Abstract
Appropriate leaf rolling enhances erect-leaf habits and photosynthetic efficiency, which consequently improves grain yield. Here, we reported the novel lateral organ boundaries domain (LBD) gene OsLBD3-7, which is involved in the regulation of leaf rolling. OsLBD3-7 works as a transcription activator and its protein is located on the plasma membrane and in the nucleus. Overexpression of OsLBD3-7 leads to narrow and adaxially rolled leaves. Microscopy of flag leaf cross-sections indicated that overexpression of OsLBD3-7 led to a decrease in both bulliform cell size and number. Transcriptional analysis showed that key genes that had been reported to be negative regulators of bulliform cell development were up-regulated in transgenic plants. These results indicated that OsLBD3-7 might acts as an upstream regulatory gene of bulliform cell development to regulate leaf rolling, which will give more insights on the leaf rolling regulation mechanism.
Introduction
Rice leaf shape or three-dimensional architecture plays important roles in the leaf’s function, including photosynthesis, respiration and transpiration [1, 2]. Moderate leaf rolling can improve photosynthetic efficiency, reduce sunlight damage to leaves and enhance leaf drought stress resistance, therefore strongly increasing yield performance. In contrast, serious leaf rolling could hinder plant development and severely reduce crop yield.
Most rolled leaves are related to leaf adaxial-abaxial polarity establishment and bulliform cell development [2–10]. Bulliform cells are groups of large and highly vacuolated cells that mainly exist in monocots and play a crucial role in the modulation of leaf rolling. It has been shown that the shrinking or expanding of bulliform cells contribute to the leaf rolling up or opening under different water stress conditions [11]. To date, several genes have been identified as leaf rolling genes in rice, which are associated with bulliform cell development. Rice outermost cell-specific gene 5 (OsRoc5) is homologous to the Arabidopsis homeodomain leucine zipper class IV gene GLABRA2 [3]. Suppression of OsRoc5 causes abaxially rolled leaves and a reduced number and size of bulliform cells, whereas overexpression of Roc5 results in adaxially rolled leaves and an increased number and volume of bulliform cells [3]. SEMI-ROLLED LEAF1 (OsSRL1) encodes vacuolar H+-ATPase subunits and a H+-pyrophosphatase that regulates leaf rolling by repressing the development of bulliform cells [9]. Mutant srl1 shows adaxially rolled leaves and an increased number of bulliform cells [9]. Rice NARROW AND ROLLED LEAF1 (OsNRL1) encodes the cellulose synthase-like protein D4 (OsCslD4) [8]. nrl1 mutant in rice shows semi-rolled leaves by reducing the size of bulliform cells [8]. ADAXIALIZED LEAF1 (OsADL1) encodes a plant-specific calpain-like cysteine proteinase. adl1 mutant plant results in abaxially rolled leaves due to the ectopic growth of bulliform cells [12, 13]. ABAXIALLY CURLED LEAF1 (OsACL1) encodes an unknown protein of 116 amino acids [5]. Overexpression of OsACL1 produces abaxial leaf curling and increases the number and size of bulliform cells [5]. ROLLING-LEAF 14 (OsRL14) encodes a 2OG-Fe (II) oxygenase [7]. Mutant rl14 exhibits incurved leaves and the shrinkage of bulliform cells on the adaxial side [7]. SHALLOT-LIKE1 (OsSLL1) / ROLLED LEAF 9 (OsRL9) encodes a SHAQKYF class MYB family transcription factor [2, 6]. sll1 mutant displays an adaxialized leaves phenotype, while OsSLL1 overexpression shows abaxial leaf features and suppresses bulliform cell development.
LBD gene family is a plant-specific transcription factor family, with 43 and 35 members in the Arabidopsis and rice genomes, respectively [14, 15]. LBD genes are involved in embryonic shoot apical meristem (SAM) formation and have an important influence on the formation and development of plant lateral organs, such as leaves, flowers and roots [14, 16, 17]. LBD proteins contain a characteristic LOB domain composed of a C-motif that is required for DNA-binding, a conserved glycine residue, and a leucine-zipper-like sequence that is required for protein-protein interactions [16]. The first identified LBD gene, AtLOB, was isolated from Arabidopsis in 2002 [14]. Ectopic expression of AtLOB exhibits smaller plant types and sterility. AtAS2 (AtLBD6) is the second identified LBD gene in Arabidopsis. AtAS2 is involved in symmetric flat leaf formation by inhibiting cell proliferation in the adaxial layer [18–20]. AtAS2 is homologous to OsAS2 (OsLBD1-9) in rice; OsAS2 overexpression could inhibit shoot differentiation, promote cell division, delay cell differentiation and lead to aberrant twisted leaves [21].
