Abstract
Spithioneines A and B (1 and 2), two new bohemamine-type pyrrolizidine alkaloids possessing an unusual ergothioneine moiety, were isolated from a marine-derived Streptomyces spinoverrucosus. Their structures were elucidated by spectroscopic analysis, CD spectra, and chemical degradation and synthesis. Compounds 1 and 2 are rare natural products that incorporate the amino acid ergothioneine.
Bacteria are prolific producers of structurally complex natural products, often incorporating multiple biosynthetic pathways to produce molecules of complex structure.1 Over the past two decades there has been increased interest in exploiting the complex chemical diversity from marine bacteria, leading to the discovery and biological characterization of more than 400 new secondary metabolites with cytotoxicity and antimicrobial activity from this resource.2 Among these novel structures are the abyssomycins,3 lomaiviticin,4 salinosporamide A,5 and the marinomycins,6 all of which have garnered the interest of chemical and biological investigations into the potential role of these molecules as therapeutics.
In our ongoing research efforts to identify bioactive metabolites from marine-derived bacteria based on a combination of structural novelty and biological activity, we have combined our natural product fraction library with high-throughput screening and analytical characterization by LC–MS and NMR profiling. In this process, some bioactive compounds with novel structures have been identified.7 Although we are typically focused on active molecules, we occasionally observe unusual structural features that are further investigated.
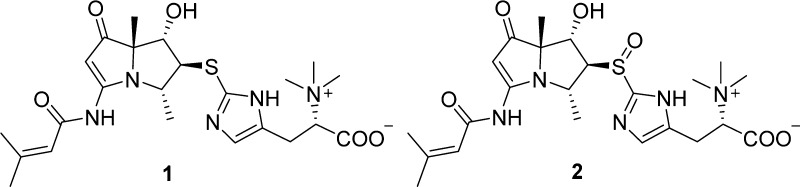
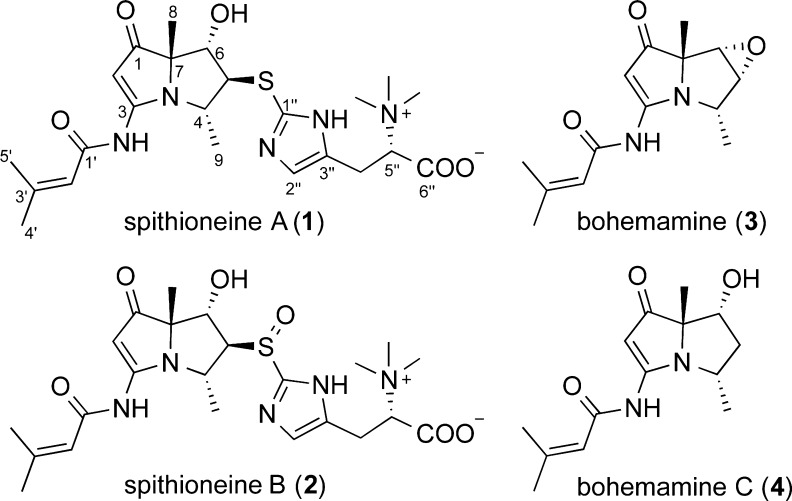
As part of these efforts, we were attracted to a series of olefinic and aromatic 1H NMR signals observed in extracts from the Streptomyces spinoverrucosus strain SNB-048. Further analysis of the extract by LC–UV–MS revealed that it contained metabolites with similar UV absorptions to those of the pyrrolizidine-containing bohemamines (3 and 4).8 Chemical investigation resulted in the isolation of two new bohemamine-type pyrrolizidine alkaloids, spithioneines A and B (1 and 2) (Figure 1). Structurally, compounds 1 and 2 possess an ergothioneine moiety, which is rare in natural products.9
Figure 1.
Structures of spithioneines (1 and 2) and bohemamines (3 and 4).
