ++ | 21 | 9 |
| +++ | 6 | 3 |
ITPA Variants Protected Against Week 4 Anemia
Week 4 anemia was considered as both a continuous variable and a categorical variable to replicate the original description of ITPA variants and anemia in genotype 1 HCV patients. Predicted ITPase deficiency was associated with protection against anemia in week 4 (Figs. 1 and 2). Moderate to severe ITPase deficiency was associated with additive protection in week 4, although a statistical comparison was limited by the low numbers of patients with the extreme phenotype (Figs. 1 and 2). With multivariate linear regression modeling, both ITPA variants (rs1127354 and rs7270101) were found to be strongly and independently associated with quantitative week 4 Hb reduction, with the minor alleles being protective (Tables 5 and 6). The variants were also protective against week 4 Hb reductions greater than 3 g/dL. The combined ITPase deficiency variable was more strongly associated with protection against week 4 anemia (quantitative/qualitative; Tables 5 and 6) than either polymorphism alone and was estimated to explain 13% of the variability in quantitative Hb reduction in week 4 after adjustments for other clinical variables (Supporting Information Table 2). Neither the ITPA variants nor the ITPase deficiency variable was associated with the baseline Hb level (data not shown).
Fig. 1.
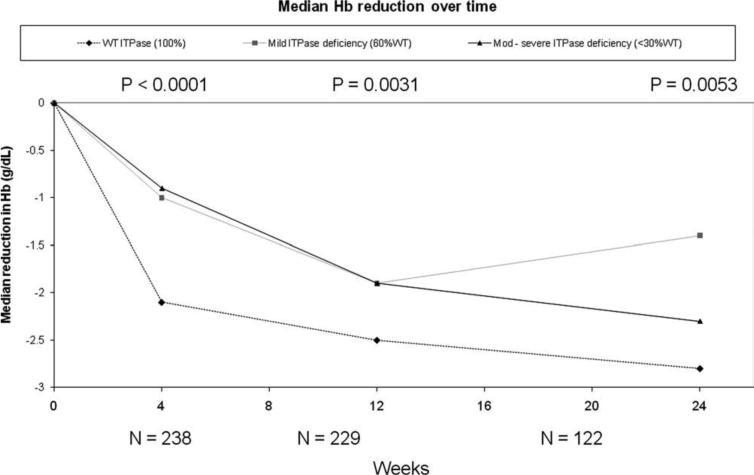
Quantitative Hb reduction from the baseline. The predicted ITPase deficiency was associated with a less absolute reduction in the median Hb levels at all time points during treatment. Only six patients were predicted to have severe (+++) ITPase deficiency; therefore, patients with moderate or severe ITPase deficiency were combined into a single variable. Data are presented as medians and interquartile ranges. The statistical analysis compares patients with normal ITPase activity to the composite group of patients with mild or moderate to severe ITPase deficiency. P values are listed above the relevant time points.
Fig. 2.
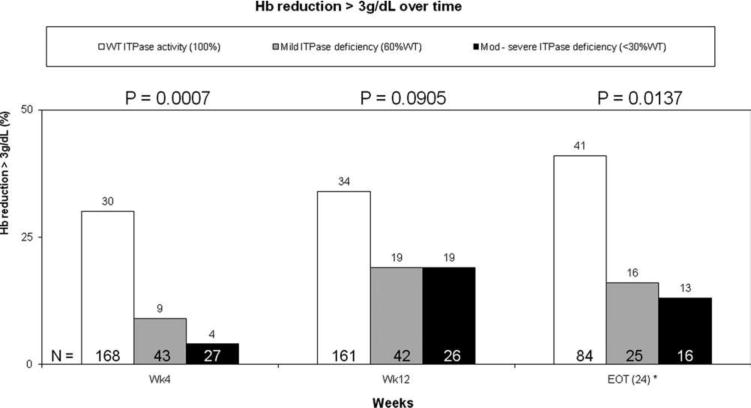
Hb reduction > 3 g/dL from the baseline. The predicted ITPase deficiency was associated with protection against clinically significant reductions (>3 g/dL) in Hb throughout the course of PEG-IFN/RBV combination therapy. Only six patients were predicted to have severe (+++) ITPase deficiency; therefore, patients with moderate or severe ITPase deficiency were combined into a single variable. The statistical analysis compares patients with normal ITPase activity to the composite group of patients with predicted ITPase deficiency. P values for each time point are listed above the columns.
Table 5.