In this study, we identified a novel rice rolled leaf gene that encodes a LBD family transcription factor. OsLBD3-7 overexpression promoted narrow and adaxially rolled leaves by decreasing the size and number of bulliform cells. OsLBD3-7 could also up-regulate bulliform cell negative regulators, which implies that OsLBD3-7 acts as a suppressor of bulliform cell development.
Methods
Plant Materials and Growth Conditions
The rice cultivar Kita-ake (Oryza sativa japonica cv. Kita-ake) was used as the wild type line for phenotypic observation, histological analysis and gene expression analysis. In 2014 and 2015, all of the materials were grown in a field of the Experiment Station of Chinese Academy of Agricultural Sciences in Beijing (39°54′N, 116°23′E) under natural conditions from May to October of each year.
Generation of Transgenic Rice Plants
To generate the OsLBD3-7V and OsLBD3-7 constructs, full length OsLBD3-7 was amplified from Nipponbare cDNA libraries and inserted into the gateway entry vector pDONR-201 (Invitrogen). Then, OsLBD3-7 was recombined into destination vector pBCV [22] and pCAMBIA1301-Bar-FLAG using the Gateway cloning system (Invitrogen). The primers are listed in Table A in S1 File. The constructs were introduced into Agrobacterium tumefaciens strain EHA105 and then transformed into Kita-ake wild type plants[23].
OsLBD3-7 Bioinformatics Analysis
The OsLBD3-7 gene locus identifier is LOC_Os03g57670.1. Protein sequences were taken from the Arabidopsis (https://www.arabidopsis.org/) and rice (http://rice.plantbiology.msu.edu/) databases. Protein alignment was completed with ClustalX 2.0. A phylogenetic tree was constructed with MEGA5.1 using the Neighbor-Joining (NJ) method. The bootstrap values for nodes in the phylogenetic tree were from 1000 replications. The handling gap option was pairwise deletion, and the numbers at the branching points indicated the bootstrap values. The accession numbers and protein sequences were shown in Table B and C in S1 File.
Leaf Rolling Index (LRI) Measurement
The mature flag leaves were measured in three replicates and each replicate included 10 individuals for each material. Lw was the greatest flag leaf width. Ln was the natural distance of the flag leaf margins at the same site. Leaf rolling index (LRI) was calculated with the following formula: LRI = (Lw-Ln)/Lw [10].
Histological Analysis
The first and the second fully expanded leaves of 21 day seedling were collected, fixed in a FAA solution (60% (v/v) ethanol, 5% (v/v) glacial acetic acid and 5% (v/v) formaldehyde) and vacuum pumped for 40 minutes. Then, the leaves were dehydrated with a series of ethanol solutions (70% (v/v) ethanol, 80% (v/v) ethanol, 85% (v/v) ethanol, 90% (v/v) ethanol, 95% (v/v) ethanol, and anhydrous ethanol) and destained with a series of xylene solutions (3:1 ethanol: xylene, 1:1 ethanol: xylene, 1:3 ethanol: xylene, and pure xylene). The leaf tissues were soaked in each ethanol and xylene solution for two hours. Finally, the leaves were embedded in paraffin. Tissue sections were cut with a Leica rotary microtome, fixed on a glass slide, and stained with 0.05% Toluidine Blue O (Sigma).
Subcellular Localization
Full length OsLBD3-7 was inserted into the PA7-YFP vector, which had been digested with BamHI and SmaI using the In-fusion system (Clontech). OsLBD3-7-YFP was transiently expressed in Arabidopsis mesophyll cells [24]. The AtAHL-RFP fusion protein was used as a nuclear marker. The fluorescence signal was observed under a confocal microscope (Zeiss LSM700) after 16 h of transformation at room temperature in the dark.
Transactivation Activity Assays in Yeast
The full length OsLBD3-7 was inserted into the pGBKT7 vector, which had been digested with EcoRI, and then was transformed into the yeast strain AH109. The pGBKT7 empty vector and BD-VP64 vector were used as controls. Transformed yeast was dropped onto SD/-W (-Trp) and SD/-W-H-Ade (-Trp/-His/-Ade) plates and allowed to grow for 48 hours before taking photographs. β-galactosidase activity was measured according to the Yeast Protocols Handbook (Clontech) using chlorophenol red-β-D-galactopyranoside as a substrate (CPRG, Roche Biochemical).