Spithioneine A (1) was isolated as a colorless oil that gave a HRESIMS [M + H]+ ion of m/z 492.2274, corresponding to a molecular formula of C23H33N5O5S (calcd for C23H34N5O5S, 492.2275). The IR spectrum showed stretches indicative of an amide group (3409 cm–1) and a α,β-unsaturated ketone (1649 cm–1). The UV spectrum displayed absorptions at 250, 278, and 330 nm, similar to those reported for bohemamines.8 The 1H NMR spectrum of 1 in DMSO-d6 indicated the presence of two methyl groups at δ 1.37 (s) and δ 1.30 (d, J = 6.7 Hz), two vinyl methyl groups at δ 2.13 (d, J = 1.3 Hz) and δ 1.87 (d, J = 1.3 Hz), two olefinic proton signals at δ 6.01 (m) and δ 5.52 (s), and an amide exchangeable proton at δ 10.48 (s). Three methine signals at δ 3.91 (d, J = 1.4 Hz), δ 3.76 (dd, J = 2.2, 1.4 Hz), and δ 3.89 (qd, J = 6.7, 2.2 Hz) could also be observed. The 13C NMR spectrum in DMSO-d6 showed two carbonyl signals at δ 201.0 and δ 164.0, assignable to a ketone and amide, respectively. The NMR signals (Table 1) were consistent with those of bohemamine C (4) and 5-chlorobohemamine C,8c indicating spithioneine A is composed of a pyrrolizidine core.
Table 1. 1H (600 MHz) and 13C (100 MHz) NMR Data for Compounds 1 and 2 in DMSO-d6.
|
1 |
2 |
|||
|---|---|---|---|---|
| no. | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) |
| 1 | 201.0, C | 200.2, C | ||
| 2 | 94.4, CH | 5.52, s | 94.4, CH | 5.51, s |
| 3 | 166.7, C | 166.2, C | ||
| 4 | 61.3, CH | 3.89, qd (6.7, 2.2) | 54.4, CH | 4.44, q (6.7) |
| 5 | 62.8, CH | 3.76, dd (2.2, 1.4) | 81.6, CH | 3.90, s |
| 6 | 76.7, CH | 3.91, d (1.4) | 73.1, CH | 3.47, s |
| 7 | 77.6, C | 77.4, C | ||
| 8 | 24.6, CH3 | 1.37, s | 23.6, CH3 | 1.17, s |
| 9 | 18.6, CH3 | 1.30, d (6.7) | 19.4, CH3 | 1.34, d (6.7) |
| 1′ | 164.0, C | 163.9, C | ||
| 2′ | 117.7, CH | 6.01, m | 117.6, CH | 6.01, m |
| 3′ | 156.2, C | 156.5, C | ||
| 4′ | 27.3, CH3 | 1.87, d (1.3) | 27.3, CH3 | 1.90, s |
| 5′ | 20.0, CH3 | 2.13, d (1.3) | 20.0, CH3 | 2.15, s |
| 1″ | 136.4, C | 144.2, C | ||
| 2″ | NDa | 6.92, s | ND | 7.13, s |
| 3″ | ND | 136.6, C | ||
| 4″ | 25.9, CH2 | 3.11, dd (14.4, 4.0) | 25.5, CH2 | 3.18, dd (14.0, 3.6) |
| 3.02, dd (14.4, 9.6) | 3.08, dd (14.0, 10.4) | |||
| 5″ | 77.4, CH | 3.79, dd (9.5, 4.0) | 77.4, CH | 3.84, dd (10.2, 3.8) |
| 6″ | 167.3, C | 166.7, C | ||
| 3-NH | 10.48, s | 10.55, brs | ||
| 5″-NCH3 | 51.2, CH3 | 3.15, s | 51.1, CH3 | 3.15, s |
ND: not detected.
