Abstract
Diacylglycerol (DG) lipase, which hydrolyses 1-stearoyl-2-arachidonyl-sn-glycerol to produce an endocannabinoid, 2-arachidonoylglycerol, was purified from the soluble fraction of rat brain lysates. DG lipase was purified about 1,200-fold by a sequential column chromatographic procedure. Among proteins identified by mass spectrometry analysis in the partially purified DG lipase sample, only DDHD domain containing two (DDHD2), which was formerly regarded as a phospholipase A1, exhibited significant DG lipase activity. Rat DDHD2 expressed in Chinese hamster ovary cells showed similar enzymatic properties to partially purified DG lipase from rat brain. The source of DG lipase activity in rat brain was immunoprecipitated using anti-DDHD2 antibody. Thus, we concluded that the DG lipase activity in the soluble fraction of rat brain is derived from DDHD2. DDHD2 is distributed widely in the rat brain. Immunohistochemical analysis revealed that DDHD2 is expressed in hippocampal neurons, but not in glia.
Keywords: 2-arachidonoylglycerol, diacylglycerol, endocannabinoid, lipase, phospholipase
Diacylglycerol (DG) lipase is a key enzyme in the pathway of 2-arachidonoylglycerol (2-AG) biosynthesis (1–3). 2-AG, an endocannabinoid, induces various effects by acting on cannabinoid receptors, CB1 and CB2 (1–3). CB1 is highly expressed in the central nervous system (CNS) (4, 5). An important physiological function of 2-AG is the modulation of neurotransmission through CB1 in the CNS. 2-AG has been shown to modulate long-term potentiation, a model for learning and memory (6). Depolarization-induced suppression of inhibition (DSI) is a well-studied electrophysiological process involving endocannabinoids, especially 2-AG (6–8). DSI is the phenomenon whereby post-synaptic depolarization induces inhibition of the release of the neurotransmitter gamma aminobutyric acid. DSI is thought to be one of the mechanisms of short-term synaptic plasticity in the hippocampus.
Endocannabinoid signalling also contributes to a wide range of processes including axonal growth and guidance during development, adult neurogenesis and many behavioural responses (9–11).
The main pathway for the biosynthesis of 2-AG is thought to be the hydrolysis of arachidonic acid-containing DG at the sn-1 position. DG is produced from inositol phospholipids by phospholipase C (PLC) through the activation of metabotropic glutamate receptor (12, 13). Bisogno et al. reported the cDNA cloning of two sn-1 DG lipases, DGLα and DGLβ (14). These enzymes are membrane proteins and cleave the fatty acid at the sn-1 position of DG. Analysis of DGLα-deficient mice revealed that 2-AG produced by DGLα is required for DSI in hippocampal neurons (15, 16). It is unknown whether other DG lipases are involved in the modulation of neurotransmission. DG lipase activity in bovine aorta was observed in the soluble fraction, but not in the membrane fraction (17, 18), which indicated that the activity was not derived from DGLα or DGLβ. Attempts have been made to purify the soluble DG lipase, but it has not yet cloned.
In this study, we purify DG lipase in the soluble fraction of rat brain using sequential column chromatography and show that DDHD domain containing two (DDHD2), also known as phospholipase A1 (PLA1), is responsible for the DG lipase activity.
Materials and Methods
Materials
1-Stearoyl-2-[1-14C]arachidonoyl-sn-glycerol ([14C]SAG) was purchased from GE Healthcare (Little Chalfont, UK). 1-Stearoyl-2-[1-14C] arachidonoyl-sn-glycero-3-phosphocholine was purchased from Muromachi Technos (Tokyo, Japan). 1-Stearoyl-2-arachidonoyl-sn-glycerol (SAG), 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphocholine, 2-arachidonoylglycerol (2-AG), Bacillus cereus PLC and tetrahydrolipstatin (THL) were products of Sigma-Aldrich (St. Louis, MO, USA). 1-Stearoyl-2-arachidonoyl-sn-glycerol 3-phosphate was purchased from Avanti Polar Lipids (Alabaster, AL, USA). RHC 80267 was purchased from BIOMOL (Plymouth, PA, USA). Thin-layer chromatography (TLC) plates (Silica gel 60) were purchased from Merck (Darmstadt, Germany). POROS HP2 and POROS HQ columns were obtained from Applied Biosystems (Foster City, CA, USA). Bio-Scale CHT 5-I columns were purchased from Bio-Rad (Hercules, CA, USA). Superdex 200 10/300 GL and PD-10 columns were obtained from GE Healthcare. Polymerase chain reaction (PCR) primers were purchased from Operon Biotechnologies (Tokyo, Japan). HEPES and sodium deoxycholate (DOC) were purchased from Dojindo (Kumamoto, Japan). cDNA clones were purchased from Openbiosystems (Huntsville, AL, USA): acyl-peptide hydrolase in pCMV-SPORT6.ccdb (clone ID: 7937766), anti-depressant-related protein ADRG123 in pExpress-1 (clone ID: 7103903), dihydropyrimidinase-like two in pExpress-1 (clone ID: 7107549) and TUC-4b in pCMV-SPORT6 (clone ID: 6177053). Human DDHD2 cDNA was purchased from Kazusa DNA Research Institute (clone ID: pF1KA0725). SYPRO Ruby was purchased from Life Technologies. All other reagents were purchased from Wako Pure Chemicals (Osaka, Japan).
Preparation of SAG
[1-14C] Arachidonoyl-sn-glycero-3-phosphocholine was evaporated under a stream of N2 and resuspended in a reaction solution containing 2 ml of diethyl ether and 1 ml of 0.1 M Tris–HCl pH 7.0 by sonication. After adding 25 units of Bacillus cereus PLC, the reaction mixture was stirred for 2 h at room temperature. The organic phase was dried under a stream of N2 and resolved in chloroform/methanol (2:1) (v/v).
Measurement of DG lipase activity
A total of 4.5 nmol [14C]SAG was evaporated under a stream of N2 and resuspended in 400 µl of assay buffer (0.125 M NaCl, 0.125 M Tris–HCl pH 8.0, 1.25 mM EDTA, 6.25 mM DOC) by sonication and vortexing. After 2 min of preincubation at 37°C, 100 µl of the enzyme solution was added. The mixture was incubated at 37°C for 10 min. Reactions were stopped by adding 2.1 ml of chloroform/methanol (1:2) (v/v). Lipids were extracted by the Bligh and Dyer method (19). The organic phase was dried using a centrifugal evaporator. The lipids were dissolved in 50 µl of chloroform/methanol (2:1) (v/v) and were separated by TLC using benzene/diethylether/ethanol/ammonia (40:140:4:0.2, v/v/v/v) as a developing solvent. TLC plates were exposed to imaging plates (BAS-MS2040; Fuji Film, Tokyo, Japan) overnight. The radioactivity was quantified using a Typhoon 9210 imager (GE Healthcare). The kinetic parameters were calculated using Prism software (GraphPad, La Jolla, CA, USA).
For the screening of DG lipase in Chinese hamster ovary (CHO) cells, the same assay conditions were applied except for using 40 µM cold SAG. The detection of SAG and 2-AG was performed using iodine vapour.
Animals
The animal study was approved by Gunma University Animal Committee (Permit Number: 13-018), and the rats were treated in accordance with the Gunma University guidelines for the care and use of laboratory animals. All efforts were made to minimize their suffering.
Purification of DG lipase from rat brain
All purification steps except for column chromatography were carried out at 4°C. A total of 20–40 Wistar rats (male, 6 weeks old) were anaesthetized using diethylether and then decapitated. Immediately, the whole brains were removed and homogenized in 10 volumes of a homogenizing buffer (0.3 M sucrose, 50 mM Tris–HCl, pH 7.4, 1 mM EDTA, 1 mM DTT) using a Potter-Elvehjem glass-Teflon homogenizer. After the homogenates had been centrifuged at 105,000 × g and 4°C for 60 min, the supernatants were subjected to ammonium sulphate precipitation. Samples were adjusted to 20% saturation of ammonium sulphate by the addition of solid ammonium sulphate, equilibrated at 4°C for 15 min, and centrifuged at 10,000 × g for 30 min. After centrifugation, the supernatant was adjusted to 40% saturated ammonium sulphate. The protein solution was centrifuged at 10,000 × g for 30 min. Pellets were resolved in 50 mM sodium phosphate, pH 7.2, containing 1 mM EDTA, 1 mM DTT and 1.5 M ammonium sulphate. The protein solution was loaded onto a POROS HP2 hydrophobic interaction column (4.6 × 100 mm), pre-equilibrated with 50 mM sodium phosphate, pH 7.2, containing 1 mM EDTA, 1 mM DTT, 1.5 M ammonium sulphate and 0.05% Triton X-100. The protein was eluted by decreasing the ammonium sulphate concentration from 1.5 to 0 M. Pooled DG-lipase-containing fractions were desalted using a PD-10 column, pre-equilibrated with 20 mM Tris–HCl, pH 7.0, 1 mM EDTA and 1 mM DTT, and were eluted using the same buffer. The protein solution was then loaded onto a POROS HQ column (4.6 × 100 mm), pre-equilibrated with 20 mM Tris–HCl, pH 7.0, containing 1 mM EDTA, 1 mM DTT and 0.05% Triton X-100. The protein was further eluted with a linear gradient of NaCl from 0 to 1 M. Pooled DG-lipase-active fractions were desalted using PD-10 columns pre-equilibrated with 10 mM sodium phosphate pH 7.2, containing 1 mM DTT and 0.05% Triton X-100, and were eluted with the same buffer. The desalted protein solution was further applied to a Bio-Scale CHT 5-I column (10 × 64 mm), a hydroxyapatite column pre-equilibrated with 10 mM sodium phosphate pH 7.2, containing 1 mM DTT and 0.05% Triton X-100. The protein was eluted using increasing concentrations of sodium phosphate, pH 7.2, from 10 to 500 mM. After concentrating the DG lipase-active fractions using centrifugal filter devices (Microcon; Millipore, Billerica, MA, USA), DG lipase was further purified using a Superdex 200 10/300 GL gel filtration column (1 × 300 mm), pre-equilibrated with 50 mM sodium phosphate, pH 7.2, containing 150 mM NaCl, 1 mM EDTA, 1 mM DTT and 0.05% Triton X-100. All chromatography steps were performed using the BioCAD SPRINT system (Applied Biosystems) or the AKTApurifier 10 system (GE Healthcare). The fractions that showed DG lipase activity after the CHT 5-I column were used for characterization of the enzymatic properties.
