Abstract
Background
A single nucleotide polymorphism (SNP) upstream of the IL28B gene (rs12979860) predicts sustained virological response (SVR) to peginterferon–ribavirin therapy in chronic hepatitis C patients. There is scarce information regarding the influence of this IL28B SNP on early viral kinetics during therapy, particularly in patients coinfected with HIV, in whom treatment response is lower than in hepatitis C virus (HCV)-monoinfected patients.
Methods
We selected 196 HIV/HCV-coinfected individuals who had completed a course of peginterferon–ribavirin therapy, and a validated outcome for SVR. Association of IL28B SNPs with rapid, early and end-of-treatment virological responses [rapid virological response (RVR), early virological response (EVR) and end of treatment virological response, respectively] was assessed in univariate and multivariate analyses.
Results
Rate of SVR in the study population was 54%. Frequency of the IL28B CC genotype was 44%. The distribution of HCV genotypes was as follows: HCV-1 57%, HCV-2 1%, HCV-3 30% and HCV-4 12%. Compared to CT/TT, the CC genotype was associated with significantly higher rates of all on-treatment viral outcomes, after adjusting for other predictors of viral response as serum HCV-RNA, HCV genotype and liver fibrosis staging. IL28B CC genotype kept its predictive power of SVR in patients who did not achieve RVR or cEVR. The association between IL28B SNP and viral kinetics and treatment outcomes was significant only for HCV genotypes 1 and 4.
Conclusion
IL28B CC genotype is a strong predictor of virological response to therapy in HIV/HCV-coinfected patients. This effect is mediated by an increase in viral clearance during the first 12 weeks of treatment and is mainly seen in patients infected with HCV genotypes 1 and 4.
Keywords: genetic polymorphisms, HIV/hepatitis C virus-coinfection, IL28B, interferon-l, interferon, rs12979860, viral kinetics
Introduction
Hepatitis C virus (HCV) infects more than 175 million people worldwide [1]. In western countries, HCV is the leading cause of end-stage liver disease and hepatocellular carcinoma, as well as the main indication for liver transplantation [1]. HCV and HIV-1 share routes of transmission and establish chronic infections; therefore, coinfection is relatively common (15–40%) [2]. Current therapy for chronic hepatitis C is based on a combination of peginterferon-α (pegIFNα) and ribavirin (RBV) given for 6–18 months, depending on early viral kinetics and HCV genotype [3]. Unfortunately, the medications are poorly tolerated and overall only half of patients achieve sustained HCV clearance [3]. This figure is lower in HIV/HCV-coinfected patients [2]. Thus, identification of baseline predictors of treatment success is desirable for making adequate treatment decisions, encouraging therapy in those with a high likelihood of cure and deferring treatment when the chances of response are minimal and drug-related side-effects may be troublesome.
Several variables associated with sustained virological response (SVR) can be found at baseline, before beginning therapy or during treatment. The best baseline predictors of SVR are HCV genotypes 2 or 3, low serum HCV-RNA, null or minimal liver fibrosis and younger age [4]. However, none of them, either individually or in combination, may preclude the administration of therapy to any patient [3,4], as a few individuals may respond in the absence of a good profile. Among the on-treatment variables predicting SVR, the attainment of undetectable serum HCV-RNA at week 4 (rapid virological response, RVR) is the best predictor of SVR [3]. However, a limited proportion of patients who do not achieve RVR may still be cured if therapy is continued, and thus all patients are encouraged to stay on therapy until week 12 [3,4]. Then, if serum HCV-RNA do not drop more than 2 logs (EVR, early virological response) or become undetectable (cEVR, complete EVR), treatment can be discontinued given that the chances of cure are rather low [3,4].
Different genome-wide association studies have identified single nucleotide polymorphisms (SNPs) upstream of the IL28B gene coding for IFN-λ3 that are strongly associated with the chance of SVR in HCV-mono-infected individuals [5–7]. Similar findings have recently been reproduced in HCV–HIV-coinfected patients [8]. The SNP with the strongest association, rs12979860, confers more than two-fold increase in the SVR rate after adjusting for other baseline variables. Thus, IL28B genotyping has emerged as one of the single strongest baseline predictors of treatment success, and a model has been developed to predict the probability of SVR based on the interplay between these variables [9]. What is less clear is in what extent baseline predictors of treatment success, including IL28 genotyping, may overcome the predictive power of on-treatment viral kinetics, as RVR. In other words, it seems crucial to unveil the relationship between IL28B genotypes and on-treatment virological responses. A few studies have already addressed this question in HCV-monoinfected individuals [10,11] and more recently, one study has examined the effect of IL28B genotypes on very early viral kinetics (at several time points during the first 4 weeks of therapy) in 26 HIV–HCV-coinfected individuals [12]. Our study is the first exploring the influence of IL28B genotypes on several on-treatment outcomes with clinical relevance, as well as its relationship with other well known predictors of response, in a large population of HIV–HCV-coinfected patients in whom viral kinetics and treatment outcomes may differ from HCV-monoinfected patients [13].