The structure of the pyrrolizidine core was supported by the 13C NMR data (Table 1). Critical assignments were based on COSY correlations of H-9/H4/H-5/H-6, and the key HMBC correlations from H-2 to C-1/C-3/C-7, H-4 to C-3/C-9, H-5 to C-4/C-6/C-7/C-9, H-8 to C-1/C-6/C-7, 3-NH to C-2/C-1′, H-2′ to C-1′/C-3′, H-4′ to C-2′/C-3′/C-5′, and H-5′ to C-2′/C-3′/C-4′ (Figure 2). Moreover, the 1H NMR spectrum in DMSO-d6 showed an olefinic proton at δ 6.92 (s), two methylene protons at δ 3.11 (dd, J = 14.4, 4.0 Hz) and δ 3.02 (dd, J = 14.4, 9.6 Hz), a methine proton at δ 3.79 (dd, J = 9.5, 4.0 Hz), and three equivalent singlet methyl protons at δ 3.15. The correlation between methylene (H-4″) and methine (H-5″) was determined by COSY. The large methyl singlet at δ 3.15 with a corresponding 13C at δ 51.2 was highly suggestive of trimethylammonium functionality. The HMBC correlations from the δ 3.15 singlet to C-5″ (δ 77.4) further supported the proposed trimethylammonium functional group. The HMBC correlations from both H-4″ and H-5″ to a carbonyl carbon C-6″ (δC 167.3) suggested the existence of an amino acid derivative.
Figure 2.
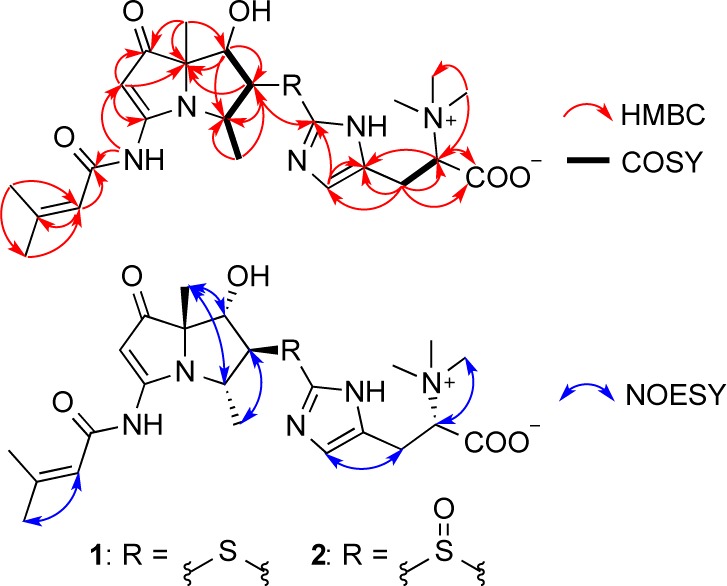
Key HMBC, 1H–1H COSY, and NOESY correlations for 1 and 2.
As not all signals of the 13C NMR spectrum in DMSO-d6 were observable, an additional set of spectra were obtained in CD3OD. The 13C NMR spectrum in CD3OD (Table S1) showed two additional olefinic carbons at δ 121.0 and δ 136.6 that corresponded to C-2″ and C-3″. The HMBC correlations (in CD3OD) from H-4″ to C-2″/C-3″ and H-2″ to C-1″ (δC 138.7)/C-3″ indicated the presence of a hercynine moiety in 1.10 This moiety shows identical NMR chemical shifts as those found in ergothioneine,11 clithioneine,9a and JBIR-73.9b The remaining sulfur, the 13C chemical shifts of C-5 and C-1″, and the HMBC correlation from H-5 to C-1″ established the connection between the hercynine unit and the bohemamine skeleton.
NOE correlations between H-5 and H-9, between H-6 and H-8, and between H-4 and H-8 were observed in the 2D NOESY, suggesting the relative configuration of 1 as shown (Figure 2) and identical to that of bohemamine (3). NOE correlations combined with the small coupling constants between H-4 and H-5 (J = 3.4 Hz) and between H-5 and H-6 (J = 2.0 Hz) also indicated the conformation of 1 as shown in Figure 3.
Figure 3.