Effects of inhibitors on the enzyme activity
THL and RHC 80267 were dissolved in dimethylsulfoxide (DMSO) to give stock solutions of 10 and 20 mM, respectively. THL and RHC 80267 were diluted with DMSO so that the volume of solvent added to the assay mixture was 0.5%. Both inhibitors were added to the assay mixture just before the start of the preincubation.
Identification of proteins in partially purified DG lipase using mass spectrometry
Partially purified DG lipase (4 µg) from rat brain was completely dried in a centrifugal evaporator (TOMY Seiko, Tokyo, Japan) and dissolved in 13 µl of 8 M urea, 0.5 M Tris–HCl, pH8.5 and 1 mM EDTA. The solution was incubated at 37°C for 90 min after the addition of DTT at a final concentration of 35 mM. Subsequently, 5 µl of 40 mg/ml iodoacetamide (final concentration 10 mg/ml) was added and incubated at 37°C for 30 min. The alkylated protein solution was diluted with 60 µl of pure water. Trypsin (Promega, final concentration 1 µg/ml) was added to the solution and incubated overnight at 37°C. The digestion reaction was terminated by adding formic acid at a final concentration of 0.5%.
The reversed-phase liquid chromatography tandem mass spectrometry (MS/MS) analysis was performed on NanoFrontier eLD (Hitachi High-Technologies, Tokyo, Japan). The digested peptides were eluted through a MonoCap for FastFlow (0.05 mm i.d. × 150 mm) C18 monolith capillary column by a linear gradient of mobile phase A/B = 98/2 to 60/40 (where mobile phase A consisted of H2O/acetonitrile/formic acid = 98/2/0.1 (v/v/v) and mobile phase B consisted of H2O/acetonitrile/formic acid = 2/98/0.1 (v/v/v)) for 90 min at a flow rate of 200 nl/min, and then subjected to online mass spectrometer by electrospray ionization. The MS/MS spectra were acquired in a positive ion mode by a collision-induced dissociation method. Proteins were identified using Mascot (Matrix Science, London, UK) (20) with the NCBI database.
cDNA cloning of rat DDHD2
Total RNA was prepared from rat brain using TRIZOL reagent (Life Technologies). Poly A+ RNA was purified from the total RNA using Oligotex-dT30 (Roche Diagnostics, Basel, Switzerland). A library of adaptor-ligated double-strand cDNA was prepared using the Marathon cDNA Amplification Kit (Clontech, Mountain View, CA, USA). Rat DDHD2 cDNA was amplified by PCR using the primers 5′-AACTCGAGGATCATCAGGGGAATCGTACCAGG-3′ and 5′-TTGCGGCCGCTTACTGTAAAGGCTGATCAAGGAAGATC-3′ and the cDNA library as template DNA. The 5′ untranslated regions (UTRs) of rat DDHD2 cDNA were also cloned and sequenced using Marathon cDNA Amplification Kit (Clontech). The primers used to amplify the 5′ UTR of rat DDHD2 transcripts were an adaptor primer (5′-CCATCCTAATACGACTCACTATAGGGC-3′) and a gene-specific primer (5′-GCCGACCTTGCTCAGTGGATGTTGAACCCC-3′). The 5′-rapid amplification of cDNA ends (RACE) PCR product was cloned into T-Vector pMD20 (Takara, Tokyo, Japan) and sequenced (accession number: AB914679).
Vector construction for the expression of rat and human DDHD2
Human DDHD2 cDNA was amplified by PCR using the primers 5′-AACTCGAGGATCATCAGTGCAGTCACAACAGG-3′ and 5′-TTGCGGCCGCTTACTGTAAAGGCTGATCAAGGAAGATACC-3′ and the plasmid vector (pF1KA0725: Kazusa DNA Research Institute) as template DNA. To prepare an expression vector of rat and human DDHD2, the PCR products were subcloned into XhoI and NotI sites of pCMVHA.
Expression of recombinant protein in CHO cells
CHO cells were maintained in Ham’s F12 medium (Life Technologies) containing 10% foetal calf serum (Sigma-Aldrich). The cells were cultured at 37°C in a humidified incubator with 5% CO2. The cells were transfected with each expression vector using Fugene6 (Roche Diagnostics). After 24 h, the cells were collected and washed with phosphate-buffered saline (PBS). The CHO cells were disrupted using a Dounce homogenizer in homogenizing buffer (20 mM HEPES–NaOH pH 7.4, 0.25 M sucrose). The soluble fraction was prepared by sequential centrifugation at 1,000 × g for 7 min, 2,000 × g for 30 min and 105,000 × g for 1 h. The resultant precipitate was resuspended with the homogenizing buffer.
Polyclonal antibody of rat DDHD2
A rabbit antiserum was obtained by immunizing a rabbit with a synthesized peptide derived from rat DDHD2 (amino acids 611–625), adding a cysteine at the N-terminus (CSEPSSEKPSDANTEE) (Scrum, Tokyo, Japan). Rabbit immunoglobulin was purified using protein G Sepharose FF beads (GE Healthcare) and eluted with 0.1 M glycine-HCl, pH 2.8. The obtained immunoglobulin solution was loaded onto a peptide affinity column made of Sulfolink Coupling gel (1 ml) (Thermo Fisher Scientific, Waltham, MA, USA) bound with the synthesized peptide. The column was washed with 1 ml of PBS three times. The DDHD2 polyclonal antibody was eluted with 3 M MgCl2 and then dialysed with PBS.
Immunoprecipitation of rat brain DDHD2
Rat whole-brain homogenates were centrifuged at 105,000 × g for 1 h. The supernatant was fractionated by ammonium sulphate precipitation. The 20–40% ammonium sulphate precipitate fraction from rat brain was dissolved in 20 mM HEPES–NaOH, pH 7.4, containing 1 mM EDTA. A 300 µl solution containing 1.3 mg protein from the 20–40% ammonium sulphate precipitate fraction, 10 µg of anti-rat DDHD2 antibody, 20 mM HEPES, pH 7.4, 1 mM EDTA, 0.05% Triton X-100 and 0.05% DOC was then incubated at 4°C for 1 h. After adding 25 µl of Protein G Sepharose FF beads, the solution was gently agitated at 4°C for 1 h. The beads were washed twice with 0.5 ml of wash buffer (20 mM HEPES–NaOH, pH 7.4, 1 mM EDTA, 0.1% Triton X-100 and 0.1% DOC). The beads then were suspended with 50 µl of the wash buffer and used for the western blotting and enzyme assay.
Purification of recombinant rat DDHD2
The soluble fraction of CHO cells expressing N-terminal HA-tagged rat DDHD2 (HA-rDDHD2) was obtained as described above. Fifteen hundred microliters of the protein solution containing 36.3 mg of protein of the soluble fraction, 120 µg of anti-rat DDHD2 antibody and 0.1% Triton X-100 was incubated for 1 h at 4°C. After adding 200 µl of Protein G Sepharose FF beads, the solution was gently agitated at 4°C for 1 h. The beads were washed twice and suspended with 750 µl of the wash buffer. HA-rDDHD2 was eluted with the wash buffer containing 2.1 mg/ml synthesized peptide that was used for immunization of the rabbit. The protein sample was used for the Km determination and inhibitor experiments.
Western blotting
Rat brain was dissected into several regions. Tissues were homogenized in 50 mM Tris–HCl, pH 7.4, containing 1 mM EDTA, 1 mM DTT and complete protease inhibitor cocktail (Roche Diagnostics).
Protein samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a polyvinylidene difluoride (PVDF) membrane (Clear Blot Membrane-P; ATTO, Tokyo, Japan). This membrane was blocked with 5% non-fat milk (Snow Brand Milk Products, Tokyo, Japan) in PBS containing 0.1% Tween 20 (PBS-T) at 4°C overnight. It was then incubated in 5% non-fat milk in PBS-T containing the purified polyclonal anti-rat DDHD2 antibody (1.3 µg/ml), anti-HA monoclonal antibody (100 ng/ml; 3F10, Roche Diagnostics) or anti-β-tubulin III antibody (200 ng/ml; Sigma) at room temperature for 1 h. Horse radish peroxidase (HRP)-conjugated anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, USA) or HRP-conjugated anti-rat IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as the secondary antibody. The protein bands were visualized using ECL-prime western blotting detection reagents (GE Healthcare) and the LAS-4010 imaging system (GE Healthcare).
Immunostaining of rat brain DDHD2
Six-micrometre-thick paraffin-embedded sagittal sections from rat brain (Zyagen, San Diego, CA, USA) were deparaffinized in xylene and then rehydrated. After washing with water for 5 min three times, the sections were heated at 98°C for 40 min in Immunosaver solution (Nisshin EM, Tokyo, Japan) for antigen retrieval. After again washing with water for 5 min three times, the endogenous peroxidase activity was blocked with 3% H2O2 for 10 min. After washing with PBS-T three times, normal goat serum was used for the blocking of non-specific binding. Anti-rat DDHD2 polyclonal antibody (1.8 µg/ml) was applied to the sections at room temperature for 30 min, followed by washing with PBS-T three times. The tissue sections were stained using the VECTASTAIN Elite ABC kit in accordance with the manufacturer’s instructions (Vector Laboratories, Burlingame, CA, USA). The sections were visualized with 3,3′-diaminobenzidine (DAB) (Sigma-Aldrich) in Tris–HCl, pH 7.4, containing 0.024% H2O2. All sections were counterstained with Mayer’s haematoxylin (Wako).