Patients and methods
Study population
Patients were recruited at Hospital Carlos III, a reference HIV clinic located in Madrid, Spain. From an initial cohort of 650 HIV/HCV-coinfected patients with regular follow-up, whose main characteristics have been described elsewhere [14], we selected 196 individuals who had completed a course of therapy with pegIFNα– RBV, had a validated outcome for SVR and had stored samples available for IL28B rs12979860 SNP genotyping. Only individuals naive for IFN were included in this analysis. Patients with HBV coinfection were excluded.
Treatment regimens included pegIFNα-2a or pegIFNα-2b at standard doses (180 µg/week or 1.5 µg/kg per week, respectively) and weight-adjusted RBV (1000 mg/day for patients weighting < 75 kg and 1200 mg/day for patients weighting > 75 kg). Following international guidelines for coinfected individuals [2], patients with HCV genotypes 1 or 4 received either 48 or 72 weeks of treatment, and patients with HCV genotypes 2 or 3 received 24 or 48 weeks of therapy, according to virological response at week 4. Early stopping rules were applied for patients with suboptimal virological response at week 12 [2].
The following on-treatment virological responses were assessed: RVR, EVR, cEVR and end of treatment virological response (EOTVR), reflecting undetectable serum HCV-RNA at the completion of therapy. SVR was defined as undetectable serum HCV-RNA 24 weeks after the end of treatment; otherwise, patients were considered as relapsers [15]. For the purpose of this study, relapsers were considered along with nonresponders, who were patients who experienced suboptimal virological response during the treatment period, and accordingly did not complete the planned duration of therapy. Patients with poor drug compliance and/or who discontinued therapy due to side-effects were excluded from this analysis.
To participate in the study, written informed consent for genetic testing was obtained from all individuals, and the study protocol was evaluated and approved by the hospital ethics committee. Both plasma and peripheral blood mononuclear cells (PBMCs) were stored for all patients.
Hepatitis C virus and HIV viremia and hepatitis C virus genotyping
Plasma HCV-RNA was measured using a real-time PCR assay (COBAS TaqMan; Roche, Barcelona, Spain), which has a lower limit of detection of 10 IU/ml. HCV genotyping was performed using a commercial real-time-PCR hybridization assay (Versant HCV Genotype v2.0 LiPA; Siemens, Barcelona, Spain), which maximally reduces the chances of HCV genotype misclassification [16]. Plasma HIV-RNA was measured using Versant HIV-1 RNA v3.0 (Siemens), which has a lower limit of detection of 50 copies/ml.
Liver fibrosis staging
The extent of liver fibrosis was measured using transient elastography by FibroScan (Echosens, Paris, France). Details about this noninvasive method, the examination procedure and correlation of liver fibrosis estimates with liver biopsy have been reported elsewhere [17,18]. The median value of all tests per patient is expressed in kilopascal (kPa) units. Advanced liver fibrosis (severe fibrosis or cirrhosis, corresponding to Metavir scores F3 and F4, respectively) was defined for liver stiffness values at least 9.5 kPa, based on results from studies conducted in both HCV-monoinfected and HIV/HCV-coinfected patients [19,20].
rs12979860 SNP genotyping
IL28 genotyping was performed at the Duke Institute for Genome Sciences and Policy and at Hospital Carlos III. It was conducted in a blinded fashion on DNA specimens collected from each individual, using the 5′ nuclease assay with allele-specific TaqMan probes (ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System; Applied Biosystems, Carlsbad, California, USA) [21]. Genotyping calls were manually inspected and verified prior to release.
Statistical analyses
The main characteristics of the study population, and the different parameters evaluated are expressed as median (interquartile range). Comparisons between groups were carried out using nonparametric analysis of variance (ANOVA). Associations between different qualitative parameters were explored using χ2 or Fisher’s exact tests, as appropriate. Univariable (χ2 test) and multivariable (logistic regression) analyses were used to assess the predictors of treatment outcome. Receiver operating characteristic (ROC) analysis was performed to estimate the cut-off values with the best positive predictive value (PPV) of treatment response. All statistical analyses were performed using the SPSS software version 13 (SPSS Inc., Chicago, Illinois, USA). All P values were two-tailed and were considered as significant only when below 0.05.