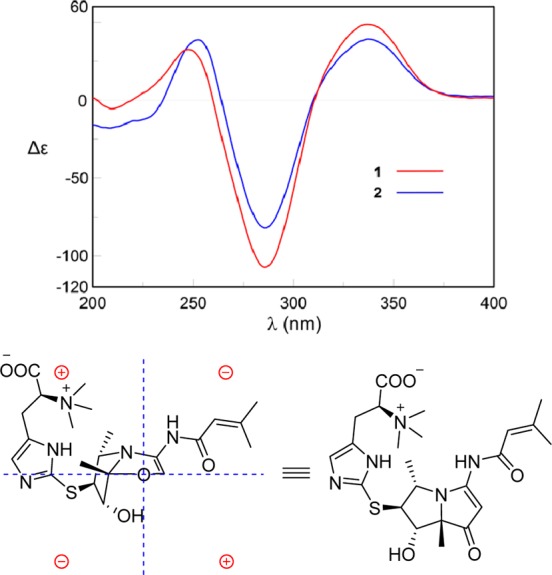
CD spectra of 1 and 2 in MeOH. Representation of the octant rule for the cyclopentenone moiety of 1.
In order to determine the absolute configuration, we utilized CD spectroscopy. On the basis of the octant rule for cyclopentenone,12 the positive Cotton effect at 337 nm (Δε +48.5) for n–π* suggested that the absolute configuration was (4S,5S,6S,7S), consistent with the core configuration of bohemamines, such as NP25302, whose absolute configuration has been determined by total synthesis.13 However, the absolute configuration of most bohemamine analogues have not been reported. The octant rule for cyclopentenone could be an effective method to determine the absolute configurations of this family of compounds (Figures 3 and S1).
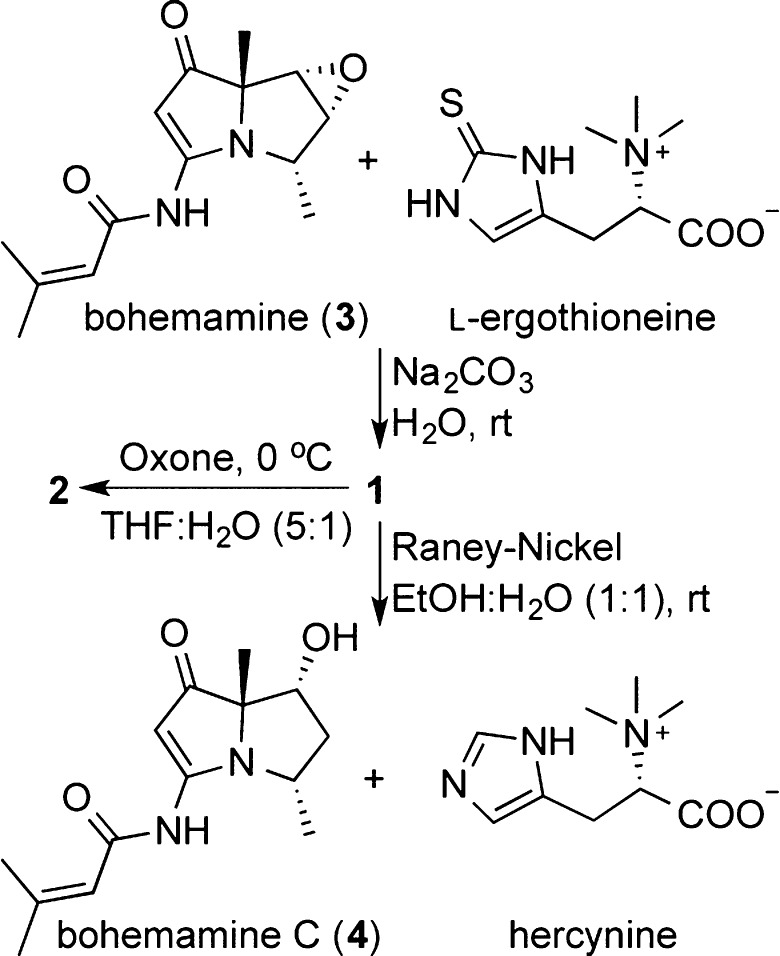
To further confirm the structure and the absolute configuration of the C-5″, we treated 1 with Raney–Nickel14 to yield bohemamine C and hercynine (Scheme 1). We were able to isolate sufficient quantities of bohemamine C (4) for analysis by MS, NMR, and CD (Figure S1). The specific rotation of isolated hercynine ([α]D23 +40.1) was determined to be consistent with literature reports for (S)-hercynine ([α]D22 +44.7).15 Thus, the absolute configuration of C-5″ was determined as S. In addition, compound 1 could be detected by LC–MS in the reaction mixture of bohemamine and l-ergothioneine under alkaline condition (Scheme 1 and Figure S2).