For the immunofluorescent staining, 6-µm-thick paraffin coronal sections No.10 from rat brain (Zyagen) were used. Blocking was performed with 1% bovine serum albumin in PBS after antigen retrieval. Then, anti-rat DDHD2 polyclonal antibody (1.8 µg/ml), anti-MAP2 antibody (5F9; Millipore) (1:100), or anti-GFAP antibody (2E1; Santa Cruz Biotechnology) (1:100) was applied to the sections at room temperature for 60 min. The secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG (H+L) antibody or Alexa Fluor 546 goat anti-mouse IgG (H+L) antibody) was then applied to the sections at room temperature for 60 min. After washing with PBS three times, all sections were counterstained with 4′,6-diamidino-2-phenylindole. Images were obtained using a BZ-9000 fluorescence microscope (KEYENCE, Tokyo, Japan)
Statistical analysis
Student’s t-test was used to analyse the significance of differences between two groups. All statistical analyses were carried out using Prism software.
Results
Purification of DG lipase
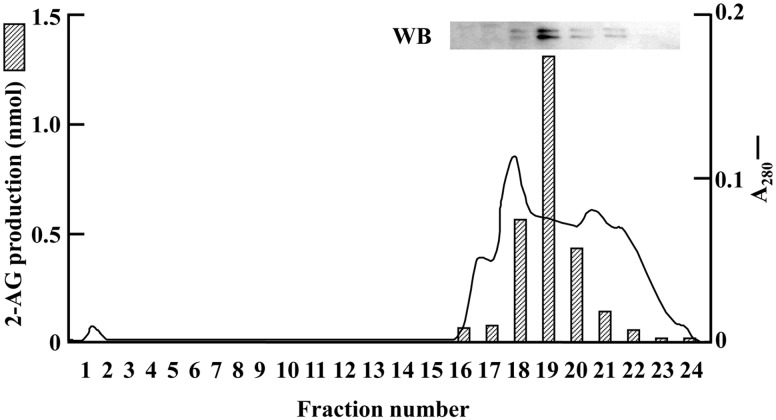
The DG lipase activity of the homogenates of whole rat brain was significantly elevated in the presence of 5 mM DOC (Supplementary Fig. S1). Thus, 5 mM DOC was added to all DG lipase assay mixtures. DG lipase activity of the subcellular fraction of the whole brain homogenates was examined. Each fraction (600 × g pellet, 10,000 × g pellet, 105,000 × g pellet and 105,000 × g supernatant) showed DG lipase activity (Table I). Among them, the specific activity was the highest in the 10,000 × g pellet fraction, whereas the greatest total activity was found in the 105,000 × g supernatant fraction. In the 10,000 × g and 105,000 × g pellet fractions, the DG lipase activities could be accounted for by DGLα and DGLβ. However, the DG lipase activity in the 105,000 × g supernatant fraction was considered to have been derived from unknown enzymes. Thus, we chose this fraction for further purification. During the column chromatography, the recovery of DG lipase activity was low at first. So, the effects of Triton X-100 on the stability of DG lipase in rat brain were examined. The soluble fraction of rat brain was incubated with 0.01–1.0% Triton X-100 for 20 min at 4°C. Then, the DG lipase activity was measured. Triton X-100 did not inhibit the DG lipase activity at concentrations of 0.01–0.05% (Supplementary Fig. S2). Therefore, 0.05% Triton X-100 was added to the buffers for column chromatography. The recovery of the DG lipase activity was significantly improved during the purification procedure. The results of protein purification are summarized in Table II. The 105,000 × g supernatant fraction was subjected to ammonium sulphate precipitation. By this procedure, the source of approximately 75% of the total activity was collected in the 20–40% fraction. The source of almost all of the DG lipase activity bound to a POROS HP2 column, and was eluted with a decreasing gradient of ammonium sulphate. The DG lipase activity from eluent from the POROS HQ column showed a single peak on the chromatogram (Fig. 1). From the elution profile of gel filtration chromatography, we estimated the molecular weight of the protein with the DG lipase activity to be around 150 kD. After the final step of purification, specific activity had increased about 1,200-fold with a yield of 6.7% compared with that for the 105,000 × g supernatant (Table II). In the early purification steps, both 2-AG and free [14C]arachidonic acid were produced from [14C]SAG (Supplementary Fig. S3), suggesting that these samples were contaminated with monoacylglycerol lipase producing [14C]arachidonic acid from [14C]2-AG, or other enzymes directly hydrolysing [14C]SAG to release [14C]arachidonic acid. Using the DG lipase obtained from CHT 5-I column chromatography, no free arachidonic acid was produced from [14C]SAG (Supplementary Fig. S3). These results revealed that the DG lipase purified during these purification procedures was specific for the sn-1 position, and did not possess 2-AG hydrolysing activity.
Table I.
Subcellular localization of DG lipase from a rat brain
| Fraction | Total protein (mg) | Specific activity (nmol/10 min/mg) | Total activity (nmol/10 min) |
|---|---|---|---|
| 600 × g ppt | 3.8 | 6.8 | 25.8 |
| 10,000 × g ppt | 2.8 | 12.6 | 35.2 |
| 105,000 × g ppt | 0.6 | 4.4 | 2.6 |
| 105,000 × g sup | 18.4 | 2.1 | 38.6 |
DG lipase activity in subcellular fractions from rat brain was measured in the presence of 5 mM DOC as described in Materials and Methods. Enzyme activity was represented as the production of 2-AG in a 10-min reaction. Values are from one typical measurement among several similar experiments.
Table II.
Partial purification of DG lipase from rat brains
| Purification steps | Total protein (mg) | Total activity (nmol/ 10 min) | Specific activity (nmol/ 10 min/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| 105,000 × g sup | 534 | 909 | 1.7 | 100 | 1 |
| (NH4)2SO4 ppt | 285 | 685 | 2.4 | 75 | 1.4 |
| POROS HP2 | 138 | 649 | 4.7 | 71 | 2.8 |
| POROS HQ | 23 | 553 | 25 | 61 | 14 |
| CHT 5-I | 0.36 | 366 | 1,013 | 40 | 596 |
| Superdex 200 | 0.03 | 61 | 2,037 | 6.7 | 1,198 |
DG lipase was purified from 24 rat brains as described in Materials and Methods. Enzyme activity was represented as the production of 2-AG in a 10-min reaction. Values are from one typical measurement among several similar experiments.
Fig. 1.
Anion exchange column chromatography for the purification of DG lipase from rat brain. A typical elution profile of POROS HQ column chromatography in the purification step of rat brain DG lipase is shown. Western blot of fractions 16–23 using anti-rat DDHD2 antibody is also shown. Absorbance at 280 nm is shown as a solid line. The enzyme activity (hatched bars) of each fraction was determined as described in Materials and Methods. Data are representative of three independent experiments.
Identification of DG lipase protein by mass spectrometry
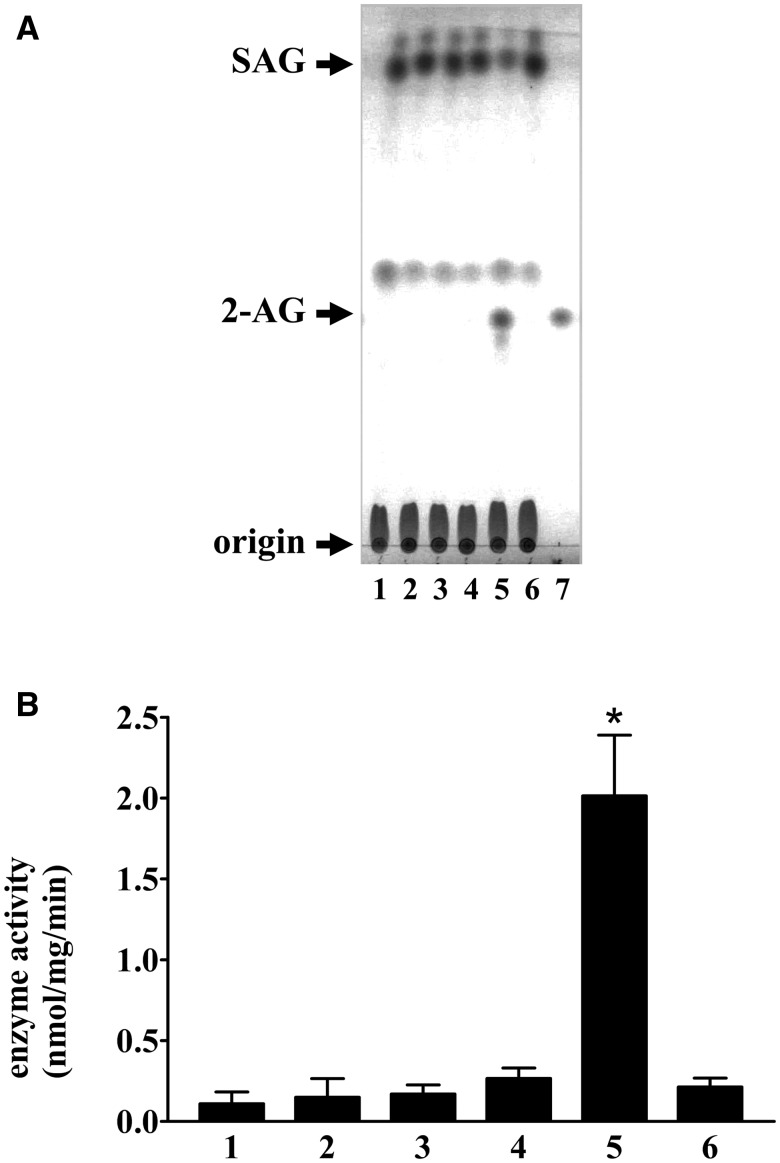
Even the DG lipase sample from the final purification step showed many protein bands detected by SDS-PAGE (Supplementary Fig. S4). We attempted to identify the protein species in the sample by mass spectrometry. More than 100 proteins were identified in the partially purified DG lipase sample as a result of a MASCOT search. We focused on the top 30 proteins in terms of the MASCOT score, as listed in Table III. Considering the annotation of these proteins, five proteins regarded as hydrolases were picked out as candidate proteins for screening DG lipase enzyme activity. These five proteins were separately expressed in CHO cells using corresponding expression vectors, and their DG lipase activities were examined (Fig. 2). The cDNA of human DDHD2 was used instead of the rat DDHD2 cDNA in this screening experiment. Among the five proteins, only the expression of human DDHD2 imparted the soluble fraction of the expressing CHO cells with elevated DG lipase activity. The amount of 2-AG increased and the amount of SAG decreased only in the enzymatic reaction using the soluble fraction of CHO cells expressing human DDHD2.