RNA Isolation and qRT-PCR Analysis
Plants were grown under continuous light till four leaves stage and total RNA was extracted using the Trizol reagent (Invitrogen). The first complementary DNA was synthesized from DNase-treated total RNA (2 μg) with oligo (dT) primers using TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). qRT-PCR was performed using SYBR Premix Ex Taq (Takara) on a Roche Light Cycler 480 instrument. Quantitative reverse transcription PCR (qRT-PCR) experiments were performed with three replicates using the Actin1 gene (LOC_Os05g36290.1) as the internal standard to normalize the expression of OsLBD3-7 and the other tested genes. The gene expression levels (GEL) were calculated using the formula: [GEL = 2-ΔCt, ΔCt = Ct (Genes)-Ct (Actin1); Ct (cycle threshold) represents the number of cycles required for the fluorescence signal to exceed background level.
Immunoblots
Immunoblot analysis was performed using one-week-old seedlings as described previously [25].
Results
OsLBD3-7 Overexpression Displayed Rolled Leaf Phenotype
In our previous report [26], we utilized a hybrid transcription factor (HTF) approach that mimics the naturally occurring “exon shuffling” process [27] to fuse 1,500 rice transcription factors with the transcription activation module VP64 or repression module 4EAR, transforming them into rice accession Kita-ake and obtaining 50,000 transgenic rice lines. Among them, we identified an HTF, OsLBD3-7V, whose overexpression led to a rolled leaf phenotype. Three independent transgenic lines, OsLBD3-7V-7, OsLBD3-7V-10 and OsLBD3-7V-13, which displayed different RNA and protein expression levels, were selected for further phenotypic analysis (Fig 1d and 1e). We first measured the LRI. The average LRI values for OsLBD3-7V-7, OsLBD3-7V-10 and OsLBD3-7V-13 flag leaves were 0.84, 0.83 and 0.78, respectively, while the wild type flag leaf LRI value was 0.05 (Fig 1f). Next, we measured the widths of the flag leaves. The results showed that OsLBD3-7V-7, OsLBD3-7V-10 and OsLBD3-7V-13 flag leaf widths were 1.24 cm, 1.15 cm and 1.25 cm, respectively, which were significantly narrower than the 1.42 cm wild type leaf (Fig 1g).
Fig 1. Phenotypic analysis of OsLBD3-7V overexpressing plants.
a Gross morphology of WT and OsLBD3-7V. Bars = 10 cm. b The flag leaf morphology of WT and OsLBD3-7V. Bars = 5 mm. c Cross-sections of mature flag leaves. Bars = 5 mm. d Immunoblot analysis of wild type and OsLBD3-7V plants. e OsLBD3-7V expression level analysis by qRT-PCR. f LRI statistical analysis of flag leaves. g Flag leaf width statistical analysis. ab, abaxial; ad, adaxial. Data are shown as means ± s.d. (Student’s t tests, **P < 0.01, n = 3).
However, the additional VP64 activation domain fused to OsLBD3-7 might lead to an artifactual phenotype. Thus, we constructed an OsLBD3-7 overexpression vector under the control of the maize ubiquitin promoter (Pubi) and obtained 41 independent OsLBD3-7 overexpression lines using an Agrobacterium-mediated transformation method. All of the transgenic lines also exhibited varying degrees of adaxially rolled leaf phenotypes (Fig 2). Taken together, these data show that OsLBD3-7 overexpression induced narrow and adaxially rolled leaf phenotypes, while other phenotypic parameters remain unchanged compared with WT plants.
Fig 2. Phenotypic analysis of OsLBD3-7 overexpressing plants.
a Gross morphology of WT and OsLBD3-7. Bars = 10 cm. b The flag leaf morphology of WT and OsLBD3-7. Bars = 5 mm. c Cross-sections of mature flag leaves. Bars = 5 mm. d Immunoblot analysis of wild type and OsLBD3-7 plants. e OsLBD3-7 expression level analysis by qRT-PCR. f LRI statistical analysis of flag leaves. g Flag leaf width statistical analysis. ab, abaxial; ad, adaxial. Data are shown as means ± s.d. (Student’s t tests, **P < 0.01, n = 3).
We also obtained OsLBD3-7-RNAi transgenic rice. None of these transgenic plants showed any obvious phenotypic changes during the entire rice growth period in comparison with wild type plants, although their mRNA levels were significantly reduced (data not shown).