Scheme 1. Chemical Transformation of 1 and 2.
The molecular formula of spithioneine B (2) was found to be C23H33N5O6S based on the HRESIMS (Supporting Information), which was only one oxygen more than that of 1. The 1H and 13C NMR data of 2 (Tables 1 and S1) were similar to those of 1 except for some differences observed for the chemical shifts of C-4, C-5, C-6, and C-1″, all locations near the sulfide moiety. Thus, we deduced the sulfide in 1 was oxidized to give the sulfoxide in 2. This was confirmed by treatment of 1 with oxone in THF/H2O at 0 °C to yield 2 (Scheme 1) and analysis of resultant compound.16 The configuration of the chiral sulfoxide moiety was not determined.
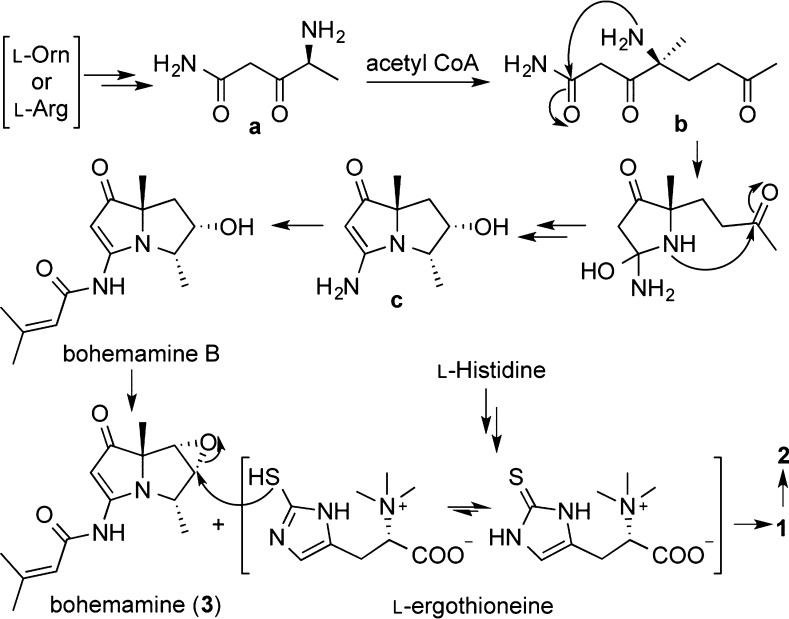
A plausible biosynthetic pathway for spithioneines A and B (1 and 2) is postulated (Scheme 2) to originate from amino acid and polyketide biosynthesis. l-Ornithine or l-arginine undergoes oxidation and reduction to yield intermediate a, which can form intermediate b by condensation with acetate.17 Formation of the bohemamine core would be generated from intermediate b, which undergoes dual nucleophilic addition of the amine and dehydration to form the pyrrolizidine ring system (intermediate c). Amidation of intermediate c would give rise to bohemamine B, which can be converted to the epoxide to yield bohemamine (3). The sulfur atom of ergothioneine, which is known to be derived from histidine,18 would carry out nucleophilic attack from the backside of the epoxide to give 1, and further oxidize to generate 2. Recently, the draft genome sequencing of the marine-derived Streptomyces sp. TP-A0873, a producer of bohemamine, was carried out. The genome contained at least 14 gene clusters for polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS). However, biosynthetic gene clusters for bohemamine were not identified.19
Scheme 2. Plausible Biosynthetic Pathway of Spithioneines A and B (1 and 2).
