Abstract
The evolutionarily conserved small G-protein Ran plays important role in nuclear translocation of proteins, cell cycle regulation, and nuclear envelope maintenance in mammalian cells and yeast. Arabidopsis Ran proteins are encoded by a family of four genes and are highly conserved at the protein level. However, their biological functions are poorly understood. We report here that AtRAN1 plays an important role in vegetative growth and the molecular improvement of stress tolerance in Arabidopsis. AtRAN1 overexpression promoted vegetative growth and enhanced abiotic tolerance, while the atran1 atran3 double mutant showed higher freezing sensitivity than WT. The AtRAN1 gene is ubiquitously expressed in plants, and the expression levels are higher in the buds, flowers and siliques. Subcellular localization results showed that AtRAN1 is mainly localized in the nucleus, with some present in the cytoplasm. AtRAN1 could maintain cell division and cell cycle progression and promote the formation of an intact nuclear envelope, especially under freezing conditions.
Introduction
The small soluble GTP-binding protein Ran has been shown to be important for the nuclear translocation of proteins and cell cycle regulation in mammalian and yeast cells [1–3]. Ran is mainly localized in the nucleus. Ran exerts its biological function by binding and hydrolyzing GTP. However, intrinsic nucleotide exchange and GTP hydrolysis involving Ran are very low [4–5]. Many types of regulatory proteins have been identified that interact specifically with Ran and stimulate its nucleotide exchange and GTP hydrolysis rates for thousands of times [6]. The identification of RanBP1 (Ran binding protein) and RanGAP (Ran GTPase-activating protein) made the function of Ran more clear in plants [5, 7]. RanGAP and its co-activator RanBP are excluded from the nucleus and deplete RanGTP from the cytoplasm.
Four Ran GTPases (AtRAN1, AtRAN2, AtRAN3, AtRAN4) have been identified in the Arabidopsis genome by comparing amino acid similarities [8–9]. Ran GTPases interact directly with AtKPNB1 (a homolog of human KPNB1), which is involved in abscisic acid response and drought tolerance [10]. MOS14 (modifier of snc1-1, 14) interacts with AtRAN1 and is required for the proper splicing of resistance genes and plant immunity [11]. RAN1 mediates seed development through its parental ratio by affecting the onset of endosperm cellularization [12]. The interaction of AtRAN2 with PHIP1 (Phragmoplastin interacting protein 1) may play a unique role in mRNA polarization [13]. With the aid of AtRAN3, AtMBD5 becomes localized to the vicinity of chromosomes during cell division and may play an important role in the maintenance of chromatin structures [14]. Wheat RAN1 is involved in the regulation of cell division and alters the primordial meristem, mitotic progression, and sensitivity to auxin in rice and Arabidopsis [15]. Decreased ATP levels induced by oxidative stress lead to decreased Ran-GTP levels and disordered Ran distribution [16]. In rice, the expression of OsRAN1 is induced by jasmonic acid [17]. The overexpression of OsRAN2 affects the sensitivity to salt stress in rice [18]. OsRAN1 and OsRAN2 play important roles in cold tolerance [19–20]. In this report we indicate that AtRAN1 not only involved in vegetative growth and seed yield but also regulates stress tolerance partly redundant with AtRAN1. AtRAN1 could promote cell cycle progression and maintain nuclear envelop under freezing stress. The sweet potato Ran genes show differential transcriptional regulation in response to various environmental stresses with tissue specificity [21]. Lepidium latifolium L. (LlaRan) is involved in cold stress [22]. Therefore, RAN proteins not only have a major impact on plant development but also mediate plant responses to the environment.
Abiotic stresses often affect plant growth and crop productivity, resulting in great reductions in output [23]. The identification of stress response genes is very important to plant survival and yields under stress conditions [24]. It is well known that nucleo-cytoplasmic trafficking of proteins can affect post-transcriptional regulation, such as mRNA processing, RNA import/export to/from the nucleus, and other processes. [25]. The nucleo-cytoplasmic trafficking of proteins through Ran-dependent pathways may play an important role in the response to abiotic stress [26]. For example, AtKNB1, an Arabidopsis homolog gene of human importin β1, is required for ABA response and drought tolerance [27]. SAD2 (super sensitive to ABA and drought 2) encodes an importin beta-domain family protein likely to be involved in nuclear transport in ABA signaling and UV stress [28].
Normally, plants in temperate regions can increase their freezing tolerance through exposure to chilling temperatures in a process known as cold acclimation [29–30]. This process can result in greatly altered gene expression, biomembrane lipid composition, and other qualities [31]. Unpredictable cold snaps can result in pollen sterility at the flowering stage [32]. The maintenance of cell division, especially under cold stress, is important to plant survival and growth. In this study, we explored the roles of AtRAN1 in the cell cycle progression and regulation of stress tolerance in Arabidopsis.
Plant growth is an important yet poorly understood biological process [33–34]. The investigation of the AtRAN1 gene is therefore helpful for our comprehension of plant growth mechanisms. By using Arabidopsis AtRAN1-knockdown and AtRAN1-overexpressing lines, we demonstrate that AtRAN1 controls plant growth, stress resistance and yield. Our results imply that plant Ran GTPase plays an important role in the link between environmental cues and growth processes in plants.
Materials and Methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana L., Heynh. ecotype Columbia (Col-0)) was used in this study. The T-DNA mutants atran1-1 (SALK_138680), atran1-2 (SALK_067649), atran2 (SALK_123620C) and atran3 (SALK_074683) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/) and genotyped by polymerase chain reaction (PCR) using primers flanking the insertions (S3 Table). To generate the double mutant, we crossed atran3 with atran1-1 and atran1-2, atran3 with atran2, and the genotypes of the F2 plants were identified by PCR. The Arabidopsis seeds were surface-sterilized and planted in MS medium (Murashige & Skoog 1962) containing 1% sucrose. Plates were maintained in the dark at 4°C for 2 d to synchronize germination and then transferred to a growth chamber under a 16/8 h photoperiod. The flowering time was determined by counting the days and rosette leaves once a flower bud was visible.