Results
Study population
Table 1 summarizes the main characteristics of the HIV/HCV-coinfected study population. Of note, all patients were of European ancestry and no Asians or Africans were present in this group. Up to 83% reported prior intravenous drug use as the primary route of HCV transmission. Most patients were men and only a quarter of the population presented low baseline serum HCV-RNA (<600 000 IU/ml). The median CD4 cell count was 483 cells/µl, with 74% of patients on antiretroviral therapy and undetectable plasma HIV-RNA. The proportion of patients with undetectable HIV viremia did not vary according to HCV genotype (76, 69 and 74% for HCV genotypes 1, 3 and 4, respectively). In untreated HIV-infected individuals, median plasma HIV-RNA was 3.4 (2.2–3.9) log copies/ml and the median CD4 cell count was 475 (370–652) cells/µl. HCV genotype distribution was as follows: HCV-1 57%, HCV-2 1%, HCV-3 30% and HCV-4 12%. Most patients (70%) had no evidence of significant liver fibrosis.
Table 1.
Baseline characteristics of the study population.
| Variable | All patients (n = 196) | CT/TT carriers (n = 110) | CC carriers (n = 86) | P |
|---|---|---|---|---|
| Age (years) | 42 (38–45) | 43 (38–45) | 41 (39–45) | 0.9 |
| Male sex | 75% (147) | 79% (87) | 70% (60) | 0.14 |
| Plasma HCV-RNA (log IU/ml) | 6.63 (5.87–7.05) | 6.68 (6.02–7.06) | 6.49 (5.56–7.04) | 0.12 |
| Plasma HCV-RNA < 600 000 IU/ml | 25% (48) | 20% (22) | 30% (26) | 0.1 |
| HCV genotypes | 0.001 | |||
| 1 | 57% (112) | 68% (74) | 44% (38) | |
| 2 | 1% (2) | 0% (0) | 2% (2) | |
| 3 | 30% (58) | 19% (21) | 43% (37) | |
| 4 | 12% (23) | 13% (14) | 11% (9) | |
| Liver stiffness (kPa) | 6.8 (5.1–10) | 7 (5.1–10) | 6.2 (4.8–11) | 0.28 |
| Significant liver fibrosis (≥9.5 kPa) | 30% (59) | 31% (34) | 27% (23) | 0.53 |
| CD4 cell count (cells/µl) | 483 (378–692) | 502 (398–720) | 480 (359–646) | 0.23 |
| Plasma HIV RNA < 50 copies/ml | 74% (145) | 72% (78) | 78% (66) | 0.39 |
| Liver enzymes | ||||
| AST IU/ml | 56 (43–85) | 49 (41–73) | 64 (48–97) | 0.02 |
| ALT IU/ml | 75 (52–112) | 64 (48–99) | 85 (61–136) | 0.02 |
| GGT IU/ml | 66 (40–131) | 76 (43–153) | 60 (36–99) | 0.08 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl transpeptidase. For continuous variables, all data are given as median (interquartile range).
In the whole population, the prevalence of the CC rs12979860 genotype was 44% (86 individuals). There were no significant differences between CC and non-CC patients for most baseline characteristics, except for the distribution of HCV genotypes and liver enzymes levels. The proportion of patients infected with HCV genotype 1 was higher in non-CC than CC patients (68 vs. 44%), whereas conversely HCV genotype 3 was more frequent in CC than non-CC patients (43 vs. 19%). Liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were significantly increased in CC compared to non-CC patients.
Impact of rs12979860 IL28B genotypes on virological responses
The proportion of patients achieving virological response at different time points is recorded in Table 2. The overall SVR rate in patients completing the planned duration of therapy was 54% (105/196). Information regarding RVR, EVR, cEVR and EOTVR was available for 139, 175, 175 and 189 patients, respectively. The overall rate of these on-treatment virological responses was 31% (43/139), 80% (140/175), 59% (104/175) and 65% (123/189), respectively. When patients were stratified according to IL28B genotype, the subset carrying the CC genotype showed a significantly greater rate of RVR, EVR, cEVR, EOTVR and SVR than those harboring CT/TT genotypes. Of note, half of CC patients achieved RVR and all attained EVR, whereas RVR only was seen in 17% of non-CC patients and only 62% of them attained EVR. Moreover, cEVR was achieved by 82% of CC patients but only in half (41%) of non-CC patients. Finally, EOTVR and SVR were also significantly higher in CC compared to non-CC patients (87 vs. 48% and 76 vs. 36%, respectively).
Table 2.