Table III.
Proteins detected in the final purification sample by mass spectrometry
| Rank in MASCOT score | Annotation of protein | Hydrolase | This study |
|---|---|---|---|
| 1 | Dihydropyrimidinase-like 2 | Yes | Yes |
| 2 | rCG35703 | ||
| 3 | Neural cell adhesion molecule 1 | ||
| 4 | Microtubule-associated protein 1A | ||
| 5 | Ncam1 protein | ||
| 6 | Tenascin R | ||
| 7 | Neural cell adhesion molecule 1 | ||
| 8 | Acyl-peptide hydrolase | Yes | Yes |
| 9 | Similar to DDHD domain containing 2 | Yes | Yes |
| 10 | Poly(A) binding protein, cytoplasmic 1 | ||
| 11 | Gephyrin | ||
| 12 | TUC-4b | Yes | Yes |
| 13 | Deaminase 3 | Yes | |
| 14 | Dynactin 2 | ||
| 15 | Microtubule associated protein 1A | ||
| 16 | Adenosine monophosphate deaminase 2 (isoform L) | Yes | |
| 17 | Ubiquilin 2 | ||
| 18 | Similar to talin 2 isoform 2 | ||
| 19 | Poly(A) binding protein, cytoplasmic 3 | ||
| 20 | Glial fibrillary acidic protein | ||
| 21 | rCG31475 | ||
| 22 | Ceruloplasmin | ||
| 23 | Poly A binding protein, cytoplasmic 2 | ||
| 24 | Amylo-1,6-glucosidase | Yes | |
| 25 | Collapsin response mediator protein | Yes | |
| 26 | Beta actin | ||
| 27 | Spectrin, beta, non-erythrocytic 2 | ||
| 28 | Beta-adducin | ||
| 29 | Antidepressant-related protein ADRG123 | Yes | Yes |
| 30 | Similar to CG11448-PA |
The top 30 proteins in terms of the MASCOT score are listed. Nine proteins were regarded as hydrolases. Among them, five proteins were selected as candidate proteins for screening of DG lipase enzyme activity.
Fig. 2.
Screening of DG lipase activity. (A) DG lipase activity in the soluble fraction of CHO cells expressing the candidate proteins (100 µg) was measured using 40 µM cold SAG as a substrate. The assay mixture was incubated at 37°C for 30 min. Lipids were separated using TLC and visualized using iodine vapour. Lane 1: acyl-peptide hydrolase, lane 2: anti-depressant-related protein ARG123, lane 3: dihydropyrimidinase-like 2, lane 4: TUC-4b, lane 5: human DDHD2, lane 6: vector control, lane 7: 5 nmol 2-AG. (B) The amount of 2-AG shown in panel (A) was quantified using ImageJ software. Data are representative of three independent experiments. Data are the means of triplicate measurements. Error bars represent standard errors of the mean. *P < 0.05 for human DDHD2 versus vector control.
cDNA cloning of rat DDHD2
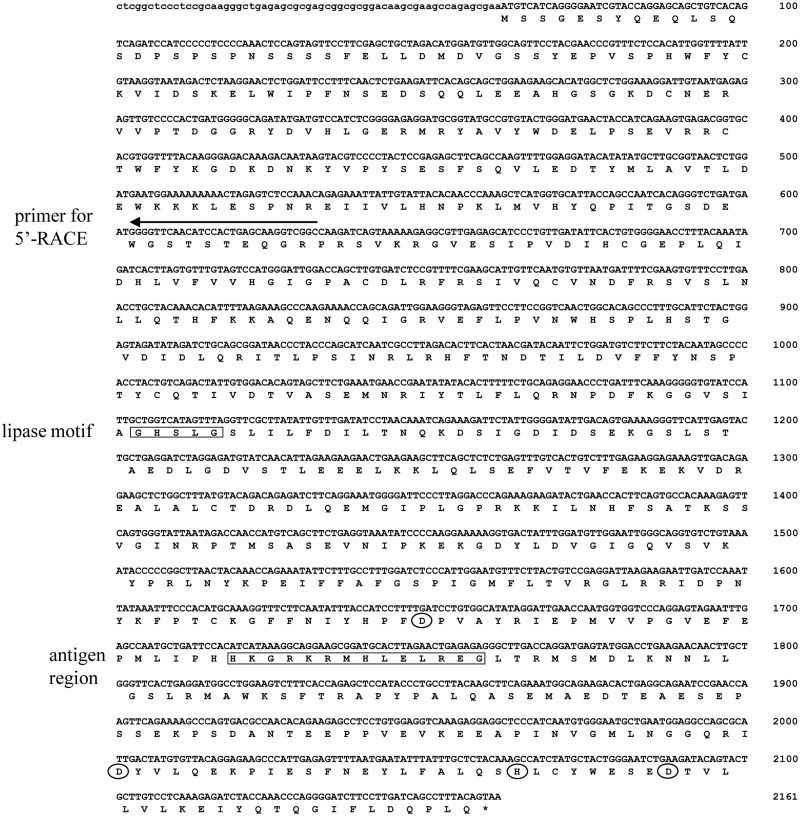
As the overexpression of human DDHD2 led to increased DG lipase activity in the soluble fraction of CHO cells, it needed to be confirmed that rat DDHD2 also exhibits DG lipase activity. The open-reading frame of rat DDHD2 was cloned by PCR using a cDNA library from rat brain (Fig. 3). Rat DDHD2 protein is composed of 699 amino acids, whereas human DDHD2 has 711. The amino acid identity between human and rat DDHD2 is 88%. The 5′ UTR of the rat DDHD2 cDNA was also cloned and sequenced (Fig. 3). The detected 5′ UTR was composed of 61 nucleotides. Although there was no in-frame stop codon in the 5′ UTR, the position of the first methionine of rat DDHD2 was assumed to be the same as that of human DDHD2.
Fig. 3.
Nucleic acid sequence of rat DDHD2 cDNA. The arrow indicates the nucleotides for 5′-RACE. The region of the antigen peptide used for the generation of the anti-DDHD2 antibody is enclosed by a square. The lipase motif is also enclosed by a square. The four amino acids contained in the DDHD domain are surrounded by circles.
The lipase motif, Gly-His-Ser-Leu-Gly, was conserved between rat and human DDHD2. The serine residue in the motif is considered to be essential for lipase activity (21). The DDHD domain in rat DDHD2 is 216 residues long (484–699). It contains four conserved residues, Asp530, Asp648, His699 and Asp677. The domain is conserved in several phosphoesterases (22).
DG lipase activity of rat DDHD2 expressed in CHO cells
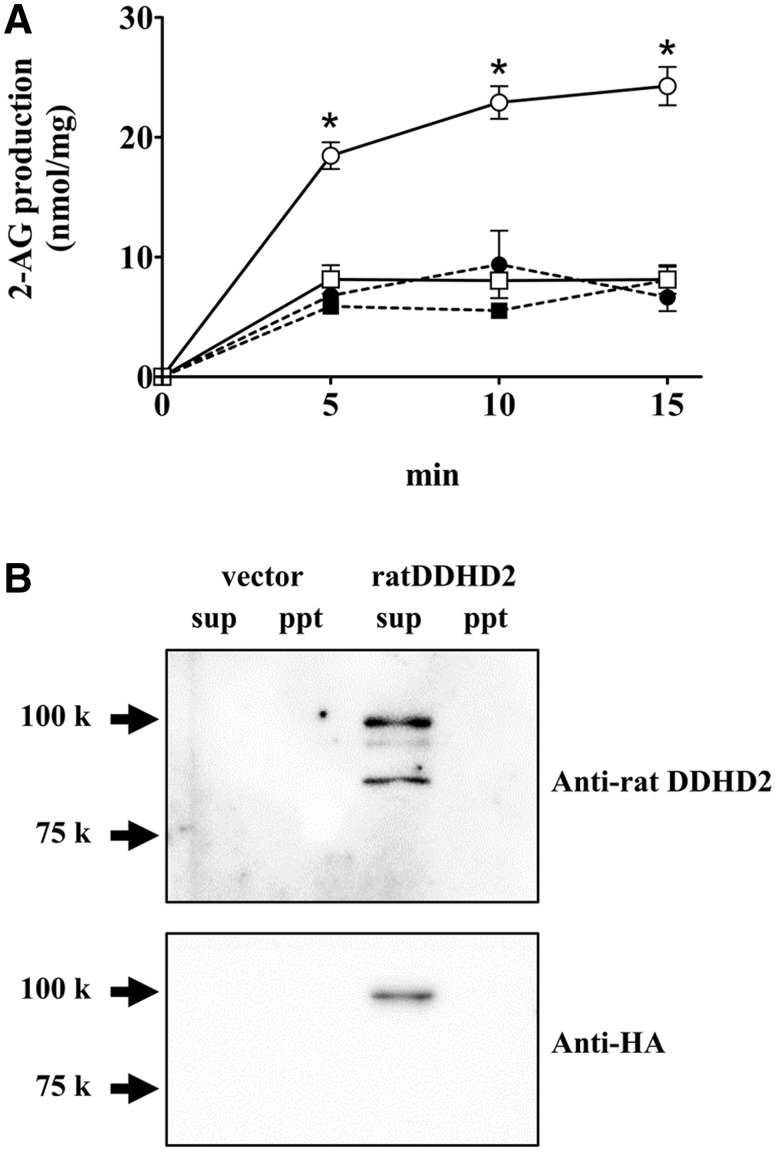
The DG lipase activity in the soluble and membrane fractions of CHO cells expressing HA-rDDHD2 was measured using [14C]SAG as a substrate (Fig. 4A). The amount of 2-AG increased in a time-dependent manner in the case of the soluble fraction of CHO cells expressing HA-rDDHD2; however, the activity was quite low in the control CHO cells transfected with empty vector. The specific activity in the soluble fraction of CHO cells expressing HA-rDDHD2 was 3.7 nmol/min/mg. No difference in enzyme activity was observed between the membrane fractions of cells transfected with HA-rDDHD2 and those with empty vector. The expression of HA-rDDHD2 was examined using anti-DDHD2 and anti-HA antibodies. DDHD2 was expressed mostly in the soluble fraction of HA-rDDHD2-transfected CHO cells (Fig. 4B). The calculated molecular weight of HA-tagged rat DDHD2 is 79 kD including the HA tag (3 kD). However, a protein band was observed at 100 kD using anti-HA antibody (Fig. 4B). On the other hand, using the anti-DDHD2 antibody, two protein bands at 85 and 95 kD were observed in addition to 100 kD band. Thus, it was suggested that these smaller bands are attributed to the N-term truncated form of DDHD2. DDHD2 in rat brain were also detected as two protein bands at 80 and 90 kD using the anti-DDHD2 antibody (Fig. 5A). However, the estimated size of HA-rDDHD2 expressed in CHO cells (100 kD) seemed bigger than the estimated size of the DDHD2 in rat brain (90 kD). This discrepancy might be due to unknown effect of the HA tag or some post-transcriptional modifications.