Plasmid constructions and generation of transgenic plants
The full-length cDNA of AtRAN1 and AtRAN3 was amplified from Col-0 wild-type cDNA. The modified green fluorescent protein (GFP) gene was amplified using the pCAMBIA1302 vector. To construct the plasmid for gene overexpression, AtRAN1 and AtRAN1::GFP were cloned into a pHB vector [35] to generate 35S::AtRAN1 and 35S::AtRAN1::GFP transgenic lines. All reagents and enzymes used here for PCR amplification or restriction digestion were purchased from Takara, Japan. Arabidopsis plants (ecotype Columbia) were transformed by the floral-dipping method [36]. Hygromycin (Roche) resistance was used to screen positive transgenic plants. The concentration of hygromycin used for screening the plants was 50 mg/L. Genomic PCR was used to confirm transgenic plants with primers specific for the hygromycin phosphotransferase (HPT) gene. Semi-quantitative reverse transcription (RT)-PCR and quantitative PCR (qPCR) were performed to detect the gene expression levels in transgenic Arabidopsis. The primers used are shown in Supplementary S1 and S2 Tables at Plos one online.
Subcellular localization study
The GFP signal was observed in the roots of 1-week-old transgenic Arabidopsis seedlings, which were constitutively transformed with constructs of double 35S::AtRAN1::GFP and analyzed by using confocal microscopy (Zeiss LSM510; Jena, Germany). In addition, transient expression of the double 35S::AtRAN1::GFP construct was observed in tobacco epidermal cells.
Freezing, Salt and ABA treatment assay
Freezing treatment was performed as previously described [37]. The atran1-1, atran1-2, atran2, atran3, atran1-1 atran3, atran1-2 atran3, AtRAN1-OE1, AtRAN1-OE3 and wild-type plants were grown in a growth chamber under 16-h-light (23°C)/8-h-dark (21°C) cycles until treatment. For freezing treatment, the seedlings were placed in a controlled temperature chamber and subjected to freezing stress. Soil-grown plants (4 weeks old) or 10-d-old seedlings grown on MS medium plates were subjected to a temperature drop from 4°C to -2°C in the growth chamber. When the temperature reached -2°C, ice crystals were placed on the plates or soil to induce crystallization. After one hour at -2°C, the temperature was lowered to the final temperature. After treatment at the final temperature, the plants or seedlings were thawed at 4°C overnight. Following recovery for 14 d under 16-h-light (23°C)/8-h-dark (21°C) cycles, the plants were photographed. Salt treatment was performed using seedlings in MS medium containing 100 mM NaCl. ABA treatment was performed using seedlings in MS medium containing 0.5 μM ABA. The plants were photographed after treatment.
Ion leakage assay
The electrolyte leakage test was performed as previously described [38]. The electrolyte leakage assay is a widely used means to assess the extent of plant injury in relation to temperature stress. The test is based on the principle that damage to cell membranes results in enhanced leakage of solutes into the apoplastic water [39].
RT-PCR and real-time PCR
The synthesis of cDNA was performed using the ReverTra Ace qPCR Master Kit (FSQ-201) as previously described [40–42]. An aliquot of 2 μL of ten-fold diluted cDNA was used as the RT-PCR template in a 20 μL reaction system. All PCR products were loaded onto a 1% agarose gel to visualize the amplified cDNAs. RT-PCR was repeated three times. Actin2 was used as a control for 24 cycles. The fluorescence intensity of the DNA bands was quantified using Bio-Rad's ChemiDocTM MP Imaging System. For real-time PCR, the cDNA samples were diluted to 2 ng μl−1. Triplicate quantitative assays were performed using 1 μL of cDNA dilution with the SYBR GreenMaster mix and an ABI fast sequence detection system according to the manufacturer's protocol (Applied Biosystems, Foster City, CA, USA). The amplification of Actin2 was used as an internal control to normalize all data. The primers for gene expression are listed in Supplementary S2 Table at Plos one online.
Nuclear Envelope Observation
Ten-day-old Arabidopsis transgenic and wild-type seedlings were treated for 0 h and 3 h at -4°C. Arabidopsis root tips (4–8 mm) were fixed for 5–6 h in fixation buffer (3% glutaraldehyde in 0.1 m PBS, pH 7.2). The materials were washed three or four times with 0.1 m PBS and fixed in 1% osmic acid at 4°C overnight. The materials were then washed three or four times with 0.1 m PBS and dehydrated with an ethanol series of 30, 50, 70% (4°C, overnight), followed by 80, 90, 95, and 100% (30 min for every concentration). Ethanol was then replaced with acetone (1:1) and infiltrated with a mixture of acetone: resin = 2:1 followed by mixture having a ratio of 1:1; 1:2 for 3 hours each and 100% resin for 12 h. Root tips were cut into 50–70 nm pieces using an ultramicrotome, and the nuclear envelope was observed by transmission electron microscopy (HITACHI H-7650, Japan).
Data analysis
GraphPad primer 5 software and MS Office tools were used to create graphs and analyze the data as required. Statistical analysis was performed using Student’s t-test. P values<0.05 were considered significant.
Results
Molecular characterization of AtRAN1 gene
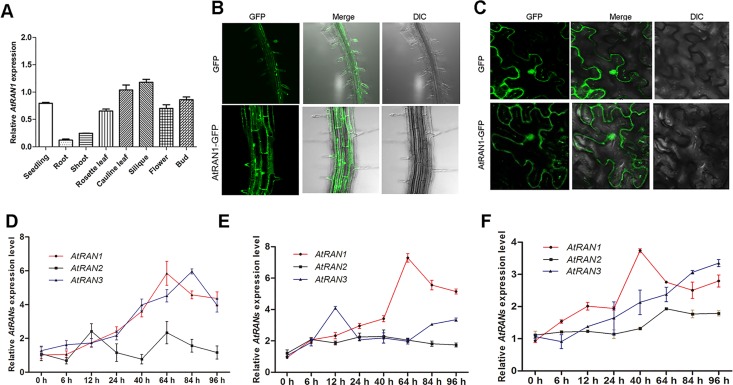
We characterized the expression levels of the AtRAN1 gene. The expression patterns of the AtRAN1 gene in seedlings, roots, shoots, rosette leaves, cauline leaves, siliques, and flowers were investigated by qPCR. We found that the gene was ubiquitously expressed in plants, and the AtRAN1 expression levels were higher in the buds, flowers, leaves, and siliques, than in the other organs examined (Fig 1A). Next, the subcellular localization of AtRAN1::GFP was traced to the root cells of transgenic Arabidopsis steadily overexpressing the AtRAN1::GFP gene. The green fluorescent signal of AtRAN1::GFP was detected mainly in the nucleus (Fig 1B), while in the control, the green fluorescent signal was randomly distributed in the cells. In addition, this localization pattern was observed through the transient expression of AtRAN1::GFP in tobacco epidermal cells, with identical results (Fig 1C).