The Number and Size of Bulliform Cells in OsLBD3-7V Transgenic Plants Were Decreased
To confirm that how overexpression of OsLBD3-7 result in rolled leaf phenotype, we observed the detailed anatomy of wild type (WT) and OsLBD3-7V plant stretched leaves. The paraffin section results indicated that the OsLBD3-7V leaves’ abaxial epidermis exhibited no obvious differences compared with the WT. However, the number of bulliform cells in the adaxial epidermis of OsLBD3-7V leaves were remarkably reduced compared with WT leaves (Fig 3a). We counted the number of bulliform cells in the first and the second fully expanded leaves. Statistical analysis indicated that the number of OsLBD3-7V bulliform cells was 21% less than in the WT (Fig 3b). Furthermore, we measured the size of the bulliform cells in the OsLBD3-7V paraffin sections. The average values showed that the OsLBD3-7V bulliform cell size were 80% smaller compared with the WT (Fig 3c). These results indicated that the decreasing both bulliform cell size and number led to the rolled leaf phenotype.
Fig 3. Histological observation of OsLBD3-7V seeding leaves.
a Cross-sections of the first and second fully expanded leaves from 21-day-old wild type and OsLBD3-7V seedlings. ab, abaxial; ad, adaxial. Bars = 100 μm. b and c Bulliform cell number and area statistical analysis from wild type and OsLBD3-7V seeding leaves. Data are shown as means ± s.d. (Student’s t tests, **P < 0.01, n = 10).
Rice OsLBD3-7 Encodes a Typical LBD Family Transcription Factor
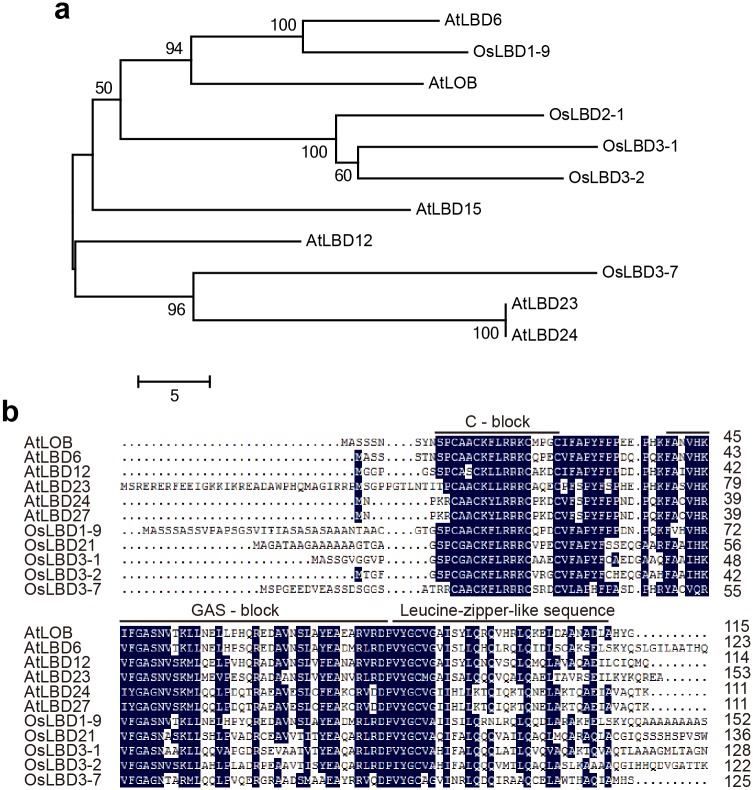
Rice genome annotation analysis (http://rice.plantbiology.msu.edu/) showed that OsLBD3-7 is located on chromosome 3 and encodes a 171 amino acid residues protein. Protein structural analysis revealed that OsLBD3-7 contains a C motif that has a 22 amino acid CX2CX6CX3C zinc finger-like domain, a GAS motif that starts with YX2VQ and ends with DP amino acids and a VX6IX6IX6L leucine zipper-like domain at the C-terminal (Fig 4b). Phylogenetic tree analysis showed that OsLBD3-7 shares high similarity with other homologues in rice (OsLBD1-9, OsLBD2-1, OsLBD3-1, and OsLBD3-2) and Arabidopsis (AtLOB, AtLBD6, AtLBD12, AtLBD23, AtLBD24 and AtLBD15) (Fig 4a). Amino acid alignment showed that OsLBD3-7 shares 52% to 57% identity with Arabidopsis homologues, while sharing less than 45% identity with rice homologues (Fig 4b). Together, these data suggest that OsLBD3-7 possesses the typical features of a Class I LBD family gene.