Compounds 1 and 2 showed no cytotoxicity to the nonsmall cell lung cancer cell lines HCC366, A549, HCC44, and HCC1171 or antibacterial activities against Pseudomonas aeruginosa and Bacillus subtilis.
In summary, we identified two new bohemamine-type pyrrolizidine alkaloids from a marine-derived Streptomyces spinoverrucosus. Their interest lies in the incorporation of the ergothioneine moiety into a polyketide. Ergothioneine itself has a variety of interesting biological activities. Past studies have implicated ergothioneine to be an inhibitor of oxidative stress,20 promoter of neuronal differentiation,21 and metal ion chelator.22 Ergothioneine accumulates in higher organisms up to millimolar levels via active transport with organic cation transporter (OCTN1).23 The physiological role of ergothioneine has yet to be established.
Acknowledgments
We acknowledge the following grants for funding this project: Welch Foundation I-1689 and NIH R01CA1499833. J.B.M. is a Chilton/Bell Foundation Endowed Scholar.
Supporting Information Available
Experimental details, CD spectra of some bohemamine analogues, the NMR data in CD3OD, and the HRESIMS and NMR spectra of 1 and 2. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.5b01328.
The authors declare no competing financial interest.
Supplementary Material
References
- a Bérdy J. J. Antibiot. 2005, 58, 1–26. [DOI] [PubMed] [Google Scholar]; b Challis G. L. Microbiology 2008, 154, 1555–1569. [DOI] [PubMed] [Google Scholar]; c Zhang W.; Tang Y. J. Med. Chem. 2008, 51, 2629–2633. [DOI] [PubMed] [Google Scholar]; d Demain A. L.; Sanchez S. J. Antibiot. 2009, 62, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bull A. T.; Stach J. E. Trends Microbiol. 2007, 15, 491–499. [DOI] [PubMed] [Google Scholar]; b Molinski T. F.; Dalisay D. S.; Lievens S. L.; Saludes J. P. Nat. Rev. Drug Discovery 2009, 8, 69–85. [DOI] [PubMed] [Google Scholar]; c Subramani R.; Aalbersberg W. Microbiol. Res. 2012, 167, 571–580. [DOI] [PubMed] [Google Scholar]; d Manivasagan P.; Venkatesan J.; Sivakumar K.; Kim S. K. Microbiol. Res. 2014, 169, 262–278. [DOI] [PubMed] [Google Scholar]
- Bister B.; Bischoff D.; Strobele M.; Riedlinger J.; Reicke A.; Wolter F.; Bull A. T.; Zahner H.; Fiedler H. P.; Sussmuth R. D. Angew. Chem., Int. Ed. 2004, 43, 2574–2576. [DOI] [PubMed] [Google Scholar]
- He H.; Ding W.; Bernan V. S.; Richardson A. D.; Ireland C. M.; Greenstein M.; Ellestad G. A.; Carter G. T. J. Am. Chem. Soc. 2001, 123, 5362–5363. [DOI] [PubMed] [Google Scholar]
- Feling R. H.; Buchanan G. O.; Mincer T. J.; Kauffman C. A.; Jensen P. R.; Fenical W. Angew. Chem., Int. Ed. 2003, 42, 355–357. [DOI] [PubMed] [Google Scholar]
- Kwon H. C.; Kauffman C. A.; Jensen P. R.; Fenical W. J. Am. Chem. Soc. 2006, 128, 1622–1632. [DOI] [PubMed] [Google Scholar]