Fig. 4.
DG lipase activity of rat DDHD2 expressed in CHO cells. (A) The DG lipase activity was measured using 9 µM [14C]SAG as a substrate. The protein samples used for the enzyme assay were the soluble (open circles) and the membrane (closed circles) fractions of CHO cells expressing HA-rDDHD2, and were the soluble (open squares) and the membrane (closed squares) fraction of vector-transfected CHO cells. (B) The soluble fraction (25 µg) and membrane fraction (25 µg) from the CHO cells were used for western blotting. An anti-rat DDHD2 antibody (upper panel) and an anti-HA antibody (lower panel) were used. Data are the means of triplicate measurements. Error bars represent standard error of the mean. Data are representative of three independent experiments. *P < 0.01 for the soluble fraction of CHO cells expressing HA-rDDHD2 versus the soluble fraction of vector-transfected CHO cells.
Fig. 5.
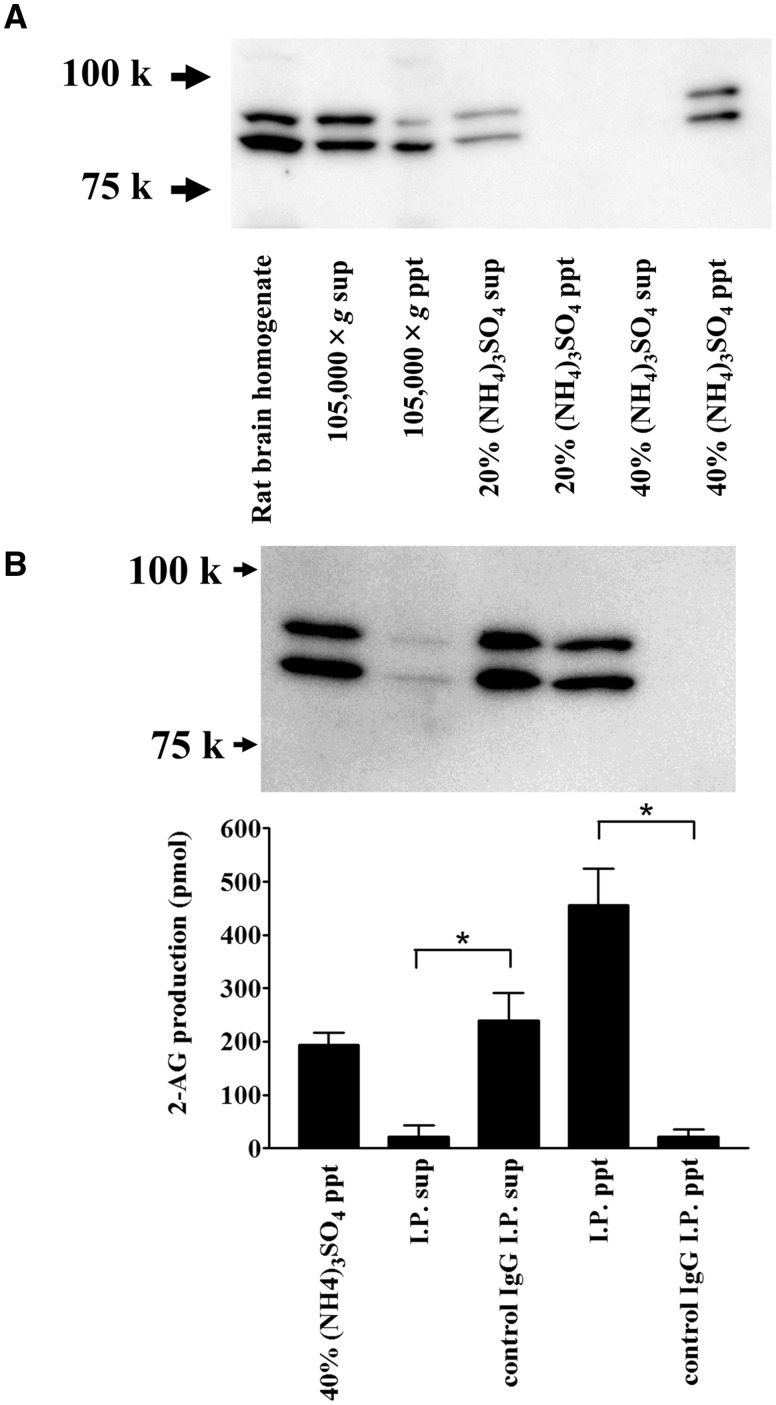
Ammonium sulphate precipitation and immunoprecipitation of rat brain DDHD2. (A) Ammonium sulphate fractionation of rat brain DDHD2. The soluble fraction of rat brain homogenates (105,000 × g supernatant) was further fractionated by ammonium sulphate precipitation. The same proportion of each sample was loaded onto an SDS-PAGE and analysed by western blotting using an anti-rat DDHD2 antibody. (B) Immunoprecipitation of DDHD2 in rat brain. Forty percent ammonium sulphate precipitate was used for the experiment. Ten micrograms of the anti-DDHD2 or control IgG was used. Aliquots of the samples were loaded onto an SDS-PAGE gel and used for the DG lipase assay. DG lipase activity in the immunoprecipitated beads or in the supernatant was measured. [14C]SAG was used as a substrate. Data are the means of triplicate measurements. Error bars represent standard error of the mean. Data are representative of three independent experiments. *P < 0.01 for the anti-DDHD2 IgG samples versus control IgG samples.
Immunochemical identification of rat brain DG lipase
To examine whether soluble DG lipase activity in rat brain is derived from DDHD2, we performed an immunoprecipitation (IP) experiment using an anti-rat DDHD2 antibody. DDHD2 was detected in rat whole-brain homogenates by western blotting (Fig. 5). Both 80 and 90 kD protein bands were observed with anti-rat DDHD2 antibodies. These protein bands appeared in the lane of 40% ammonium sulphate precipitate (Fig. 5A), which is consistent with the observation that approximately 75% of total DG lipase activity of the soluble fraction of rat brain homogenates also exhibited for the ammonium sulphate fraction. Rat DDHD2 protein in the 20–40% ammonium sulphate fraction was almost completely precipitated using an anti-rat DDHD2 antibody, whereas it was not precipitated at all using preimmune IgG prepared from preimmune rabbit serum (Fig. 5B).Additionally, the source of almost all DG lipase activity was precipitated using the anti-DDHD2 antibody, whereas very little of it was precipitated using control IgG. The DG lipase activity in IP precipitate (ppt) sample was double that in the IP control supernatant (sup) or 40% (NH4)3SO4 ppt samples. This suggests that 40% (NH4)3SO4 ppt sample contained some inhibitory factors or non-radioisotope substrates for DDHD2. In the purification of DG lipase from rat brains by POROS HQ column chromatography, the eluted source of the DG lipase activity corresponded to the DDHD2 protein detected by western blotting (Fig. 1). Therefore, the high level of DG lipase activity observed in the soluble fraction in rat brain was likely derived from DDHD2.
Enzymatic properties of the partially purified DG lipase from rat brain and rat DDHD2
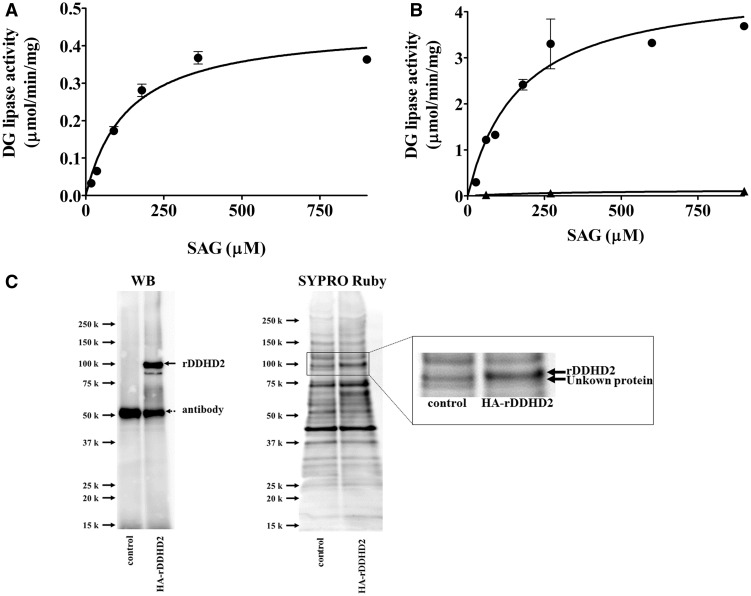
The DG lipase activity of partial purified DG lipase from rat brain was measured using [14C]SAG as a substrate (Fig. 6). The effect of substrate concentration on the enzyme activity was examined (Fig. 6A). The profile of DG lipase activity versus substrate concentration fit the Michaelis–Menten equation. Vmax and Km values were 0.45 ± 0.027 µmol/min/mg and 135 ± 24 µM, respectively. The DG lipase activity of immunopurified HA-rDDHD2 expressed in CHO cells was measured using [14C]SAG as a substrate (Fig. 6B). The profile of DG lipase activity versus substrate concentration also fitted the Michaelis–Menten equation. The Km value was 169 ± 47 µM. The value for HA-rDDHD2 was comparable to that of the partially purified DG lipase from rat brain (135 µM). Control measurements showed a very low level of DG lipase activity using the same aliquot of protein sample obtained from vector transfected cells (Fig. 6B). The whole protein contained in the immunopurified HA-rDDHD2 sample and the control sample was analysed by western blotting and SYPRO Ruby staining following SDS-PAGE (Fig. 6C). Purification of HA-rDDHD2 (arrow with a solid line) was successfully performed despite the contamination of the anti-DDHD2 antibody (arrow with dotted line) that was leakily eluted and detected by western blotting at 50 kD. Many protein bands were seen in the SYPRO Ruby-stained gel (Fig. 6C). In contrast to the control sample, the HA-rDDHD2 was detected at 100 kD in the SYPRO Ruby-stained gel (Fig. 6C). These results show that the DG lipase activity in the immunopurified HA-rDDHD2 sample originated from DDHD2, not from other proteins that might have been purified by associating with DDHD2.