ITPA Variants and ITPase Deficiency Are Associated With a Reduction in the Week 4 Hb Level: Multivariate Linear Regression Modeling of Week 4 Anemia (Absolute Reduction in Hb)
| ITPA Variant | Estimate | Standard Error | P Value |
|---|---|---|---|
| rs1127354 | −1.02 | 0.24 | 4.73 × 10−5 |
| rs7270101 | −0.96 | 0.19 | 1.05 × 10−6 |
| ITPA Variant | Adjusted Estimate* | Standard Error | Adjusted P Value* |
| rs1127354 | −1.10 | 0.23 | 2.90 × 10−6 |
| rs7270101 | −1.01 | 0.18 | 6.78 × 10−8 |
| ITPase Deficiency Variable | Estimate | Standard Error | P Value |
| Grade 0/1/2/3† | −0.75 | 0.11 | 3.82 × 10−11 |
ITPA variants were strongly and independently associated with protection from quantitative Hb reduction during week 4 of therapy. The composite ITPase deficiency variable was more strongly associated with protection from a week 4 quantitative Hb reduction than either individual ITPA variant (the complete multivariate linear regression model, showing estimates for all included variables, is shown in Supporting Information Table 1). The covariates were age, sex, body mass index, baseline Hb level, baseline glomerular filtration rate, METAVIR F3-F4 versus F0–F2, and RBV starting dose (mg/kg). The HCV RNA level, HCV genotype, and sex were not significantly associated with Hb reduction in univariate analyses.
Adjusted estimates and P values were calculated with models in which the other functional variant was already included.
The ITPase deficiency was graded as follows: 0 = normal activity, 1 = mild deficiency, 2 = moderate deficiency, and 3 = severe deficiency.
Table 6.
ITPA Variants and ITPase Deficiency Are Associated With a Reduction in the Week 4 Hb Level: Multivariate Logistic Regression Modeling of Qualitative Week 4 Anemia (Hb Reduction > 3 g/dL)
| ITPA Variant | Odds Ratio | 95% CI | P Value |
|---|---|---|---|
| rs1127354 | 0.051 | 0.006–0.46 | 0.0082 |
| rs7270101 | 0.141 | 0.04–0.46 | 0.0012 |
| ITPA Variant | Adjusted Odds Ratio* | 95% CI | Adjusted P Value* |
| rs1127354 | 0.033 | 0.003–0.33 | 0.0039 |
| rs7270101 | 0.106 | 0.03–0.38 | 0.0006 |
| ITPase Deficiency Variable | Odds Ratio | 95% CI | P Value |
| Grade 0/1/2/3 | 0.137 | 0.05–0.35 | 3.52 × 10−5 |
The two ITPA variants were strongly and independently associated with protection from Hb reduction > 3 g/dL in week 4. The strength of association of the ITPase deficiency variable with week 4 anemia was greater than that for either ITPA variant alone (the complete multivariate logistic regression model, including odds ratios for all variables, is shown in Supporting Information Table 2). The covariates were age, sex, body mass index, baseline Hb level, baseline glomerular filtration rate, METAVIR F3–F4 versus F0–F2, and RBV starting dose (mg/kg). Neither the HCV RNA level nor the HCV genotype was significantly associated with Hb reduction in univariate analyses.
Adjusted odds ratios and independent P values were calculated with models in which the other functional variant was already included.
ITPA Variants Were Associated With Protection Against Anemia Throughout the Course of Anti-HCV Therapy
The association between the ITPase deficiency variable and anemia was evaluated in week 12 for all patients and in week 24 for those patients remaining on treatment. Hb levels progressively decreased over the course of therapy. ITPase deficiency was associated with a lower absolute reduction in Hb at 12 and 24 weeks in comparison with patients predicted to have normal ITPase function (Figs. 1 and 2). At these later time points, moderate to severe ITPase deficiency was not associated with additive protection from anemia in comparison with patients with mild deficiency, although the number of patients predicted to have moderate to severe deficiency was low, and the rate of RBV dose reduction increased over time.