Fig 1. Expression Patterns and Subcellular Localization of AtRAN1.
The results of qRT-PCR reveal. (A) Real-time PCR analysis the AtRAN1 gene expression pattern. (B) Subcellular localization of the vector control and AtRAN1 in transgenic Arabidopsis root cells. (C) Subcellular localization of the vector control, AtRAN1 in tobacco epidermal cells. DIC, differential interference contrast, referring to bright-field images of the cells. Time course analysis of AtRAN1, AtRAN2, and AtRAN3 expression during (D) cold acclimation (4°C). (E) Salt and (F) ABA treatment conditions. Arabidopsis seedlings were germinated and grown for 7 d before they were subjected to treatment. Actin2 was used as an internal control. The error bars show SD, and are from three independent replications. And the results were repeated three times. Asterisk (*) indicates significant difference (P < 0.05).
We investigated the effects of abiotic stress on AtRAN gene expression by using qRT-PCR to monitor the expression patterns. As shown in Fig 1D, AtRAN1 expression gradually increased after 12 h of 4°C acclimation, with the highest expression at 64 h (approximately a 6-fold increase). AtRAN3 expression began increasing after 6 h of 4°C acclimation and reached approximately 6-fold at 84 h. The expression of AtRAN1 increased to more than 4-fold higher after NaCl treatment, which also increased the expression of AtRAN3 after 24 h and increased it further thereafter (Fig 1E). In addition, 0.5 μM abscisic acid was used for 96 h, with AtRAN1 levels beginning to increase after 24 h of treatment and peaking at 64 h, with a 7-fold increase. The AtRAN3 levels increased 4-fold 12 h after treatment and decreasing slightly thereafter (Fig 1F). In conclusion, the data suggest that AtRAN1 and AtRAN3 respond to cold, salt and ABA treatment in different degrees.
To address the function of the AtRAN genes, T-DNA insertion mutants were obtained from the SALK T-DNA insertion collection. Seeds of AtRAN1-1 (SALK_138680), AtRAN1-2 (SALK_067649), AtRAN2 (SALK_123620c), AtRAN3 (SALK_074683) plants were obtained, and homozygous mutants were verified using diagnostic PCR screening and DNA sequencing. AtRAN gene expression in the mutants was checked by qRT-PCR. Our data verified the insertion and a large reduction in AtRAN1 expression in the SALK_138680 line and the SALK_067649 line. Furthermore, AtRAN2 gene expression decreased in the SALK_123620c mutant. The SALK_074683 mutant showed no AtRAN3 expression (S1 Fig). Because the AtRAN1 and AtRAN2 genes are linked on the same chromosome, the atran1 and atran2 mutants were subsequently crossed with atran3 to obtain homozygous atran1 atran3, atran2 atran3 double mutants. We overexpressed AtRAN1 in the wild-type under the control of the CaMV 35S promoter (S1 Fig).
AtRAN1 overexpression promotes vegetative growth and increases yields
To characterize the AtRAN1 gene function in Arabidopsis, we first obtained many lines of highly overexpressing transgenic plants. The AtRAN1-OE plants showed many phenotypic differences during plant development. Under long-day conditions, the transgenic seeds exhibited a quicker germination rate, the transgenic 7-day seedling hypocotyls were much longer than in wild-type. Moreover, AtRAN1-OE plants exhibited larger leaf size and longer siliques (Fig 2, Table 1). In root development, the AtRAN1-overexpressing plants had approximately 1.5 cm longer primary roots and approximately 5 more lateral roots than the 16-day-old wild-type seedlings (Table 1). Thus, AtRAN1 overexpression promotes plant vegetative growth. Furthermore, we isolated the atran1-1, atran1-2, and atran1-1 atran3 mutants. However, no obvious differences were manifested in the leaf development in comparison with the wild-type plants. This rather weak mutant developmental phenotype may be due to the unattainability of the AtRAN1 knockout mutant or/and the functional redundancy with other Ran genes.
Fig 2. Pleiotropic Phenotype of AtRAN1 overexpressing Plants.
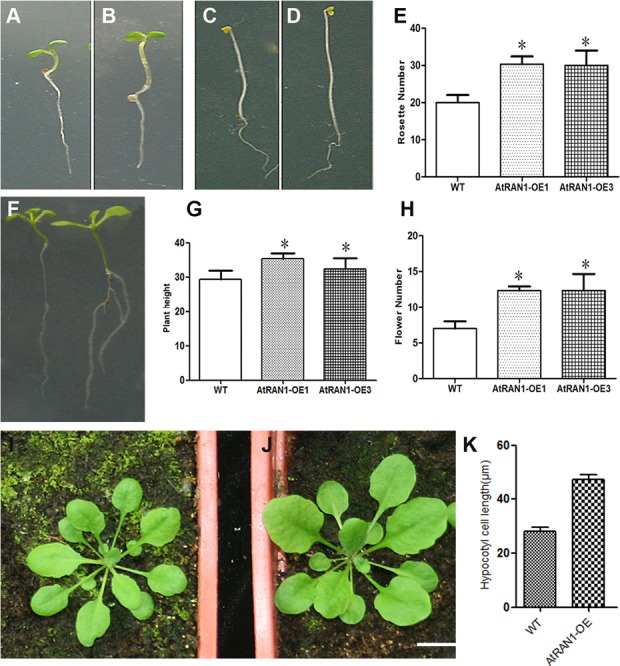
Seven-day seedlings hypocotyls in Wild-type (A, C) and AtRAN1-OE1 plants (B, D) transgenic lines plants under white light and dark conditions; Analysis of (E) Rosette leaf number; (F) Root phenotype of WT and transgenic plants. (G) Plant height; (H) Flower number; Five-week seedlings of Wild-type (I) and AtRAN1-OE1 plants (J). (K) Hypocotyl cell length in dark grown seedlings. Flower number; Figures I, J Bars = 0.5 cm. The error bars show SD.