Fig. 6.
Substrate concentration-dependence. (A) Various concentrations of [14C]SAG were incubated with the partially purified rat brain DG lipase from the CHT 5-I column (15 µg per assay). Except for the substrate concentrations, the assay conditions were the same as those for the standard assay. (B) Various concentrations of [14C]SAG were incubated with HA-rDDHD2 (closed circles) prepared using anti-rat DDHD2 antibody. Control (closed triangles) means the DG lipase activity using the same aliquot of protein sample obtained from vector transfected cells. (C) Control sample and HA-rDDHD2 sample were analysed by western blotting using anti-rat DDHD2 antibody and SYPRO Ruby staining following SDS-PAGE. The rectangular area in SYPRO Ruby staining is magnified in the right panel. Data are the means of triplicate measurements. Error bars represent standard errors of the mean. Data are representative of three independent experiments.
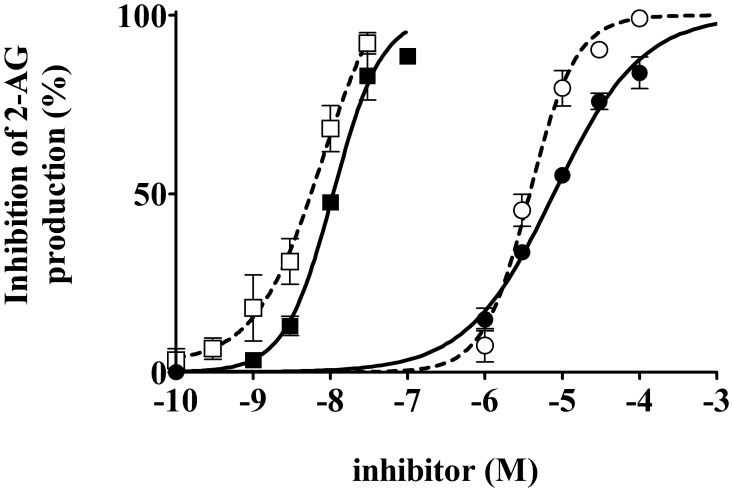
DG lipase activity was also measured in the presence of DG lipase inhibitors, such as THL, which is an irreversible inhibitor of various lipases. THL potently inhibited DG lipase activity in the partially purified enzyme with an IC50 of 10 ± 1.1 nM (Fig. 7). RHC80267, a potent inhibitor of canine platelet DG lipase, DGLα, and DGLβ (14, 23), was also applied. Partially purified rat DG lipase was also inhibited by RHC80267 in a dose-dependent manner with an IC50 value of 8 ± 1.1 µM (Fig. 7). The DG lipase activity of HA-rDDHD2 expressed in CHO cells was also measured. In the presence of the above-mentioned lipase inhibitors, THL and RHC20267, HA-rDDHD2 was also inhibited with IC50 value of 7.8 ± 1.4 nM and 3.8 ± 1.0 µM, respectively (Fig. 7). The values were comparable to those for the partially purified DG lipase from rat brain. These data are also consistent with the hypothesis that the soluble DG lipase activity observed in rat brain is due to DDHD2.
Fig. 7.
Effects of inhibitors on the partially purified rat brain DG lipase and HA-rDDHD2 expressed in CHO cells. DG lipase activity was measured in the presence of various concentrations of RHC80267 (closed circles for rat brain DG lipase, open circles for CHO-rDDHD2) and THL (closed squares for rat brain DG lipase, open squares for CHO-rDDHD2). The lipase inhibitors were added to the mixture before preincubation. Data are the means of triplicate measurements. Error bars represent standard error of the mean. Data are representative of three independent experiments.
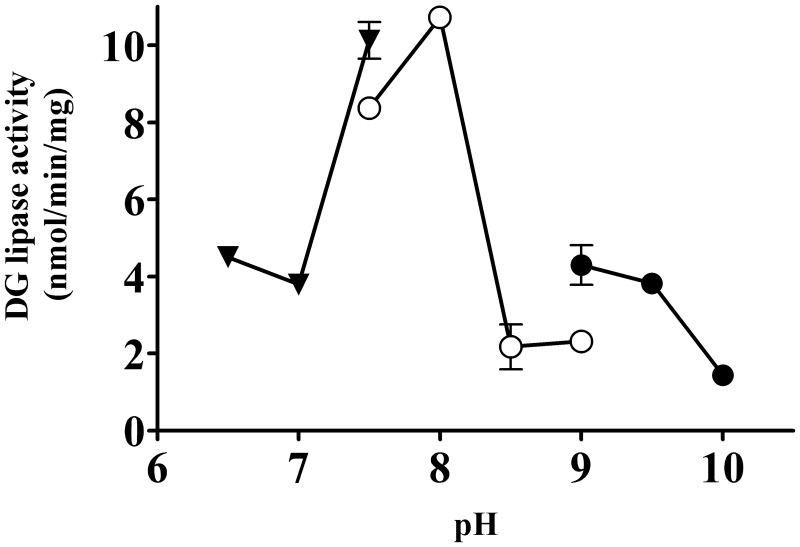
Next, the effect of pH on the enzyme activity was examined. The optimal pH for the partially purified DG lipase from rat brain was 8.0 (Fig. 8).
Fig. 8.
pH dependency of the partially purified DG lipase. DG lipase activity in the active fractions (20 µg per assay) from the CHT 5-I column was measured at various pH levels. Buffers used for the experiments were MOPS-NaOH (closed triangles), Tris-HCl (open circles) and glycine-NaOH (closed circles). Except for the buffer used, assay conditions were the same as those for the standard assay. Data are the means of triplicate measurements. Error bars represent standard errors of the mean. Data are representative of three independent experiments.
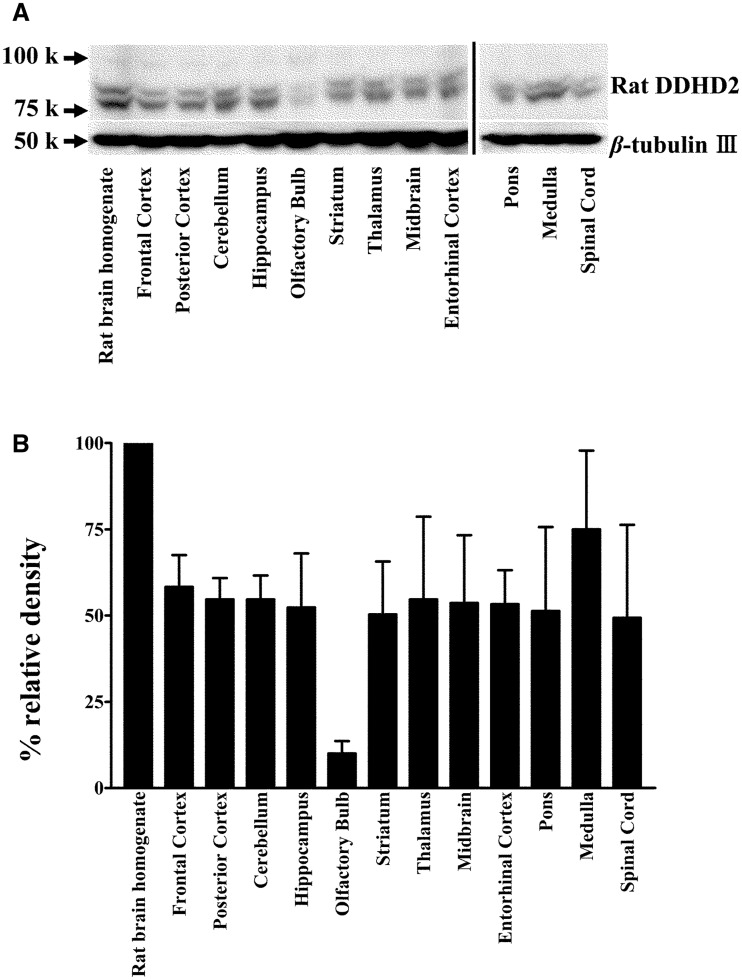
Distribution of DDHD2 in rat brain
To examine the brain regions where DDHD2 is expressed, rat brain tissue was dissected and analysed by western blotting (Fig. 9). Two major protein bands observed at 80 and 90 kD were regarded as DDHD2 considering the calculated mass and the results of column chromatography (Figs 1B and 9). Rat DDHD2 was detected widely in the brain regions tested.
Fig. 9.
Distribution of DDHD2 in rat brain. (A) Fifty micrograms of protein of homogenates from normal rat brain region tissues was loaded onto each lane. The expression of DDHD2 was analysed by western blotting using anti-rat DDHD2 antibody. The same membrane was re-probed with an anti-β-tubulin III antibody. (B) Histograms showing relative DDHD2 ratios against β-tubulin. The DDHD2 expression was quantified using ImageJ software. Data are the means of triplicate measurements. Error bars represent standard errors of the mean. Data are representative of three independent experiments.