Treatment responses according to IL28B SNP genotypes in the study population.
| All patients | Total (n = 196) |
CT/TT carriers (n = 110) |
CC carriers (n = 86) |
P |
|---|---|---|---|---|
| RVR (n = 139) | 31% | 17% | 51% | <0.0001 |
| EVR (n = 175) | 80% | 62% | 100% | <0.0001 |
| cEVR (n = 175) | 59% | 41% | 82% | <0.0001 |
| EOTVR (n = 189) | 65% | 48% | 87% | <0.0001 |
| SVR (n = 196) | 54% | 36% | 76% | <0.0001 |
| HCV GT1/4 patients | (n = 135) | (n = 88) | (n = 47) | |
| RVR (n = 97) | 22% | 11% | 42% | <0.0001 |
| EVR (n = 119) | 71% | 54% | 100% | <0.0001 |
| cEVR (n = 119) | 45% | 29% | 72% | <0.0001 |
| EOTVR (n = 132) | 57% | 41% | 85% | <0.0001 |
| SVR (n = 135) | 42% | 28% | 68% | <0.0001 |
| HCV GT2/3 patients | (n = 61) | (n = 22) | (n = 39) | |
| RVR (n = 41) | 54% | 41% | 63% | 0.18 |
| EVR (n = 55) | 96% | 90% | 100% | 0.047 |
| cEVR (n = 55) | 91% | 84% | 94% | 0.22 |
| EOTVR (n = 56) | 84% | 75% | 89% | 0.18 |
| SVR (n = 60) | 78% | 67% | 85% | 0.10 |
cEVR, complete early virological response; EOTVR, end of treatment virological response; HCV, hepatitis C virus; RVR, rapid virological response; SVR, sustained virological response.
Given the large size of our cohort, a comparison of treatment outcomes in patients infected with HCV genotypes 1, 3 and 4 was performed. Significant differences were found between genotype 3 and 1 or 4, but no significant differences were found comparing genotypes 1 and 4. For this reason, genotypes 1 and 4 were grouped together in subsequent analyses.
These differences observed according to rs12979860 IL28B genotypes were maintained when the analysis was applied only to patients infected with HCV genotypes 1 or 4 (GT1/4 patients), whereas tended to vanish in patients infected with HCV genotypes 2 or 3 (GT2/3 patients). Thus, the beneficial effect of the CC genotype was mainly exerted in the difficult-to-treat GT1/4 patients, whereas in GT2/3 patients, the additional benefit of this genotype was only marginal.
Influence of rs12979860 IL28B genotypes on viral kinetics during the first 12 weeks of treatment
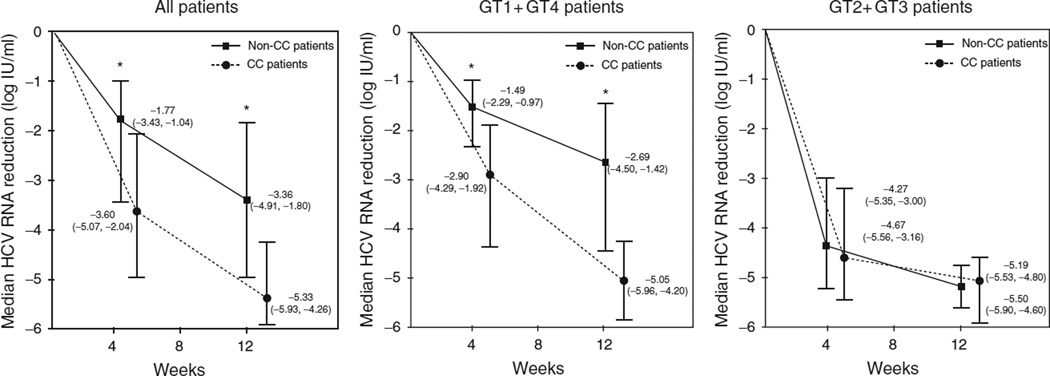
The strong association of IL28B CC genotype with on-treatment virological responses prompted us to explore the mechanism behind it. In a first step, the variation in serum HCV-RNA levels at weeks 4 and 12 with respect to baseline was calculated for each patient, and then the median variation at these two time points was compared in CC vs. non-CC patients. Interestingly, CC patients showed significantly greater declines in serum HCV-RNA at weeks 4 and 12 of treatment than non-CC patients (median decrease: 3.60 vs. 1.77 log IU/ml at week 4 and 5.33 vs. 3.36 log IU/ml at week 12, P < 0.0001 for both, Fig. 1). Whereas the same results were reproduced in GT1/4 patients, no significant differences in viral kinetics according to IL28 genotypes were seen in GT2/3 patients. Furthermore, most of the decrease in serum HCV-RNA in GT2/3 patients was attained within the first 4 weeks of therapy with only a marginal decrease between weeks 4 and 12. In contrast, in GT1/4 patients, serum HCV-RNA decreased to a much lesser extent during the first 4 weeks of therapy and continued to decline thereafter until week 12.
Fig. 1. Viral kinetics on the basis of IL28B genotype.
Viral kinetics on the basis of IL28B genotype in all patients, in patients infected with hepatitis C virus (HCV) genotypes 1 or 4 and in patients infected with HCV genotypes 2 or 3. Data show median (interquartile range) reduction of HCV viral load from baseline to 4 and 12 weeks post-HCV therapy. (*) P < 0.0001 for comparison between non-CC and CC patients using Mann-Whitney nonparametric test.