ITPA Variants Were Not Associated With the Rate of RBV Dose Modification or Treatment Outcome
RBV dose reduction was defined as a decrease in RBV dosing in any patient (step-down or discontinuation). Thirty-five patients (15%) had their RBV dose modified during therapy. Fewer patients who were treated for 12 weeks required RBV dose reduction in comparison with patients who were treated for 24 weeks (10% versus 19%, P = 0.06). There was no significant association between ITPA variants or ITPase deficiency and the need for RBV dose reduction either when the overall cohort was considered [26/171 (15%) with WT ITPase activity, 7/43 (16%) with mild ITPase deficiency, 1/21 (5%) with moderate ITPase deficiency, and 1/7 (14%) with severe ITPase deficiency were dose-reduced; P = 0.6175] or when the 12- and 24-week arms were considered separately (data not shown). We then used survival analysis to investigate the relationship between the predicted ITPase deficiency and the time to RBV dose reduction. To increase the power of the survival analysis, we combined patients with any degree of predicted ITPase deficiency and compared them to patients with normal ITPase activity. This plan was felt to be reasonable because of the limited size of the cohort and the observation that the protective effect of mild predicted ITPase deficiency was similar to that of moderate to severe predicted ITPase deficiency, particularly beyond week 4. Despite being associated with less severe anemia, ITPase deficiency was not associated with a lower rate of RBV dose reduction [hazard ratio = 0.80, 95% confidence interval (CI) = 0.38–1.71, P = 0.5679; Supporting Information Fig. 2A]. We repeated the survival analysis for patients treated for 12 or 24 weeks separately, but in neither case was an association with the rate of RBV dose reduction noted (data not shown). A relatively low Hb level of 9.5 g/dL was tolerated before dose reduction was indicated according to the study protocol. Therefore, we used a second survival analysis to plot the time to Hb < 10 g/dL; we censored at Hb < 10 g/dL (censor = 1) or at the end of follow-up/RBV dose reduction for an indication other than anemia (i.e., Hb > 10 g/dL, censor = 0). In this analysis, ITPase deficiency was associated with a delayed time to RBV dose reduction (hazard ratio = 0.25, 95% CI = 0.08–0.84, P = 0.0245; Supporting Information Fig. 2B).
rs1127354, rs7270101, and the combined ITPase deficiency variable were tested for an association with SVR in logistic regression models that included the IL-28B genotype (rs12979860), age, sex, body mass index, baseline HCV RNA level, HCV genotype, METAVIR fibrosis stage, standard/variable-duration therapy, and RBV dose. No association with SVR was observed (data not shown). There was also no association noted in this cohort between week 4 anemia and SVR whether it was considered as a quantitative trait or a categorical trait (Hb reduction > 3 g/dL). The IL-28B genotype was strongly and independently associated with SVR in this cohort, as recently reported.13
Discussion
To the best of our knowledge, this is the first report of an association between the recently described ITPA genetic polymorphisms and RBV-induced hemolytic anemia in the setting of chronic infection with genotype 2 or 3 HCV. We have confirmed that the ITPA variants are associated with protection from anemia during a shorter course of therapy with PEG-IFN plus RBV for genotype 2/3 disease.
Almost one-third of the cohort was predicted to have at least mild ITPase deficiency according to the ITPA haplotype, and these patients were observed to experience lesser degrees of hemolysis. The frequency of the protective variants was similar to that previously observed in a North American genotype 1 HCV population.10 In week 4, there was a trend toward an additive benefit in the small number of patients predicted to have moderate to severe ITPase deficiency, and this was consistent with previous observations.10,11 Thereafter, the degree of protection was similar in all patients harboring minor alleles for the ITPA variants, and this perhaps reflected the impact of RBV dose modifications.
The protective effect against anemia did not translate into differences in the overall need for RBV dose modification, however. In the original study protocol, a relatively low Hb level of 9.5 g/dL was tolerated before dose reduction was indicated. When we analyzed for the time to an Hb level of 10 g/dL, the current label indication for dose reduction in North America, ITPase deficiency was associated with a statistically significant benefit. One possible explanation for the lack of an association with RBV dose reduction was that the treating physicians had a higher tolerance for anemia in patients being treated for only 12 to 24 weeks, which masked an overall effect. Because ITPase deficiency did not predict RBV dose reduction, it was perhaps not surprising that the ITPA variants were not associated with treatment outcome. Similarly, no association between the ITPA variants, anemia, and SVR was demonstrated in two previous studies of genotype 1 HCV–infected patients.10,11
Therefore, despite the association between ITPA variants and protection from anemia, the potential clinical utility of ITPA genotyping in patients with genotype 2/3 HCV remains unclear on the basis of this data set. Furthermore, the standard recommended dose of RBV therapy is 800 mg daily and not 1000/1200 mg, which was used in this trial investigating variable-duration therapy, and ITPA variants might be expected to be less relevant in this setting. One potential role might be the pretreatment screening of individuals predicted to be at high risk for anemia or related morbidity, such as older patients or patients with chronic renal impairment or hemoglobinopathies. Prospective studies will be required to evaluate these specific groups of patients.