Fig 4. OsLBD3-7 encodes a LBD family protein.
a Phylogenetic tree analysis of the OsLBD3-7 protein with its rice and Arabidopsis homologues. b Amino acid alignment of the OsLBD3-7 protein and its Arabidopsis and rice homologues. Conserved amino acids are highlighted in blue.
Rice OsLBD3-7 Subcellular Localization and Expression Pattern
To examine OsLBD3-7 subcellular localization, we fused YFP to the OsLBD3-7 C-terminal. Transient expression in Arabidopsis mesophyll protoplasts showed that OsLBD3-7-YFP was exclusively located on the plasma membrane and in the nucleus. In contrast, the control protein YFP was only detectable in the intracellular region (Fig 5a).
Fig 5. Subcellular localization and expression pattern analysis of OsLBD3-7 in rice.
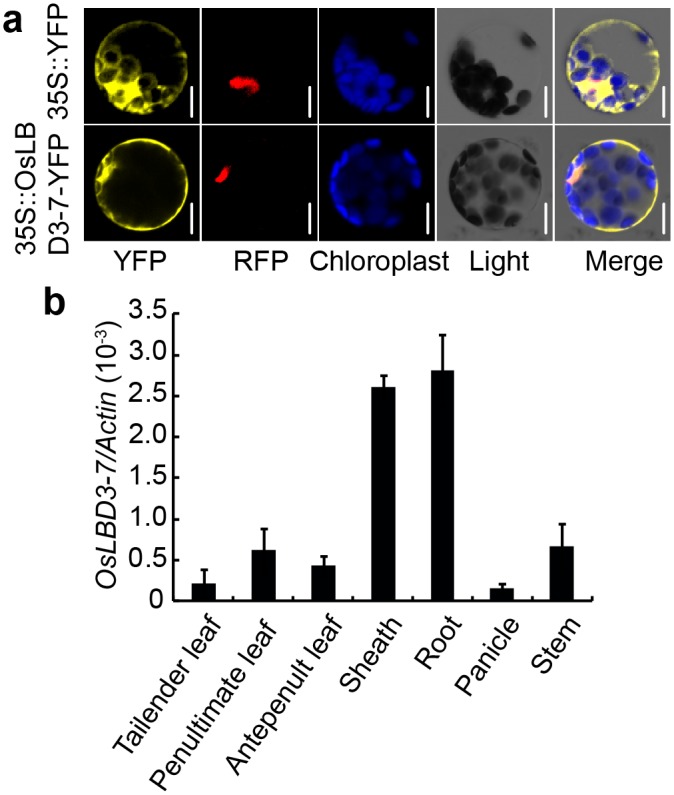
a Subcellular localization of OsLBD3-7. The OsLBD3-7-YFP fusion proteins were located on the plasma membrane and in the nucleus. YFP was used as a control. AHL-RFP fusion protein was used as a nuclear marker. Bars = 10 μm. b Expression pattern analysis of OsLBD3-7 in various vegetative organs by quantitative RT-PCR. Data are shown as means ± s.d. (n = 3).
To investigate the expression pattern of OsLBD3-7, we monitored its expression throughout its life period (from seeding to mature caryopses) with qRT-PCR. Our results showed that OsLBD3-7 was ubiquitously expressed in almost all of the detected tissues, with especially high expression in the sheath and roots (Fig 5b).
Rice OsLBD3-7 Works as a Transcription Activator
Most characterized LBD genes have been reported to be transcription factors. To determine the transcriptional activity of OsLBD3-7, OsLBD3-7 and OsLBD3-7V were fused with a BD domain and transformed into the yeast cell AH109; the pGBKT7 empty vector and the BD-VP64 vector were used as controls. As shown in Fig 6, all of the yeast transformants grew well on the (-Trp) SD medium. When moved to (-Trp/-His/-Ade) SD medium, only BD-VP64 and OsLBD3-7V grew well, while OsLBD3-7 performance was a little better than the empty vector despite growth retardation. We also quantitatively monitored the β-galactosidase activity of all of the transformants using CPRG as substrate. The observed values showed that OsLBD3-7 possessed weak transcriptional activity, which was only two times higher than the empty control. However, when fused with the VP64 activation domain, OsLBD3-7 transcriptional activity increased dramatically (Fig 6). These results indicated that both OsLBD3-7 and OsLBD3-7V work as transcription activators, although OsLBD3-7V has a much higher activity than OsLBD3-7.