Table 1. Seed production and root phenotypes in different genotype plants.
| Genotype | Seed weight (mg/plant) | Silique length (cm) | Main root (cm) | Lateral root number |
|---|---|---|---|---|
| Col-0 | 366 ± 55 | 1.6 ± 0.2 | 7.3 ± 0.8 | 13.3 ± 1.2 |
| atran1-1 | 342 ± 29 | 1.44 ± 0.4 | 6.8 ± 0.7 | 10.3 ± 1.7a |
| atran1-2 | 338 ± 42 | 1.47 ± 0.6 | 7.1 ± 1.3 | 11.0 ± 0.8a |
| atran1-1 atran3 | 321 ± 38a | 1.35 ± 0.1 | 6.4 ± 0.6 | 9.6 ± 0.7a |
| atran1-2 atran3 | 324 ± 31a | 1.38 ± 0.2 | 6.6 ± 0.4 | 9.1 ± 0.9a |
| AtRAN1-OE1 | 431 ± 46a | 1.89 ± 0.3a | 8.9 ± 1.1a | 16.8 ± 1.3a |
| AtRAN1-OE3 | 433 ± 41a | 1.87 ± 0.3a | 8.8 ± 0.7a | 17.9 ± 0.9a |
Plants were grown to full maturity in greenhouse conditions. The weight of seeds harvested was measured for each plant. The main root length and lateral root number were measured in 16-day-old seedlings on the MS medium. The mean values ± SD of seven independent plants are shown.
a indicates value that are significantly different from the respective controls (P<0.05). ND; not determined.
In addition to the promotion of vegetative growth by AtRAN1, we also characterized the seed yields in the mutants and overexpression lines. We observed that seed production and germination rate (S4 Fig) were much higher in the plants overexpressing AtRAN1. This difference may be due to increased plant growth and fitness and the resulting increases in both seed and silique size (Table 1). It has previously been reported that decreased levels of AtRAN1 affect seed body size in Arabidopsis [12]. Thus, the AtRAN1 gene both promoted plant growth and increased seed production in Arabidopsis.
AtRAN1 regulates salt tolerance and ABA insensitivity redundantly with AtRAN3
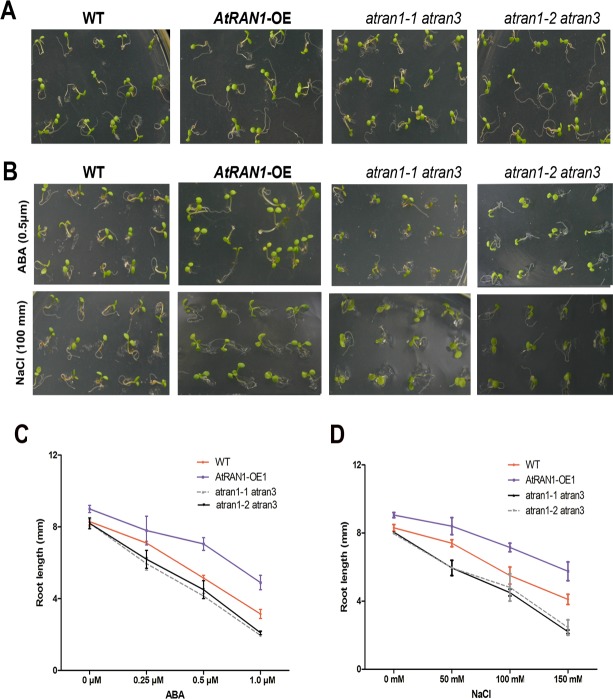
Because the AtRAN1 and AtRAN3 genes are responsive to abiotic stress, we wanted to determine whether the atran1 atran3 double mutant plants show the growth phenotype under the condition of salt stress and exogenous ABA treatment. Under normal conditions, atran1 atran3 seed germination was identical to wild-type (Fig 3A). Under salt and ABA treatment, compared with the atran1, atran2, atran3 and wild-type plants, both the atran1-1 atran3 and atran1-2 atran3 double mutant seedlings showed higher sensitivity than the wild-type (Fig 3B and S3 Fig). Since the overexpression plants are already faster to grow and bigger than wild-type and mutants, we also do a dose response curve for post germination growth in the presence of ABA and salt treatment (Fig 3C and 3D). The results showed that the inhibition effects of ABA were smaller on AtRAN1-OE plants than on atran1atran3 double mutant and wild-type. So we get the conclusion that AtRAN1 overexpression increased the tolerance to ABA treatment. Thus, we concluded that AtRAN1 was involved in salt tolerance and the ABA signaling pathway. The results suggested that AtRAN1 was required for the ABA-mediated response to general osmotic stress during early seedling growth.
Fig 3. Analysis of salt stress and ABA tolerance of RAN1 overexpression plant and atran1 atran 3 double mutant.
(A) atran1-1 atran3, atran1-2 atran3 mutants, AtRAN1-OE1, and the wild-type grow on MS medium supplemented with 1% sucrose. Photos were taken 7 days after germination. (B) Plants were grown on MS medium supplemented with 1% sucrose and 0.5 μM ABA (upper panel) or 100 mM NaCl (lower panel), respectively. Photos were taken 5 days after treatment. (C) and (D) Dose response curve of root growth under ABA (C), or salt stress conditions (D). Plants were germinated on MS medium supplemented with 1% sucrose. Root lengths were measured 5 days after ABA or NaCl treatment.
AtRAN1 overexpression altered freezing tolerance in Arabidopsis
Enhanced sensitivity to freezing was also observed in the atran1 atran3 double mutant. The freezing tolerance of the atran1, and atran3 single mutants, the atran1-1 atran3 and atran1-2 atran3 double mutants and AtRAN1-OE were compared with wild-type plants after 4°C acclimation in growth chambers. Fig 4A shows the status of each leaf in a pool of plants of each genotype. Representative plants of each population were used to visualize the phenotype before and after freezing treatment. The figure clearly shows that the atran1-1 atran3 and atran1-2 atran3 double mutants exhibited greater freezing susceptibility, with 36% and 39% survival rates. In contrast, the survival rates of the atran1, atran3 single mutants were not significantly different from the wild-type after the -14°C freezing treatment. The ion leakage was higher in the atran1 atran3 mutant background (Fig 4C). Meanwhile, AtRAN1-OE plants exhibited increased freezing tolerance under cold acclimated conditions compared to the wild-type plants (Fig 4B). Specifically, the survival rate of the wild-type plants treated after 4°C acclimation was 52%, but AtRAN1-OE lines showed a survival rate of 74% after freezing treatment. In addition, the ion leakage was lower in the AtRAN1-OE lines. Freezing tolerance was also determined for a range of different freezing temperatures for each transgenic line, and the LT50 (i.e., the temperature at which 50% of plants survive) was determined (S2 Fig). These data supported the hypothesis that AtRan1 positively regulates freezing tolerance.