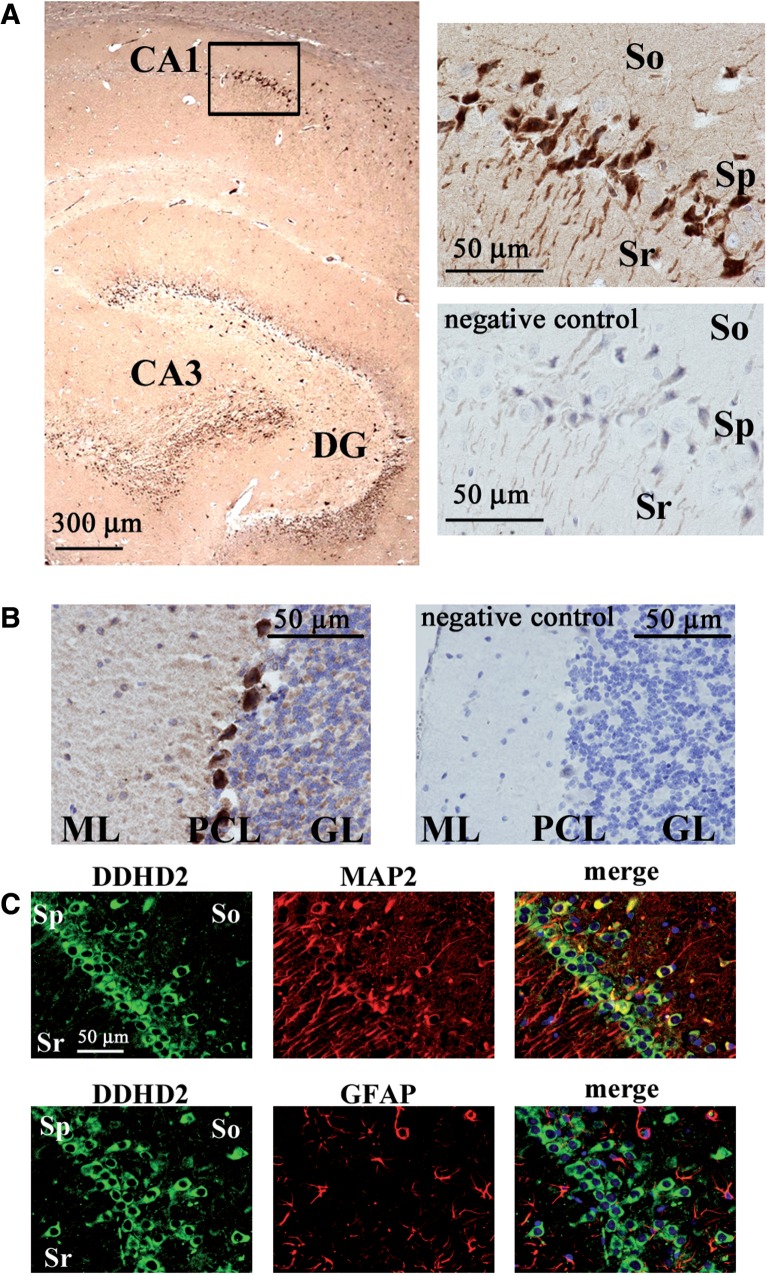
The expression of DDHD2 was further analysed by immunostaining (Fig. 10). In a sagittal section of the rat hippocampus, neuronal cells at the CA1 and CA3 regions and at the dentate gyrus were remarkably stained by the anti-DDHD2 antibody (Fig. 10A). The magnified image (upper right) indicates that the CA1 neuronal cell bodies and axons at the pyramidal cell layer expressed DDHD2. The lower right panel shows that DDHD2 staining was abolished using absorbed antibody. Thus, the regions stained by the anti-DDHD2 antibody denote the expression of DDHD2 protein. It was also shown that the expression of DDHD2 was dominant in MAP2 positive neuronal cells, but not GFAP positive glial cells at the CA1 region (Fig. 10C). Purkinje cell bodies in the rat cerebellum were remarkably stained by the anti-DDHD2 antibody (Fig. 10B). The molecular layer region where dendrites from Purkinje cells are distributed was also stained by specific antibody.
Fig. 10.
Immunostaining of DDHD2 in rat brain. (A) The distribution of DDHD2 is shown using a sagittal section of the hippocampus in a rat brain. DDHD2 was stained by the anti-DDHD2 antibody with DAB (brown). Nuclei were stained by with hematoxylin (blue). The rectangular area is magnified in the upper right panel. The lower right panel is from a negative control experiment using the excessive peptides used for the immunization (16 µg/ml) to absorb primary antibody. (B) The distribution of DDHD2 is shown using a sagittal section of the cerebellum in a rat brain. The right panel is from a negative control experiment. ML, molecular layer; PCL, Purkinje cell layer; GL, granule cell layer. (C) The distribution of DDHD2 is shown using a coronal section of a rat brain. Double staining of DDHD2 and MAP2 (upper panels) or DDHD2 and GFAP (lower panels) at the CA1 region in the hippocampus shows that DDHD2 is localized in the cell body of hippocampal neurons. Sr, stratum radiatum; So, stratum oriens; Sp, stratum pyramidale. Data are representative of three independent experiments.
Discussion
We have determined several enzymatic properties of partially purified DG lipase from rat brain. The Km value towards SAG (135 µM) was assumed to be within the range of possible physiological concentration of SAG, because the Km values of human DGLα, human DGLβ and two rat DG kinases towards SAG are 154, 74, 70 and 90 µM, respectively (14, 24). Thus, we believe that the soluble DG lipase in rat brain can hydrolyse DG under physiological conditions.
Our partially purified DG lipase from rat brain is thought to be identical to DDHD2. Although, we could not completely purify the DG lipase protein, all of the subsequent experimental data such as the Km value (Fig. 6), IC50 value for RHC 80267 (Fig. 7), ammonium sulphate fractionation (Fig. 5A), IP (Fig. 5B) and column chromatography (Fig. 1) indicated that our partially purified DG lipase was DDHD2.
Several attempts have been made to purify soluble DG lipase from bovine aorta (17, 18); however, it has not yet cloned. The enzymatic properties of DG lipases from rat brain (this study) and bovine aorta are quite similar in terms of the dependence of their enzyme activity on pH, their Km values and the effects of an inhibitor (THL) on them. Thus, it is possible that the soluble DG lipase in bovine aorta is also DDHD2. The DG lipase activity was optimal in the presence of 5 mM DOC (Supplementary Fig. S2). The critical micelle concentration of DOC is around 5 mM. It is supposed that the structure of micelles composed of SAG and DOC is critical for the attack by DDHD2 in vitro. DDHD2 is reported to be localized at the Golgi apparatus and contribute to membrane trafficking (25, 26). Further investigation is needed to determine how DDHD2 interacts with membrane lipids and acts as DG lipase. Human DDHD2 is known as PLA1, which hydrolyses phosphatidic acid and other phospholipids (21). The physiological substrates and the roles of DDHD2 have to be elucidated.
We purified DG lipase from the soluble fraction of rat brain. However, DDHD2 in rat brain was localized in both the membrane and the soluble fraction (Fig. 5A). Because DG can potentially be produced in the membrane by PLC β, membrane-bound DDHD2 might be a functional form in vivo. The binding of DDHD2 to membrane can be explained by its affinity to phosphoinositides (22). Because HA-rDDHD2 expressed in CHO cells exhibited unusual localization mainly in the cytosol (Fig. 4B), it is also possible that the processing or modification of DDHD2 is needed for the localization of membranes.
We consider it interesting to estimate how large a fraction of DG lipase activity in rat brain is derived from DDHD2. A sample of 20–40% (NH4)3SO4 exhibited activity that was 75% of soluble DG lipase (Table II), so most of the activity appeared to have been derived from DDHD2 (Fig. 5). By assuming that all of the soluble DG lipase activity is derived from DDHD2, the DG lipase activity of DDHD2 can account for at least 38% of the total DG lipase activity in rat brain. It is unclear whether DDHD2 plays a role in producing 2-AG as a retrograde messenger in vivo. CB1 is expressed in the neurons of hippocampal CA1 and the molecular layer in the cerebellum (27). In addition, DGLα is distributed in hippocampal neurons and dendrites of Purkinje cells in the molecular layer of the cerebellum (14, 28). In the hippocampal neurons and the Purkinje cells of cerebellum, endocannabinoids have been shown to participate in DSI (8, 29). Here, we showed that DDHD2 was also localized to hippocampal neurons, Purkinje cell bodies and the molecular layer of the cerebellum (Fig. 10). Thus, we suggest that DDHD2 might be involved in 2-AG production in the hippocampus and cerebellum. It should also be elucidated whether DDHD2 is involved in DSI in vivo. Human DDHD2 is the causative gene for a complex hereditary spastic paraplegia called SPG54 (30, 31). Very recently, Inloes et al. (32) reported that DDHD2−/− mice presented a dysfunction of locomotion, and the accumulation of triacylglycerol (TG) in the brain, which resembled SPG54 caused by DDHD2 mutation in humans. They documented that lysates of HEK293T cells expressing mouse DDHD2 exhibited TG lipase activity. A lack of the TG lipase activity of DDHD2 led to the dysfunction of locomotion following to the TG accumulations (32). In addition, they reported that mouse DDHD2 had DG lipase activity. Independent of their studies, we showed DG lipase activity of rat DDHD2 (Fig. 6B). Thus, we consider that our data are convincing. However, the relationship between 2-AG produced by DG lipase activity of mouse DDHD2 and the disorders observed in DDHD2−/− mice is unknown.
Here, we reported the purification and cloning of a soluble DG lipase from rat brain. Studies on the enzymatic character of the partially purified DG lipase revealed that this enzyme is identical to DDHD2. Our studies also showed that DDHD2 was expressed in hippocampal neuron (Fig. 10). Further studies might elucidate whether 2-AG produced by DDHD2 is related to endocannabinoid signalling in the brain.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgement
The authors thank the Laboratory for Analytical Instruments, Education and Research Support Center, Gunma University Graduate School of Medicine.
Funding
This work was supported in part by JSPS KAKENHI Grant Numbers 23592275, 24390069 and 23659106.
Conflict of Interest
None declared.