A linear regression analysis confirmed the association between the CC genotype and viral kinetics in GT1/4 patients, either at week 4 or 12. In this analysis, the IL28B genotype was the only variable significantly associated with the magnitude of serum HCV-RNA decrease at these two time points after controlling for baseline serum HCV-RNA, liver fibrosis staging, age and sex. The beta coefficients in the regression model for the CC genotype at weeks 4 and 12 were β = −1.20 and β = −1.79, respectively (P < 0.0001 for both).
Multivariable logistic regression analysis
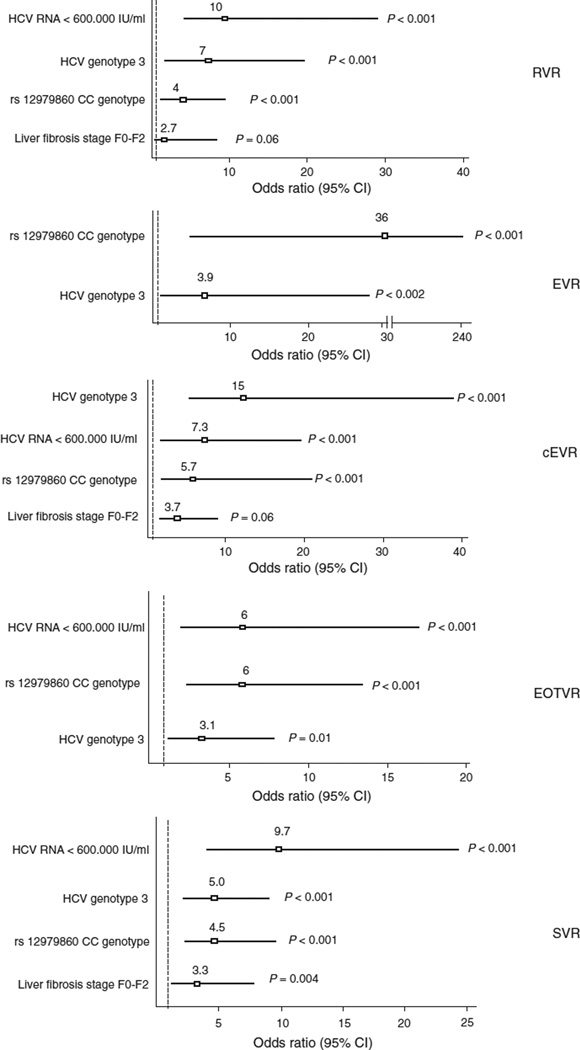
Given that multiple variables, including age, sex, BMI, serum HCV-RNA, detectable plasma HIV-RNA, HCV genotype and liver fibrosis staging have previously shown to influence HCV treatment outcomes, we examined the independent contribution of the IL28B genotype on virological responses. For this purpose, a multivariable logistic regression analysis, including all baseline predictors of virological response in the univariate analysis (baseline serum HCV-RNA < 600 000 IU/ml, HCV genotype 3, liver fibrosis Metavir stages F0–F2 and IL28B CC genotype) were assessed. Their respective contribution to RVR, EVR, cEVR, EOTVR and SVR is shown in Figure 2. It is noteworthy that the IL28B CC genotype remained as strong predictor of response at all different time points. Similar results were observed when this multivariable analysis was applied only to GT1/4 patients (data not shown), whereas in GT2/3 patients, there was no significant influence of the IL28B genotype on virological responses.
Fig. 2. Predictors of virological responses to peginterferon–ribavirin therapy in HIV/HCV-coinfected patients.
Horizontal axes represent adjusted odds ratio (95% confidence interval).
Interplay between IL28B genotype, early viral kinetics and sustained virological response
RVR and EVR are included in the current treatment guidelines as two therapeutic milestones that greatly influence SVR. It is unknown in what extent the impact of IL28B genotypes on SVR is mediated by early viral kinetics. In our study, overall 31% (43/139) of patients achieved RVR and almost all of them (93%, 40/43) attained SVR. Interestingly, in individuals without RVR (69%, 96/139), the presence of the IL28B CC genotype was significantly associated with more than two-fold increase in the SVR rate in comparison with non-CC carriers (64 vs. 27%, respectively). When adjustment was made for other baseline variables, the IL28 CC genotype in non-RVR patients was independently associated with SVR [odds ratio (OR) 8.7 (2.5–30), P = 0.001]. In agreement with this finding, the IL28B CC genotype predicted SVR independently of RVR, although the OR for RVR was higher [7.6 (1.8–31); P = 0.005] than for the IL28B CC genotype [3.9 (1.4–10.5), P = 0.009]. It is noteworthy that the beneficial effect of the CC genotype on SVR in the subset of patients lacking RVR was recognized not only in GT1/4 patients but also in GT2/3 patients. SVR rates were 89 and 50% in CC and non-CC carriers, respectively (P = 0.06) in non-RVR GT2/3 patients.