Fig 4. Freezing Tolerance Analysis of atran Mutants and transgenic plants.
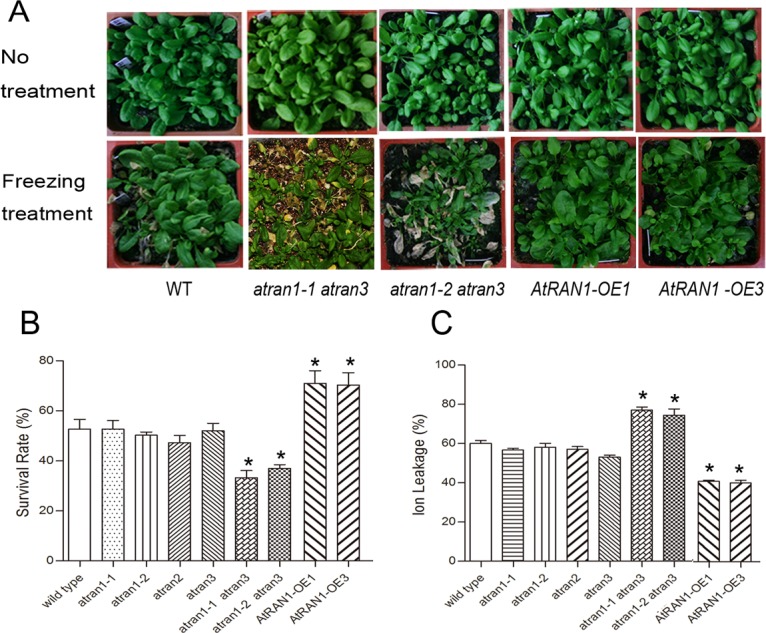
(A) Three-week-old AtRAN gene double mutants and WT plants were cold stressed at -14°C after 4-day 4°C acclimation and then transferred back to the normal condition for recovery. Photographs of representative seedlings of the WT and transgenic lines were taken after 14 d of recovery. (B) Survival rate of the mutants and transgenic plants after freezing stress. (C) Ion leakage assay of the mutants and transgenic plants after freezing stress. The error bars show SD, and were from three independent replications. And the results were repeated three times. Asterisk (*) indicates significant difference (P < 0.05).
AtRAN1 regulates cold response gene expression levels
Cold stress induced the CBF genes, which are well-known regulators of cold response. CBF proteins activated the transcription of the DRE/CRT cis-element-containing COR genes [42–44]. To determine the molecular mechanisms by which AtRAN1 regulated freezing tolerance, we tested changes in the expression of the cold responsive genes. When atran1-1 atran3, AtRAN1-OE1 and wild-type plants were treated after freezing, the expression levels of CBF1 and CBF3 were increased to identical levels, but CBF2 expression levels were lower in the atran1 atran3 mutant and slightly higher in AtRAN1-OE1 (Fig 5A–5C). The expression levels of downstream genes, including COR15A, COR47, and RD29A, were much lower in the atran1 atran3 mutant after freezing treatment but higher in the AtRAN1-OE1 plants (Fig 5D–5F). These results suggest that AtRAN1 modulates cold signaling pathway components.
Fig 5. Cold response Genes and Cell Cycle-related Genes expression Under Normal and Freezing Conditions.
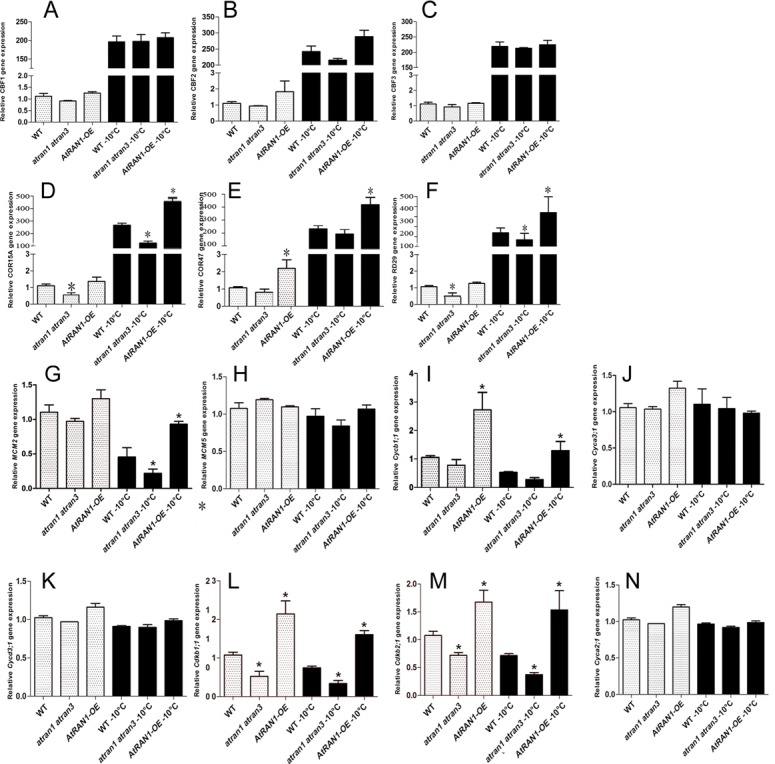
Expression levels of (A) AtCBF1 (DREB1B), (B) AtCBF2 (DREB1C) and (C) AtCBF3 (DREB1A) genes and (D) COR15A, (E) COR47 and (F) RD29A downstream genes in 7-day old AtRAN1-OE, the atran1-1 atran3 mutant and wild-type plants under before and after freezing treatment. Values are means and SD (n = 4). Expression of (G) MCM2, (H) MCM5, (I) Cycb1;1, (J) Cyca3;1, (K) Cycd3;1, (L) Cdkb1;1, (M) Cdkb2;1 and (N) Cyca2;1 cell cycle-related gene levels in wild-type, atran1-1 atran3 mutant and transgenic plants before and after 0.5h freezing stress, after 4°C acclimation, The error bars show SD, and are from three independent replications. And the results were repeated three times. Asterisk (*) indicates significant difference (P < 0.05).