Fig 6. Transcription activation analysis of OsLBD3-7 in a yeast GAL4 system.
Each transformed yeast strain was dropped onto SD/-W and SD/-W-H-Ade plates and allowed to grow for 48 hours before taking photographs. β-galactosidase activity was quantified through a liquid culture assay using CPRG as the substrate. Data are shown as means ± s.d. (Student’s t tests, **P < 0.01, n = 3).
Bulliform Cell Negative Regulation Genes Were Up-Regulated in the Rolled Leaf Plants
Our data indicated that the decrease in both bulliform cell size and number led to the rolled leaf phenotype, which suggests that OsLBD3-7 is involved in bulliform cell development regulation. To confirm this speculation, we compared the mRNA expression of several key genes involved in bulliform cell regulation between the OsLBD3-7/OsLBD3-7V transformants and wild type seedlings at the four-leaf stage. The qRT-PCR results showed that the expression of OsADL1, OsSRL1 and OsNRL1 were significantly up-regulated (2–4 folds) in the OsLBD3-7/OsLBD3-7V transgenic seedlings compared with their respective expression in the WT, while OsACL1 was significantly decreased (Fig 7). Therefore, the LBD family transcription factor OsLBD3-7 might work as an upstream regulator of bulliform cell development to regulate leaf rolling.
Fig 7. Expression analysis of bulliform cell regulation genes in OsLBD3-7V and OsLBD3-7 overexpressing plants.
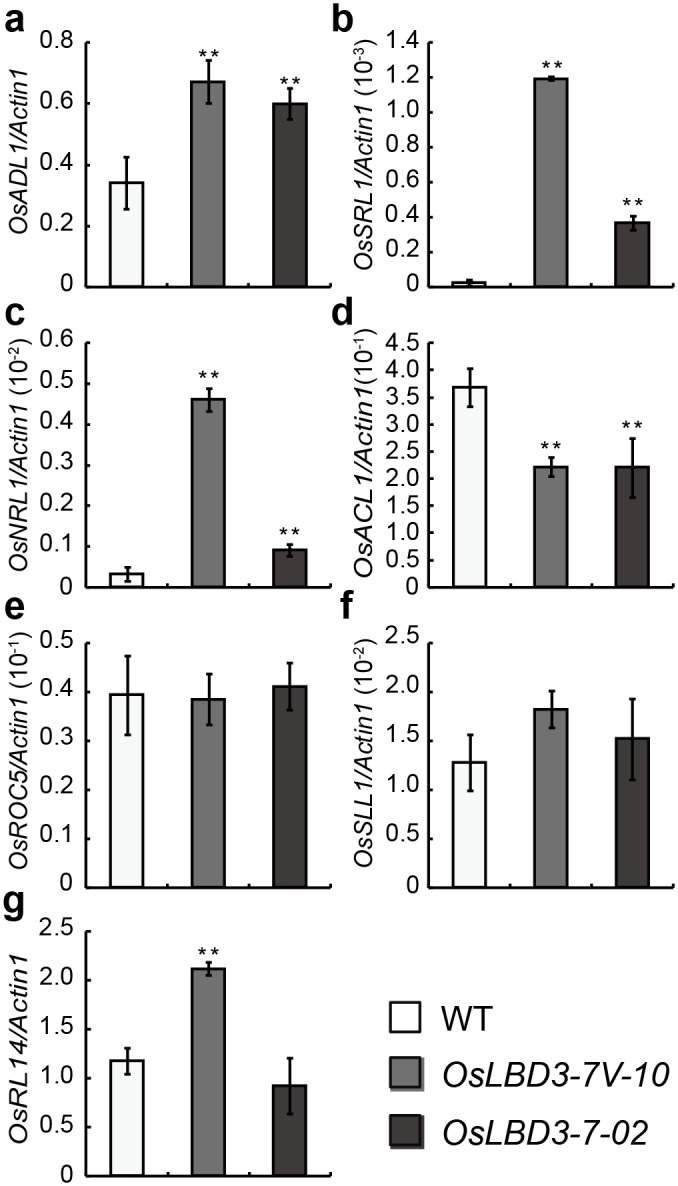
a OsADL1, b OsSRL1, c OsNRL1, d OsACL1, e OsROC5, f OsSLL1, and g OsRL14. Data are shown as means ± s.d. (Student’s t tests, **P < 0.01, n = 3).