Overall 80% (140/175) of patients reached EVR. In this subset of patients, the presence of the IL28B CC genotype was slightly associated with an increased SVR rate (75 vs. 59%; P = 0.05). The impact of the IL28B genotype on SVR was particularly recognizable in the subset of individuals with partial EVR in comparison with those who achieved cEVR. Of 34 patients with partial EVR, SVR in CC and non-CC carriers was 43% (six of 14) and 10% (two of 20), respectively (P = 0.04). In contrast, these figures were 85 and 82%, respectively, in cEVR patients.
Predictors of virological failure
As lack of achievement of EVR is considered as treatment failure and accordingly is used as criterion to interrupt treatment, the ability to predict at baseline non-EVR could have a great impact on therapeutic decisions. This is particularly true for GT1/4 patients as non-EVR was recognized in 29% of them, whereas it was seen in only 4% of GT2/3 patients. In our series, lack of EVR was only seen in non-CC IL28B genotypes. In this subset of patients, the presence of high serum HCV-RNA (>600 000 IU/ml) had only 50% PPV of non-EVR. Using a ROC analysis, the serum HCV-RNA cut-off with the highest PPV of non-EVR was established at 7.28 log IU/ml [area under the receiver operating characteristic curve (AUROC): 0.66 + 0.06, P = 0.02]. For this threshold, the PPV of non-EVR was 95%, meaning that in GT1/4 non-CC patients with viremia above 7.28 log IU/ml, the rate of virological failure at week 12 was 95%. It must be kept in mind that 10% (13/135) of our GT1/4 patients meet all these criteria.
Discussion
The discovery of the rs12979860 IL28B polymorphism is expected to largely impact HCV therapy. In addition to the influence on SVR [5–8], emerging data support that early viral kinetics on treatment in HCV-monoinfected patients are significantly modulated by IL28B polymorphisms [10,11]. Our study is the first to confirm this finding in a large cohort of HIV/HCV-coinfected individuals in whom response rates are lower and exhibit different viral kinetics [12,22]. Our results suggest that although the effect of IL28B polymorphisms on SVR is largely mediated by its impact on early viral responses on treatment, as RVR and EVR, they exert its action beyond that, influencing the SVR regardless of the achievement of RVR. Importantly, the significant association between IL28B polymorphisms and all on-treatment responses was independent of other well known predictors of treatment response, such as serum HCV-RNA and liver fibrosis staging. An interesting finding was that our results were mainly confined to GT1/4 patients, whereas the benefit of the IL28B CC genotype on virological outcomes was only marginal in GT2/3 patients. Overall, our results largely confirm and extend the original findings from Thompson et al. [10] examining a population of HCV GT1-monoinfected individuals to HIV-coinfected individuals and other HCV genotypes.
An interesting observation in our study was that the CC genotype dramatically influenced viral kinetics during the first 12 weeks of HCV therapy. This is in agreement with findings from a recent study that has examined very early viral kinetics on therapy in a small group of 26 coinfected patients [12]. Our results confirm that HCV decay at weeks 4 and 12 of therapy is largely influenced by IL28B genotypes. As a result of this enhancement of viral kinetics in CC carriers, and given that baseline serum HCV-RNA was similar in CC and non-CC patients, the likelihood of attaining RVR and/or EVR was significantly greater in CC than non-CC patients. It is noteworthy that the boosting in viral kinetics conferred by the CC genotype was mainly recognized in GT1/4 patients, whereas the much faster viral kinetics in GT2/3 patients was independent of the IL28B genotype. Altogether, these results suggest that different mechanisms of treatment-induced viral clearance are operating for distinct HCV genotypes.
The biological mechanisms involved in the boosting of viral kinetics associated with the CC genotype are not well understood. The IFN-λ3, a type III IFN, depicts strong in-vitro activity against HCV [23]. Both IFN-λ3 and IFNα share signaling cascades within the cells [24] and may exert an additive effect in their antiviral effect [25]. Thus, it may be hypothesized that individuals with the CC genotype may show a differential IL28B expression, whose antiviral effect is boosted when exogenous IFNα is given. Alternatively, immune pathways such as T-cell-mediated adaptive immunity might play a role. In this regard, it must be highlighted that the IFN-λ family upregulates the production of IFN-γ in response to viral infections [26]. Given the immunomodulatory action of IFN-γ and its role in recruiting T cells to livers in patients with chronic viral infections [27], IFN-λ could exert its action indirectly through IFN-γ, promoting the recruitment of T cells into the liver without inducing the expression of IFN-stimulated genes. As T-cell-mediated immunity has been proposed as one of the major mechanisms involved in the control of HCV replication [28–31], distinct IFN molecules might exert their effects using separate but interlinked branches of the immune network.