AtRAN1 overexpression promotes cell cycle genes expression
To further illuminate the cell cycle-related genes involved in the control of cold signaling, we also examined the expression patterns of genes related to the cell cycle. Compared with the expression levels of the type A and D cyclin genes, the expression levels of the type B cyclin gene Cycb1;1, the type B cyclin gene related kinase (Cdkb1;1, Cdkb2;1) and MCM2 were suppressed by short-term freezing treatment in both the wild-type and mutant plants. In the AtRAN1-OE line, the expression levels of Cycb1;1,Cdkb1;1,Cdkb2;1,MCM2 were much higher than in the wild type and mutant plants both before and after the freezing treatment (Fig 5I, 5L and 5M). Thus, cold responsive genes and cell proliferation-related genes are involved in the AtRAN1-mediated cold tolerance pathway.
AtRAN1 regulates nuclear envelope maintenance under cold
We observed the nuclear envelopes of root tip cells in the AtRAN1-OE, atran1 atran3 and wild-type Arabidopsis lines under normal and freezing conditions. Under the normal condition (22°C), the nuclear envelope NE was intact in all four lines, with no obvious differences in morphology (Fig 6A–6C). After freezing treatment at -4°C for 2 h, approximately 60%-70% of wild-type cells showed partially dissociated double membranes (Fig 6D). Most of the cells in the transgenic lines showed an intact nuclear envelope (Fig 6F). However, the atran1-1 atran3 double mutant showed many fragmentized nuclear envelopes, with few cells showing slightly dissociated nuclear envelopes (Fig 6E). These results suggested that AtRAN1 overexpression promotes the formation of an intact nuclear envelope under freezing conditions.
Fig 6. Morphological Changes in Nuclear Envelope under Normal and Freezing Conditions.
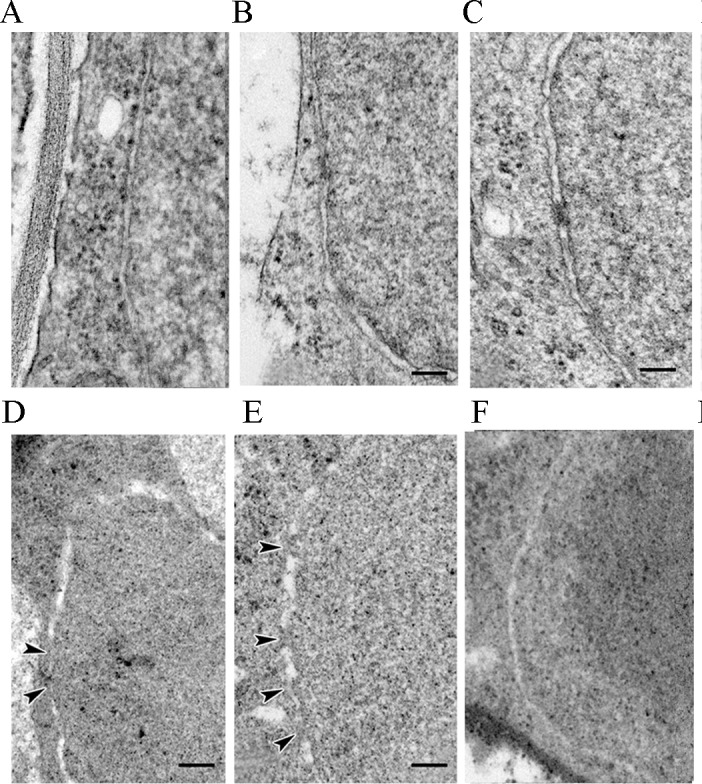
Nuclear envelope of the (A) WT, (B) atran1-1 atran3 and (C) AtRAN1-overexpressing plants under normal conditions (22°C). After 4-day 4°C acclimation, Nuclear envelope of the (D) WT, (E) atran1-1 atran3 and (F) AtRAN1-overexpressing lines were treated for 2h at -4°C. Six root tips were observed in every condition. The root tips were transversely cut in the meristematic zones. Arrows indicate the abnormal nuclear envelope. Bars = 100 nm. The error bars show SD, and are from three independent replications. The results were repeated three times.
Discussion
AtRAN1 function is well-conserved in cell cycle regulation
Ran proteins coupled with importins and exportins transport cargo proteins between the nucleus and cytoplasm. Also RanGTP plays an important role during mitosis [45]. Ran participates in various cellular processes by means of the gradient mechanism, and it concentrates around action site [46]. The nuclear envelope (NE) is a relatively isolated layer and provides a special space for gene transcription and protein translation. The nuclear envelope undergoes dynamic changes during the cell cycle progression [47–48]. Early in the mitosis stage, Ran separates from chromatin to the cytoplasm. During anaphase to telophase, coupled with nuclear envelope assembly, Ran GTPase promotes nuclear pore organization in Xenopus egg extracts [40]. In plants, despite previous reports on the function of OsRAN1 and OsRAN2 in nuclear envelope assembly, our observations indicated that AtRAN1 overexpression also promoted the formation of an intact nuclear envelope and cell cycle progression under freezing conditions (Fig 6). Therefore, plant Ran homologs may have functions that are conserved in plants, yeast, and animals during evolution.
Although AtRAN1 and AtRAN3 are both induced by cold, salt, and ABA treatment, AtRAN1 shows a high homology to AtRAN3. However, AtRAN1-OE plants exhibit many developmental phenotypes different from AtRAN3-OE plants. Consistently with this hypothesis, previous results have revealed that the expression level of the AtRAN1 gene is approximately 2-fold higher than AtRAN3. The physiological functions of the Ran genes in monocot and dicot plants are not always identical. Others have reported that OsRAN2 overexpression in rice and Arabidopsis resulted in enhanced sensitivity to osmotic stress and ABA [18]. TaRAN and OsRAN1 overexpression in Arabidopsis was found to inhibit root development [20]. Therefore, apart from the conserved role of the Ran function in cold resistance, Ran genes do not always function identically in plant development.