Glossary
Abbreviations
- 2-AG
2-arachidonoylglycerol
- CHO
Chinese hamster ovary
- CNS
central nervous system
- DAB
3,3′-diaminobenzidine
- DG
diacylglycerol
- DMSO
dimethylsulfoxide
- DOC
sodium deoxycholate
- DSI
depolarization-induced suppression of inhibition
- HA-rDDHD2
HA-tagged rat DDHD2
- HRP
horse radish peroxidase
- IP
immunoprecipitation
- MS/MS
tandem mass spectrometry
- PBS
phosphate-buffered saline
- PBS-T
PBS containing 0.1% Tween 20
- PLA1
phospholipase A1
- PLC
phospholipase C
- ppt
precipitate
- RACE
rapid amplification of cDNA ends
- RHC80267
1,6-bis(cyclohexyloximinocarbonylamino)hexane
- SAG
1-stearoyl-2-arachidonoyl-sn-glycerol
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- sup
supernatant
- TG
triacylglycerol
- THL
tetrahydrolipstatin
- TLC
thin-layer chromatography
References
- 1.Basavarajappa B.S. (2007) Critical enzymes involved in endocannabinoid metabolism. Protein Pept. Lett. 14, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J., Ueda N. (2008) Role of the endocannabinoid system in metabolic control. Curr. Opin. Nephrol. Hypertens. 17, 1–10 [DOI] [PubMed] [Google Scholar]
- 3.Sugiura T., Kishimoto S., Oka S., Gokoh M. (2006) Biochemistry, pharmacology and physiology of 2-arachidonoylglycerol, an endogenous cannabinoid receptor ligand. Prog. Lipid Res. 45, 405–446 [DOI] [PubMed] [Google Scholar]
- 4.Matsuda L.A., Bonner T.I., Lolait S.J. (1993) Localization of cannabinoid receptor mRNA in rat brain. J. Comp. Neurol. 327, 535–550 [DOI] [PubMed] [Google Scholar]
- 5.Tsou K., Brown S., Sanudo-Pena M.C., Mackie K., Walker J.M. (1998) Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience 83, 393–411 [DOI] [PubMed] [Google Scholar]
- 6.Stella N., Schweitzer P., Piomelli D. (1997) A second endogenous cannabinoid that modulates long-term potentiation. Nature 388, 773–778 [DOI] [PubMed] [Google Scholar]
- 7.Makara J.K., Mor M., Fegley D., Szabo S.I., Kathuria S., Astarita G., Duranti A., Tontini A., Tarzia G., Rivara S., Freund T.F., Piomelli D. (2005) Selective inhibition of 2-AG hydrolysis enhances endocannabinoid signaling in hippocampus. Nat. Neurosci. 8, 1139–1141 [DOI] [PubMed] [Google Scholar]
- 8.Hashimotodani Y., Ohno-Shosaku T., Kano M. (2007) Ca(2+)-assisted receptor-driven endocannabinoid release: mechanisms that associate presynaptic and postsynaptic activities. Curr. Opin. Neurobiol. 17, 360–365 [DOI] [PubMed] [Google Scholar]
- 9.Keimpema E., Barabas K., Morozov Y.M., Tortoriello G., Torii M., Cameron G., Yanagawa Y., Watanabe M., Mackie K., Harkany T. (2010) Differential subcellular recruitment of monoacylglycerol lipase generates spatial specificity of 2-arachidonoyl glycerol signaling during axonal pathfinding. J. Neurosci. 30, 13992–14007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argaw A., Duff G., Zabouri N., Cecyre B., Chaine N., Cherif H., Tea N., Lutz B., Ptito M., Bouchard J.F. (2011) Concerted action of CB1 cannabinoid receptor and deleted in colorectal cancer in axon guidance. J. Neurosci. 31, 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oudin M.J., Hobbs C., Doherty P. (2011) DAGL-dependent endocannabinoid signalling: roles in axonal pathfinding, synaptic plasticity and adult neurogenesis. Eur. J. Neurosci. 34, 1634–1646 [DOI] [PubMed] [Google Scholar]
- 12.Jung K.M., Mangieri R., Stapleton C., Kim J., Fegley D., Wallace M., Mackie K., Piomelli D. (2005) Stimulation of endocannabinoid formation in brain slice cultures through activation of group I metabotropic glutamate receptors. Mol. Pharmacol. 68, 1196–1202 [DOI] [PubMed] [Google Scholar]
- 13.Oka S., Arai S., Waku K., Tokumura A., Sugiura T. (2007) Depolarization-induced rapid generation of 2-arachidonoylglycerol, an endogenous cannabinoid receptor ligand, in rat brain synaptosomes. J. Biochem. 141, 687–697 [DOI] [PubMed] [Google Scholar]
- 14.Bisogno T., Howell F., Williams G., Minassi A., Cascio M.G., Ligresti A., Matias I., Schiano-Moriello A., Paul P., Williams E.J., Gangadharan U., Hobbs C., Di Marzo V., Doherty P. (2003) Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 163, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanimura A., Yamazaki M., Hashimotodani Y., Uchigashima M., Kawata S., Abe M., Kita Y., Hashimoto K., Shimizu T., Watanabe M., Sakimura K., Kano M. (2010) The endocannabinoid 2-arachidonoylglycerol produced by diacylglycerol lipase alpha mediates retrograde suppression of synaptic transmission. Neuron 65, 320–327 [DOI] [PubMed] [Google Scholar]
- 16.Gao Y., Vasilyev D.V., Goncalves M.B., Howell F.V., Hobbs C., Reisenberg M., Shen R., Zhang M.Y., Strassle B.W., Lu P., Mark L., Piesla M.J., Deng K., Kouranova E.V., Ring R.H., Whiteside G.T., Bates B., Walsh F.S., Williams G., Pangalos M.N., Samad T.A., Doherty P. (2010) Loss of retrograde endocannabinoid signaling and reduced adult neurogenesis in diacylglycerol lipase knock-out mice. J. Neurosci. 30, 2017–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M.W., Kraemer F.B., Severson D.L. (1995) Characterization of a partially purified diacylglycerol lipase from bovine aorta. Biochim. Biophys. Acta 1254, 311–318 [DOI] [PubMed] [Google Scholar]
- 18.Lee M.W., Severson D.L. (1994) Partial purification of a diacylglycerol lipase from bovine aorta. Biochem. J. 298, (Pt 1) 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bligh E.G., Dyer W.J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 20.www.matrixscience.com/search_form_select.html.
- 21.Nakajima K., Sonoda H., Mizoguchi T., Aoki J., Arai H., Nagahama M., Tagaya M., Tani K. (2002) A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J. Biol. Chem. 277, 11329–11335 [DOI] [PubMed] [Google Scholar]
- 22.Inoue H., Baba T., Sato S., Ohtsuki R., Takemori A., Watanabe T., Tagaya M., Tani K. (2012) Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p. Biochim. Biophys. Acta 1823, 930–939 [DOI] [PubMed] [Google Scholar]
- 23.Sutherland C.A., Amin D. (1982) Relative activities of rat and dog platelet phospholipase A2 and diglyceride lipase. Selective inhibition of diglyceride lipase by RHC 80267. J. Biol. Chem. 257, 14006–14010 [PubMed] [Google Scholar]
- 24.Kato M., Takenawa T. (1990) Purification and characterization of membrane-bound and cytosolic forms of diacylglycerol kinase from rat brain. J. Biol. Chem. 265, 794–800 [PubMed] [Google Scholar]
- 25.Morikawa R.K., Aoki J., Kano F., Murata M., Yamamoto A., Tsujimoto M., Arai H. (2009) Intracellular phospholipase A1gamma (iPLA1gamma) is a novel factor involved in coat protein complex I- and Rab6-independent retrograde transport between the endoplasmic reticulum and the Golgi complex. J. Biol. Chem. 284, 26620–26630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato S., Inoue H., Kogure T., Tagaya M., Tani K. (2010) Golgi-localized KIAA0725p regulates membrane trafficking from the Golgi apparatus to the plasma membrane in mammalian cells. FEBS Lett. 584, 4389–4395 [DOI] [PubMed] [Google Scholar]
- 27.Pettit D.A., Harrison M.P., Olson J.M., Spencer R.F., Cabral G.A. (1998) Immunohistochemical localization of the neural cannabinoid receptor in rat brain. J. Neurosci. Res. 51, 391–402 [DOI] [PubMed] [Google Scholar]
- 28.Yoshida T., Fukaya M., Uchigashima M., Miura E., Kamiya H., Kano M., Watanabe M. (2006) Localization of diacylglycerol lipase-alpha around postsynaptic spine suggests close proximity between production site of an endocannabinoid, 2-arachidonoyl-glycerol, and presynaptic cannabinoid CB1 receptor. J. Neurosci. 26, 4740–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenowitz S.D., Best A.R., Regehr W.G. (2006) Sustained elevation of dendritic calcium evokes widespread endocannabinoid release and suppression of synapses onto cerebellar Purkinje cells. J. Neurosci. 26, 6841–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuurs-Hoeijmakers J.H., Geraghty M.T., Kamsteeg E.J., Ben-Salem S., de Bot S.T., Nijhof B., van de V., II, van der Graaf M., Nobau A.C., Otte-Holler I., Vermeer S., Smith A.C., Humphreys P., Schwartzentruber J., Consortium F.C., Ali B.R., Al-Yahyaee S.A., Tariq S., Pramathan T., Bayoumi R., Kremer H.P., van de Warrenburg B.P., van den Akker W.M., Gilissen C., Veltman J.A., Janssen I.M., Vulto-van Silfhout A.T., van der Velde-Visser S., Lefeber D.J., Diekstra A., Erasmus C.E., Willemsen M.A., Vissers L.E., Lammens M., van Bokhoven H., Brunner H.G., Wevers R.A., Schenck A., Al-Gazali L., de Vries B.B., de Brouwer A.P. (2012) Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am. J. Hum. Genet. 91, 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez M., Nampoothiri S., Kornblum C., Oteyza A.C., Walter J., Konidari I., Hulme W., Speziani F., Schols L., Zuchner S., Schule R. (2013) Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur. J. Hum. Genet 21, 1214–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inloes J.M., Hsu K.L., Dix M.M., Viader A., Masuda K., Takei T., Wood M.R., Cravatt B.F. (2014) The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl Acad. Sci. U.S.A. 111, 14924–14929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.