Discussion
Leaf size and shape are important morphological traits of plant architecture that affect the yield performance. Moderate leaf rolling could enhance erect-leaf habits, therefore improving the photosynthetic efficiency and increasing the grain yield. Although several genes have been reported to regulate leaf rolling, this complex trait is not yet fully understood. In this study, we reported a new rice LBD gene, OsLBD3-7, that might function in leaf development. OsLBD3-7 is highly expressed in rice sheath and roots, and its protein is located on the plasma membrane and in the nucleus. OsLBD3-7 overexpression could lead to adaxially rolled leaves, which are caused by the suppression of bulliform cell development.
Bulliform cells are a group of large parenchyma cells that specifically exist on the upper surfaces of leaves of many monocots. Bulliform cells are involved in young leaf unfolding, mature leaf rolling and unrolling in response to water stress conditions. Most rolling leaf mutants have been reported to be associated with the changes in bulliform cell number or size. Our data proved that OsLBD3-7 overexpression could lead to cellular atrophy and a decrease in bulliform cell number. Previously, several leaf rolling/curling genes have been reported to be involved in bulliform cell development regulation. Our further investigations showed that OsADL1, OsSRL1 and OsNRL1 were up-regulated in OsLBD3-7 and OsLBD3-7V transgenic plants, while OsACL1 was significantly decreased. Among these genes, OsADL1 and OsSRL1 are bulliform cell negative regulators; mutants of these genes will lead to either bulliform cell number or size expansion. In contrast, OsACL1 is a positive regulator, as OsACL1 overexpression resulted in abaxial leaf curling and an increased number and volume of bulliform cells. Although OsNRL1 is also a positive regulator, but only the mutants have semi-rolled leaves phenotype and over-expression plants have obvious phenotype on leaves. This might imply that OsNRL1 and OsLBD3-7 share different pathway. Together, the rolled leaf phenotype do not perfectly match with the expression of these leaf rolling/curling genes, but still suggests that the LBD family transcription factor OsLBD3-7 might act as an upstream regulator of bulliform cell development to regulate leaf rolling.
The LOB domain comprises a C-domain that is presumably required for DNA-binding, which suggested that LBD genes act as transcription factors. Previous Research of ASL4 in Arabidopsis has demonstrated that ASL4 is located in the nucleus and has the DNA-binding capacity to bind the GCGGCG motif [28]. Our data have indicated that OsLBD3-7 is also located in the nucleus and has transcriptional activation activity. Thus, we further checked whether there was a GCGGCG motif in the promotor region (1,000 bps upstream) of OsADL1, OsSRL1 and OsACL1. We identified at least one GCGGCG motif in the OsSRL1 promotor regions, which suggests that OsSRL1 might be the direct targets of OsLBD3-7.
Supporting Information
Table A. Primers used in this study. Table B. Gene accession numbers in this study. Table C. The protein Sequences used to build phylogenetic tree.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work is supported by the National Natural Science Foundation of China (31571758 to TZ) and the CAAS Innovation Program.
References
- 1.Govaerts YM, Jacquemoud S, Verstraete MM, Ustin SL. Three-dimensional radiation transfer modeling in a dicotyledon leaf. Appl Opt. 1996;35:6585–9. 10.1364/AO.35.006585 [DOI] [PubMed] [Google Scholar]
- 2.Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. The Plant cell. 2009;21(3):719–35. 10.1105/tpc.108.061457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou LP, Sun XH, Zhang ZG, Liu P, Wu JX, Tian CJ, et al. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant physiology. 2011;156(3):1589–602. 10.1104/pp.111.176016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C, Li D, Liu X, Ji C, Hao L, Zhao X, et al. OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC plant biology. 2014;14:158 10.1186/1471-2229-14-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, et al. Overexpression of ACL1 (abaxially curled leaf 1) increased Bulliform cells and induced Abaxial curling of leaf blades in rice. Molecular plant. 2010;3(5):807–17. 10.1093/mp/ssq022 . [DOI] [PubMed] [Google Scholar]
- 6.Yan S, Yan CJ, Zeng XH, Yang YC, Fang YW, Tian CY, et al. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant molecular biology. 2008;68(3):239–50. 10.1007/s11103-008-9365-x . [DOI] [PubMed] [Google Scholar]