AtRAN1 links plant growth and stress tolerance with cell proliferation
To gain information about the biological role of plant RanGTPase proteins in Arabidopsis, we obtained Ran mutants and overexpression lines to reveal gene functions. The AtRAN1 -overexpressing transgenic plants manifested increased plant growth, whereas the atran1 atran3 double mutant caused some developmental defects. The hypocotyl elongation of seedlings was affected by AtRAN1 overexpression under white light and dark conditions, suggesting that the AtRAN1 control of hypocotyl cell elongation is independent of light. AtRAN1 overexpression plants hypocotyl cell were longer that the control (Fig 2K). The leaf sizes were larger in the AtRAN1 overexpression plants. We speculate that AtRAN1 is involved in plant growth through promoting both cell division and cell expansion.
Many studies have shown that cell division is closely related to stress tolerance. Overexpressing HAL3a showed improved growth as well as salt and osmotic tolerances in Arabidopsis and rice [49]. Transgenic rice lines overexpressing OsMYB3R-2 and OsCycB1;1 exhibited enhanced cold tolerance by maintaining cell division activity under cold condition [50]. Our results indicate that the expression of AtRAN1 was up-regulated under cold stress (Fig 1D). This expression pattern suggests that AtRAN1 may function as a regulator in the cold signaling pathway in Arabidopsis. In addition, AtRAN1-OE plants showed increased freezing tolerance after 4°C acclimation. Furthermore, we observed differences in freezing tolerance between the atran1 atran3 mutant and wild-type plants (Fig 5). These data demonstrated that AtRAN1 positively regulated freezing tolerance by maintaining the cell cycle. By combining our results with the findings of previous studies, we speculate that AtRAN1 is an important regulatory link between plant growth and environmental cues.
Supporting Information
(A) Structure of genomic clones encoding the AtRAN1, AtRAN2, and AtRAN3 proteins.(B) Arabidopsis T-DNA mutant screen. (C) The expression pattern of AtRAN genes in the mutant background. (D) Real-time RT-PCR analysis of the expression of AtRAN1 (E) Real-time RT-PCR analysis of the expression of AtRAN3.
(TIF)
Percent survival of acclimated three-week-old AtRAN1-OE lines, atran1 atran3 double mutant and Wild-type plants were frozen in a temperature-controlled chamber as described under Methods. At the temperatures shown, samples of plants were removed from the chamber, allowed to recover, and scored for survival. The data are means ± SE for three separate experiments.
(TIF)
Treatment conditions are same as those in Fig 3, except that plants were germinated on MS medium without sucrose. The AtRAN1-OE1 transgenic plants show higher resistance to ABA and salt treatment than the wild-type, while atran1-1 atran3 and atran1-2 atran3 show increased salt sensitivity than the wild-type.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Arabidopsis Stock Center for providing SALK T-DNA insertion lines.
Funding
This work was supported by the Strategic Priority Research Program on Space Science of the Chinese Academy of Sciences (grant No. XDA04020202-15, XDA04020415), the National Natural Science Foundation of China (No. 31570859, No. 31500236, No. 31070237 and No. 90917009) and the China Manned Space Flight Technology Project.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was supported by the Strategic Priority Research Program on Space Science of the Chinese Academy of Sciences (grant No. XDA04020202-15, XDA04020415), the National Natural Science Foundation of China (No. 31570859, No. 31500236, No. 31070237 and No. 90917009) and the China Manned Space Flight Technology Project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annual Review of Cell and Developmental Biology 1999. 15 607–660. [DOI] [PubMed] [Google Scholar]
- 2.Quimby BB, Dasso M. The small GTPase Ran: interpreting the signs. Current Opinion in Cell Biology 2003. 15 338–344. [DOI] [PubMed] [Google Scholar]
- 3.Ren MD, Drivas G, Deustachio P, Rush MG. Ran/Tc4—a Small Nuclear Gtp-Binding Protein That Regulates DNA-Synthesis. Journal of Cell Biology 1993. 120 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cellular and Molecular Life Sciences, 2007. 64 1891–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci U S A 2001. 98 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sazer S, Dasso M. The Ran decathlon: multiple roles of Ran. J Cell Sci 2000. 113, 1111–1118. [DOI] [PubMed] [Google Scholar]
- 7.Ach RA, Gruissem W. A Small Nuclear Gtp-Binding Protein from Tomato Suppresses a Schizosaccharomyces -Pombe Cell-Cycle Mutant. Proc Natl Acad Sci U S A 1994. 91; 5863–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant Journal 1997. 11; 93–103. [DOI] [PubMed] [Google Scholar]
- 9.Vernoud V, Horton AC, Yang ZB, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiology 2003. 131; 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo YJ, Wang ZJ, Ji HT, Fang H, Wang SF, Tian LN, et al. An Arabidopsis homolog of importin 1 is required for ABA response and drought tolerance. Plant Journal 2013. 75 377–389. 10.1111/tpj.12207 [DOI] [PubMed] [Google Scholar]
- 11.Xu SH, Zhang ZB, Jing BB, Gannon P, Ding JM, Xu F, et al. Transportin-SR Is Required for Proper Splicing of Resistance Genes and Plant Immunity. PLoS Genet 2011. 7 e1002159 10.1371/journal.pgen.1002159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P, Qi M, Wang Y, Chang M, Liu C, et al. Arabidopsis RAN1 Mediates Seed Development through Its Parental Ratio by Affecting the Onset of Endosperm Cellularization. Mol Plant 2014. 7 1316–1328. 10.1093/mp/ssu041 [DOI] [PubMed] [Google Scholar]
- 13.Ma L., Xie B., Hong Z., Verma D.P, Zhang Z. A novel RNA-binding protein associated with cell plate formation. Plant Physiology 2008. 148 223–234. 10.1104/pp.108.120527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano A., Kodama Y., Koike A, Shinya T., Kim H.J., Matsumoto M., et al. Interaction between methyl CpG-binding protein and Ran GTPase during cell division in tobacco cultured cells. Annals of Botany 2006. 98 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Xu Y.Y., Han Y., Bao S.L., Du J.Z., Yuan M., et al. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiology 2006. 140 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda Y., Miyamoto Y., Saiwaki T, Yoneda Y. Mechanism of the stress-induced collapse of the Ran distribution. Exp Cell Res 2006. 312 512–520. [DOI] [PubMed] [Google Scholar]
- 17.Miche L., Battistoni F., Gemmer S., Belghazi M, Reinhold-Hurek B. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol Plant Microbe Interact 2006. 19 502–511. [DOI] [PubMed] [Google Scholar]