The two functional variants in the ITPA gene on chromosome 20 represent a missense variant in exon 2 (rs1127354, P32T) and a splice-altering SNP in intron 2 (rs7270101). Both polymorphisms have previously been well characterized as functional variants in studies of patients with ITPase deficiency, a benign inherited red cell enzymopathy in which inosine triphosphate (the substrate for ITPase) accumulates in erythrocytes.5–8 However, the mechanism by which ITPase deficiency protects against hemolysis is yet to be explained. RBV hemolysis is believed to occur secondarily to RBV triphosphate–induced oxidative damage to red cells, and a reasonable hypothesis would be that accumulated inosine triphosphate depletes red cell phosphate stores and prevents the conversion of RBV to the triphosphate from. This is speculative, however, and will be an important area for future investigation.
In conclusion, genetic polymorphisms in the ITPA gene were associated with protection from RBV-associated anemia in patients being treated with a short course of PEG-IFN plus high-dose RBV (1000/1200 mg daily) for genotype 2/3 HCV. In this cohort, however, ITPA variants were not associated with the need for RBV dose modification or treatment outcome, despite a median RBV dose of 15 mg/kg. The clinical utility of ITPA genotyping in the setting of genotype 2/3 HCV, therefore, remains unclear, particularly for treatment protocols in which 800 mg of RBV is used daily. It may find a role in the pretreatment assessment of patients at high risk for RBV-associated anemia or related morbidity.
Supplementary Material
Acknowledgments
This work was supported by the Duke Clinical Research Institute (Alexander J. Thompson and Paul J. Clark), the Richard B. Boebel Family Fund (Alexander J. Thompson and Paul J. Clark), the Gastroenterology Society of Australia (Alexander J. Thompson and Paul J. Clark), the National Health and Medical Research Council of Australia (Alexander J. Thompson), and the Royal Australasian College of Physicians (Alexander J. Thompson).
Abbreviations
- CI
confidence interval
- Hb
hemoglobin
- HCV
hepatitis C virus
- IL-28B
interleukin-28B
- ITPA
inosine triphosphatase
- ITPase
inosine triphosphatase
- PEG-IFN
pegylated interferon
- RBV
ribavirin
- RVR
rapid virological response
- SNP
single-nucleotide polymorphism
- SVR
sustained virological response
- WT
wild type
Footnotes
Alexander J. Thompson and Alessandra Mangia performed the primary data analysis and wrote the first draft of the manuscript with assistance from John G. McHutchison. Kevin V. Shianna and David B. Goldstein were responsible for genotyping the inosine triphosphatase polymorphisms. All authors had full access to the data in the study and contributed to the interpretation of the results. All authors reviewed the manuscript and provided further contributions and suggestions. All authors read and approved the final manuscript.
Potential conflict of interest: Dr. Thompson advises Roche and Merck; he also serves on the speaker’s bureau of Merck. Dr. Thompson receives travel grants from Gilead. Dr. Naggie advises Vertex. Dr. Goldstein consults for Abbott and receives grants from GSK. Dr. McHutchison owns intellectual property rights in IL2bB and receives royalties on testing. Dr. McHutchison consults for, advises, serves on the speaker’s bureau of, and receives grants from Merck/Schering.
Additional Supporting Information Information may be found in the online version of this article.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 3.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Manns M, Patel K, Poynard T, Lindsay KL, Trepo C, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 5.Sumi S, Marinaki AM, Arenas M, Fairbanks L, Shobowale-Bakre M, Rees DC, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–367. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 6.Maeda T, Sumi S, Ueta A, Ohkubo Y, Ito T, Marinaki AM, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency in the Japanese population. Mol Genet Metab. 2005;85:271–279. doi: 10.1016/j.ymgme.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Shipkova M, Lorenz K, Oellerich M, Wieland E, von Ahsen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52:240–247. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 8.Atanasova S, Shipkova M, Svinarov D, Mladenova A, Genova M, Wieland E, et al. Analysis of ITPA phenotype-genotype correlation in the Bulgarian population revealed a novel gene variant in exon 6. Ther Drug Monit. 2007;29:6–10. doi: 10.1097/FTD.0b013e3180308554. [DOI] [PubMed] [Google Scholar]
- 9.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 10.Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 11.Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–1189. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangia A, Santoro R, Minerva N, Ricci GL, Carretta V, Persico M, et al. Peginterferon alfa-2b and ribavirin for 12 versus 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2005;352:2609–2617. doi: 10.1056/NEJMoa042608. [DOI] [PubMed] [Google Scholar]
- 13.Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, et al. An IL28B polymorphism determines treatment response of patients with hepatitis C genotypes 2 or 3 who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–827. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