Finally, our results have several important clinical implications. First, the beneficial effect of the IL28B CC genotype is mainly seen in the difficult-to-treat GT1/4 patients. Second, we found that the influence of the CC genotype on SVR was not only mediated by its effect on RVR, as the CC genotype still kept its predictive value of SVR in patients without RVR. In fact, patients with the CC genotype lacking RVR have a more than two-fold increase in SVR compared to non-CC genotype patients. Moreover, this beneficial effect of CC genotype in patients not achieving RVR was also present in patients infected with GT2/3. Given that RVR was a stronger predictor of SVR than the CC genotype, our data reinforce the need for assessing treatment response at week 4 in all patients regardless of the baseline profile.
Third, current guidelines for early stopping rules requires treatment exposure of at least 12 weeks in all patients, with clear impact on healthcare costs and undesirable side-effects in patients who will not ultimately benefit from treatment. Our results suggest that combination of IL28 genotyping along with week 4 response might confidently advance discontinuation of therapy in the subset of patients lacking RVR, carrying non-CC genotype and presenting high baseline HCV load. In our cohort, this subset of patients represented 10% of all GT1/4 patients.
Lastly, in the group of patients experiencing partial EVR (18% of our global study population), the presence of the IL28 CC genotype significantly increased the probability of attaining SVR (43 vs. 10% for CC and non-CC carriers, respectively). Thus, although the benefit of the CC genotype is mainly operating through its effect on viral kinetics during the first 12 weeks of therapy, it extends beyond that point. Altogether, our findings support IL28B genotyping in chronic hepatitis C patients planned to be treated with pegIFN–RBV therapy, as this information seems to be crucial to assist treatment decisions both at baseline and during therapy.
In summary, our study demonstrates an important role for IL28B genotypes in predicting on-treatment virological responses to pegIFNα–RBV therapy in HIV/HCV-coinfected individuals. The benefit of the CC genotype is largely mediated by an enhancement of viral kinetics during the first 12 weeks of therapy. Interestingly, this effect is mainly seen in GT1/4 patients and only a marginal effect is seen in GT2/3 patients. Knowledge at baseline of the IL28B genotype may helpfully assist in treatment decisions, both at the time of deciding who should be treated and during treatment.
Acknowledgments
The authors would like to thank all patients who participated in the study. This work was supported in part by grants from Fundaciόn Investigacion y Educacion en SIDA (IES), Red de Investigacion en SIDA (RIS, FIS-RD06/0006), Agencia Lain Entralgo and the European Union 6th Framework Programme (NEAT, No. LSHP-CT-2006-037570).
Footnotes
The authors acknowledge no financial conflict of interest with this work.
References
- 1.Lauer G, Walker B. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Soriano V, Puoti M, Sulkowski M, Cargnel A, Benhamou Y, Peters M, et al. Care of patients coinfected with HIV and hepatitis C virus: 2007 updated recommendations from the HCV-HIV International Panel. AIDS. 2007;21:1073–1089. doi: 10.1097/QAD.0b013e3281084e4d. [DOI] [PubMed] [Google Scholar]
- 3.Ghany M, Strader D, Thomas D, Seeff L. American Association for the Study of Liver Diseases Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau A, Vermehren J, Sarrazin C. Treatment predictors of a sustained virologic response in hepatitis B and C. J Hepatol. 2008;49:634–651. doi: 10.1016/j.jhep.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Ge D, Fellay J, Thompson A, Simon J, Shianna K, Urban T, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 6.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 8.Rallόn NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–F29. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medrano J, Keulen K, Rallόn N, Macías J, Resino S, Benito JM, Sanchez-Piedra C, et al. Modelling the probability of sustained virological response to peginterferon-ribavirin therapy in HCV-HIV coinfected patients. Clin Infect Dis. 2010;51:1209–1216. doi: 10.1086/656811. [DOI] [PubMed] [Google Scholar]