- a Hu Y.; Wang K.; MacMillan J. B. Org. Lett. 2013, 15, 390–393. [DOI] [PubMed] [Google Scholar]; b Hu Y.; Potts M. B.; Colosimo D.; Herrera-Herrera M. L.; Legako A. G.; Yousufuddin M.; White M. A.; MacMillan J. B. J. Am. Chem. Soc. 2013, 135, 13387–13392. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Fu P.; Johnson M.; Chen H.; Posner B. A.; MacMillan J. B. J. Nat. Prod. 2014, 77, 1245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Fu P.; Jamison M.; La S.; MacMillan J. B. Org. Lett. 2014, 16, 5656–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Fu P.; MacMillan J. B. J. Nat. Prod. 2015, 78, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Doyle T. W.; Nettleton D. E.; Balitz D. M.; Moseley J. E.; Grulich R. E. J. Org. Chem. 1980, 45, 1324–1326. [Google Scholar]; b Zhang Q.; Schrader K. K.; Elsohly H. N.; Takamatsu S. J. Antibiot. 2003, 56, 673–681. [DOI] [PubMed] [Google Scholar]; c Bugni T. S.; Woolery M.; Kauffman C. A.; Jensen P. R.; Fenical W. J. Nat. Prod. 2006, 69, 1626–1628. [DOI] [PubMed] [Google Scholar]
- a Konno K.; Shirahama H.; Matsumoto T. Tetrahedron Lett. 1981, 22, 1617–1618. [Google Scholar]; b Motohashi K.; Nagai A.; Takagi M.; Shin-ya K. J. Antibiot. 2011, 64, 281–283. [DOI] [PubMed] [Google Scholar]
- a Erdelmeier I.; Daunay S.; Lebel R.; Farescour L.; Yadan J. Green Chem. 2012, 14, 2256–2265. [DOI] [PubMed] [Google Scholar]; b Song H.; Leninger M.; Lee N.; Liu P. Org. Lett. 2013, 15, 4854–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Motohashi N.; Mori I.; Sugiura Y. Chem. Pharm. Bull. 1976, 24, 1737–1741. [DOI] [PubMed] [Google Scholar]; b Hu W.; Song H.; Her A. S.; Bak D. W.; Naowarojna N.; Elliott S. J.; Qin L.; Chen X.; Liu P. Org. Lett. 2014, 16, 5382–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Sundararaman P.; Barth G.; Djerassi C. J. Am. Chem. Soc. 1981, 103, 5004–5007. [Google Scholar]; b Lin S.; Shi T.; Chen K.; Zhang Z.; Shan L.; Shen Y.; Zhang W. Chem. Commun. 2011, 47, 10413–10415. [DOI] [PubMed] [Google Scholar]
- Duvall J. R.; Wu F.; Snider B. B. J. Org. Chem. 2006, 71, 8579–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S.; Wu S. C.; O’Neil S. V.; Ngu K.; Atwal K. S.. WO2001027107 A2, Apr 19, 2001.
- Reinhold V. N.; Ishikawa Y.; Melville D. B. J. Med. Chem. 1968, 11, 258–260. [DOI] [PubMed] [Google Scholar]
- Bühler S.; Goettert M.; Schollmeyer D.; Albrecht W.; Laufer S. A. J. Med. Chem. 2011, 54, 3283–3297. [DOI] [PubMed] [Google Scholar]
- Ober D.; Kaltenegger E. Phytochemistry 2009, 70, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Seebeck F. P. J. Am. Chem. Soc. 2010, 132, 6632–6633. [DOI] [PubMed] [Google Scholar]
- Komaki H.; Ichikawa N.; Hosoyama A.; Fujita N.; Igarashi Y. Genome Announce. 2015, 3, e00008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hartman P. E. Methods Enzymol. 1990, 186, 310–318. [DOI] [PubMed] [Google Scholar]; b Aruoma O. I.; Spencer J. P.; Mahmood N. Food Chem. Toxicol. 1999, 37, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Ishimoto T.; Nakamichi N.; Hosotani H.; Masuo Y.; Sugiuira T.; Kato Y. PLoS One 2014, 9, e89434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon D. P. J. Med. Chem. 1971, 14, 1084–1087. [DOI] [PubMed] [Google Scholar]
- a Brummel M. C. Med. Hypotheses 1989, 30, 39–48. [DOI] [PubMed] [Google Scholar]; b Grundemann D.; Harlfinger S.; Golz S.; Geerts A.; Lazar A.; Berkels R.; Jung N.; Rubbert A.; Schomig E. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 5256–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.