- 18.Zang A.P., Xu X.J., Neill S, Cai W.M. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. Journal of Experimental Botany 2010. 61 777–789. 10.1093/jxb/erp341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu P.P., Cai W.M. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). Journal of experimental botany 2014. 65 3277–3287. 10.1093/jxb/eru178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen N., Xu Y., Wang X., Du C., Du J., Yuan M., et al. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell and Environment, 2011. 34 52–64. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y H, Huh G H. Members of the ran family of stress-inducible small GTP-binding proteins are differentially regulated in sweetpotato plants. Journal of Plant Biotechnology, 2013. 40(1); 9–17. [Google Scholar]
- 22.Sinha V B, Grover A, Singh S, Zakwan A. Overexpression of Ran gene from Lepidium latifolium L.(LlaRan) renders transgenic tobacco plants hypersensitive to cold stress. Molecular biology reports, 2014. 41(9); 5989–5996. 10.1007/s11033-014-3476-z [DOI] [PubMed] [Google Scholar]
- 23.Xin Z, Browse J. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell and Environment 2000. 23; 893–902. [Google Scholar]
- 24.Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 2001. 13; 2063–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzucotelli E, Mastrangelo AA, Crosatti C, Guerra D, Stanca AM, Cattivelli L. Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Science 2008. 174; 420–431. [Google Scholar]
- 26.Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Molecular Cell 2004. 16; 319–330. [DOI] [PubMed] [Google Scholar]
- 27.Chinnusamy V, Gong Z, Zhu JK. Nuclear RNA Export and Its Importance in Abiotic Stress Responses of Plants. Nuclear Pre-Mrna Processing in Plants 2008. 326; 235–255. [DOI] [PubMed] [Google Scholar]
- 28.Verslues PE, Guo Y, Dong CH, Ma W, Zhu JK. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant Journal, 2006. 47; 776–787. [DOI] [PubMed] [Google Scholar]
- 29.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology 2000. 124; 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiology 1998. 118; 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu JH, Shi HZ, Lee BH, Damsz B, Cheng S, Stirm V, et al. Hasegawa P.M, Bressan R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci U S A 2004. 101; 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imin N, Kerim T, Rolfe BG, Weinman JJ. Effect of early cold stress on the maturation of rice anthers. Proteomics 2004. 4; 1873–1882. [DOI] [PubMed] [Google Scholar]
- 33.Horvath BM, Magyar Z, Zhang YX, Hamburger AW, Bako L, Visser RG, et al. EBP1 regulates organ size through cell growth and proliferation in plants. Embo Journal 2006. 25; 4909–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolai M., et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 2007. 8; 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci U S A 2005. 102; 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 1998. 16; 735–743. [DOI] [PubMed] [Google Scholar]
- 37.Zhao JS, Ren W, Zhi DY, Wang L, Xia GM. Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant Cell Reports 2007. 26; 1521–1528. [DOI] [PubMed] [Google Scholar]
- 38.Lee H, Guo Y, Ohta M, Xiong LM, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. Embo Journal 2002. 21; 2692–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thalhammer A, Hincha DK, Zuther E. Measuring freezing tolerance: electrolyte leakage and chlorophyll fluorescence assays, Plant Cold Acclimation Springer; New York, 2014. 15–24. [DOI] [PubMed] [Google Scholar]
- 40.Zhang CM, Goldberg MW, Moore WJ, Allen TD, Clarke PR. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. European Journal of Cell Biology 2002. 81; 623–633. [DOI] [PubMed] [Google Scholar]
- 41.Galbraith DW, Harkins KR, Maddox JM, Ayres N.M., Sharma D.P, Firoozabady E. Rapid Flow Cytometric Analysis of the Cell-Cycle in Intact Plant-Tissues. Science 1983. 220; 1049–1051. [DOI] [PubMed] [Google Scholar]
- 42.Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in, stress tolerance in Arabidopsis. Proc Natl Acad Sci U S A 2004. 101; 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ying L, Chen H, Cai W. BnNAC485 is involved in abiotic stress responses and flowering time in Brassica napus Plant Physiology & Biochemistry, 2014, 79(3); 77–87. [DOI] [PubMed] [Google Scholar]
- 44.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A 1997. 94; 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, et al. SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 2007. 19; 3805–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CM, Clarke PR. Roles of Ran-GTP and Ran-GDP in precursor vesicle recruitment and fusion during nuclear envelope assembly in a human cell-free system. Current Biology 2001. 11; 208–212. [DOI] [PubMed] [Google Scholar]
- 47.Di Fiore B, Ciciarello M, Lavia P. Mitotic functions of the ran GTPase network—The importance of being in the right place at the right time. Cell Cycle 2004. 3; 305–313. [PubMed] [Google Scholar]
- 48.Ellenberg J, Lippincott-Schwartz J. Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods 1999. 19; 362–372. [DOI] [PubMed] [Google Scholar]
- 49.Espinosa-Ruiz A, Belles J.M., Serrano R, Culianez-Macia F.A. Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant Journal 1999. 20; 529–539. [DOI] [PubMed] [Google Scholar]
- 50.Ma QB, Dai XY, Xu YY, Guo J, Liu YJ, Chen N, et al. Enhanced Tolerance to Chilling Stress in OsMYB3R-2 Transgenic Rice Is Mediated by Alteration in Cell Cycle and Ectopic Expression of Stress Genes. Plant Physiology 2009. 150; 244–256. 10.1104/pp.108.133454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Structure of genomic clones encoding the AtRAN1, AtRAN2, and AtRAN3 proteins.(B) Arabidopsis T-DNA mutant screen. (C) The expression pattern of AtRAN genes in the mutant background. (D) Real-time RT-PCR analysis of the expression of AtRAN1 (E) Real-time RT-PCR analysis of the expression of AtRAN3.
(TIF)
Percent survival of acclimated three-week-old AtRAN1-OE lines, atran1 atran3 double mutant and Wild-type plants were frozen in a temperature-controlled chamber as described under Methods. At the temperatures shown, samples of plants were removed from the chamber, allowed to recover, and scored for survival. The data are means ± SE for three separate experiments.
(TIF)
Treatment conditions are same as those in Fig 3, except that plants were germinated on MS medium without sucrose. The AtRAN1-OE1 transgenic plants show higher resistance to ABA and salt treatment than the wild-type, while atran1-1 atran3 and atran1-2 atran3 show increased salt sensitivity than the wild-type.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.