- 10.Thompson A, Muir A, Sulkowsky M, Ge D, Fellay J, Shianna K, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Stättermayer A, Stauber R, Hofer H, Rutter K, Beinhardt S, Scherzer T, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naïve patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–350. doi: 10.1016/j.cgh.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 12.de Araujo E, Dari D, Cottler S, Layden T, Neumann A, Melo C, et al. Pharmacodynamics of PEG-IFNα-2a and HCV response as a function of IL28B polymorphisms in HIV/HCV-coinfected patients. J Acquir Immune Defic Syndr. 2011;56:95–99. doi: 10.1097/QAI.0b013e3182020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soriano V, Nuñez M, Camino N, Barreiro P, Garcia-Samaniego J, Gonzalez-Lahoz J. Hepatitis C virus-RNA clearance in HIV-coinfected patients with chronic hepatitis C treated with pegylated interferon plus ribavirin. Antivir Ther. 2004;9:505–509. [PubMed] [Google Scholar]
- 14.Medrano J, Resino S, Vispo E, Madejόn A, Labarga P, Tuma P, et al. Hepatitis C virus (HCV) treatment uptake and changes in the prevalence of HCV genotypes in HIV-HCV coinfected patients. J Viral Hepat. 2011;18:325–330. doi: 10.1111/j.1365-2893.2010.01309.x. [DOI] [PubMed] [Google Scholar]
- 15.Medrano J, Barreiro P, Resino S, Tuma P, Rodríguez V, Vispo E, et al. Rate and timing of hepatitis C virus relapse after a successful course of pegylated interferon plus ribavirin in HIV-infected and HIV-uninfected patients. Clin Infect Dis. 2009;49:1397–1401. doi: 10.1086/630205. [DOI] [PubMed] [Google Scholar]
- 16.Chevaliez S, Bouvier-Alias M, Brillet R, Pawlotsky JM. Hepatitis C virus (HCV) genotype 1 subtype identification in new HCV drug development and future clinical practice. PLoS One. 2009;4:e8209. doi: 10.1371/journal.pone.0008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziol M, Handra-Luca A, Kettaneh A, Christidis C, Mal F, Kazemi F, et al. Noninvasive assessment of liver fibrosis by measurement of stiffness in patients with chronic hepatitis C. Hepatology. 2005;41:48–54. doi: 10.1002/hep.20506. [DOI] [PubMed] [Google Scholar]
- 18.Castera L, Vergniol J, Foucher J, Le Bail B, Chanteloup E, Haaser M, et al. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. 2005;128:343–350. doi: 10.1053/j.gastro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 19.de Ledinghen V, Douvin C, Kettaneh A, Ziol M, Roulot D, Marcellin P, et al. Diagnosis of hepatic fibrosis and cirrhosis by transient elastography in HIV/hepatitis C virus-coinfected patients. J Acquir Immune Defic Syndr. 2006;41:175–179. doi: 10.1097/01.qai.0000194238.15831.c7. [DOI] [PubMed] [Google Scholar]
- 20.Kirk G, Astemborski J, Mehta S, Spoler C, Fisher C, Allen D, et al. Assessment of liver fibrosis by transient elastography in persons with hepatitis C virus infection or HIV-hepatitis C virus coinfection. Clin Infect Dis. 2009;48:963–972. doi: 10.1086/597350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K. Allelic discrimination using fluorogenic probes and the 5’ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 22.Avidan N, Goldstein D, Rozenberg L, McLaughlin M, Ferenci P, Masur H, et al. Hepatitis C viral kinetics during treatment with pegIFN alpha-2b in HIV/HCV coinfected patients as a function of baseline CD4+ T-cell counts. J Acquir Immune Defic Syndr. 2009;52:452–458. doi: 10.1097/QAI.0b013e3181be7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robek M, Boyd B, Chisari F. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotenko S, Gallager G, Baurin V, Lewis-Antes A, Shen M, Shah N, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 25.Marcello T, Grakoui A, Barba-Spaeth G, Machlin E, Kotenko S, MacDonald M, Rice C. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 26.Ank N, West H, Bartholdy C, Eriksson K, Thomsen A, Paludan S. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalor P, Shields P, Grant A, Adams D. Recruitment of lymphocytes to the human liver. Cell Biol. 2002;80:52–64. doi: 10.1046/j.1440-1711.2002.01062.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamal S, Fehr J, Roesler B, Peters T, Rasenack J. Peginterferon alone or with ribavirin enhances HCV-specific CD4+ T-helper 1 responses in patients with chronic hepatitis C. Gastroenterology. 2002;123:1070–1083. doi: 10.1053/gast.2002.36045. [DOI] [PubMed] [Google Scholar]
- 29.Sreenarasimhaiah J, Jaramillo A, Crippin J, Lisker-Melman M, Chapman W, Mohanakumar T. Concomitant augmentation of type 1 CD4+ and CD8+ T-cell responses during successful interferon-a and ribavirin treatment for chronic hepatitis C virus infection. Hum Immunol. 2003;64:497–504. doi: 10.1016/s0198-8859(03)00041-7. [DOI] [PubMed] [Google Scholar]
- 30.Capa L, Soriano V, Garcia-Samaniego J, Nuñez M, Romero M, de Mendoza C, et al. Evolution of T cell responses to hepatitis C virus (HCV) during pegylated interferon plus ribavirin treatment in HCV-monoinfected and HCV-HIV coinfected patients. Antivir Ther. 2007;12:459–468. [PubMed] [Google Scholar]
- 31.Barnaba V. Hepatitis C virus infection: a ‘liaison a trois’ amongst the virus, the host, and chronic low-level inflammation for human survival. J Hepatol. 2010;53:752–761. doi: 10.1016/j.jhep.2010.06.003. [DOI] [PubMed] [Google Scholar]