- 7.Fang L, Zhao F, Cong Y, Sang X, Du Q, Wang D, et al. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant biotechnology journal. 2012;10(5):524–32. 10.1111/j.1467-7652.2012.00679.x . [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Zhu L, Zeng D, Gao Z, Guo L, Fang Y, et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant molecular biology. 2010;73(3):283–92. 10.1007/s11103-010-9614-7 . [DOI] [PubMed] [Google Scholar]
- 9.Xiang JJ, Zhang GH, Qian Q, Xue HW. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant physiology. 2012;159(4):1488–500. 10.1104/pp.112.199968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, Wang J, Wan X, Shen G, Wang X, Zhang J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta. 2007;226(1):99–108. 10.1007/s00425-006-0472-0 . [DOI] [PubMed] [Google Scholar]
- 11.Alvarez JM, Rocha JF, Machado SR. Bulliform cells in Loudetiopsis chrysothrix (Nees) conert and Tristachya leiostachya Nees (Poaceae): structure in relation to function. Braz Arch Biol Technol. 2008;55:113–9. [Google Scholar]
- 12.Hibara K, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, et al. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Developmental biology. 2009;334(2):345–54. 10.1016/j.ydbio.2009.07.042 . [DOI] [PubMed] [Google Scholar]
- 13.Becraft PW, Li K, Dey N, Asuncion-Crabb Y. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development. 2002;129:5217–25. [DOI] [PubMed] [Google Scholar]
- 14.Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant physiology. 2002;129(2):747–61. 10.1104/pp.010926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006;39:248–62. [DOI] [PubMed] [Google Scholar]
- 16.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends in plant science. 2011;16(1):47–52. 10.1016/j.tplants.2010.09.009 . [DOI] [PubMed] [Google Scholar]
- 17.Coudert Y, Dievart A, Droc G, Gantet P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Molecular biology and evolution. 2013;30(3):569–72. 10.1093/molbev/mss250 . [DOI] [PubMed] [Google Scholar]
- 18.Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. The Plant journal: for cell and molecular biology. 2009;58(3):525–37. 10.1111/j.1365-313X.2009.03797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohashi M, Ishiyama K, Kojima S, Konishi N, Nakano K, Kanno K, et al. Asparagine synthetase1, but not asparagine synthetase2, is responsible for the biosynthesis of asparagine following the supply of ammonium to rice roots. Plant & cell physiology. 2015;56(4):769–78. 10.1093/pcp/pcv005 . [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Wang H, Li J, Huang H, Xu L. Quantitative control of ASYMMETRIC LEAVES2 expression is critical for leaf axial patterning in Arabidopsis. Journal of experimental botany. 2013;64(16):4895–905. 10.1093/jxb/ert278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Wang F, Guo J, Zhang XS. Rice OsAS2 Gene, a Member of LOB Domain Family, Functions in the Regulation of Shoot Differentiation and Leaf Development. Journal of Plant Biology. 2009;52(5):374–81. 10.1007/s12374-009-9048-4 [DOI] [Google Scholar]
- 22.Zhao T, Liu J, Li HY, Lin JZ, Bian MD, Zhang CY, et al. Using hybrid transcription factors to study gene function in rice. Science China Life sciences. 2015;58(11):1160–2. 10.1007/s11427-015-4937-x . [DOI] [PubMed] [Google Scholar]
- 23.Hiei Y, Ohta S, Komari T, Kumashiro T. Effcient transformation of rice (Oryza-sativaL) mediated by agrobacterium and sequence-analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6: 271–82. [DOI] [PubMed] [Google Scholar]
- 24.Steffen A, Athanasios T. Transient trasformation of Arabidopsis Leaf protoplasts: a versatile experimental system to study gene expression. The Plant Journal. 1994;5(3):421–7 [DOI] [PubMed] [Google Scholar]
- 25.Meng Y, Li H, Wang Q, Liu B, Lin C. Blue Light–Dependent Interaction between Cryptochrome2 and CIB1 Regulates Transcription and Leaf Senescence in Soybean. The Plant Cell Online. 2013;25(11):4405–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao T, Liu J, Li HY, Lin JZ, Bian MD, Zhang CY, et al. Using hybrid transcription factors to study gene function in rice. Sci China Life Sci. 2015; 10.1007/s11427-015-4937-x 10.1007/s11427-015-4937-x. . [DOI] [PubMed] [Google Scholar]
- 27.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nature reviews Genetics. 2003;4(11):865–75. 10.1038/nrg1204 . [DOI] [PubMed] [Google Scholar]
- 28.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic acids research. 2007;35(19):6663–71. 10.1093/nar/gkm775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Primers used in this study. Table B. Gene accession numbers in this study. Table C. The protein Sequences used to build phylogenetic tree.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.