Abstract
This protocol describes an efficient method to site-specifically label cell-surface or purified proteins with chemical probes in two steps: PRobe Incorporation Mediated by Enzymes (PRIME) followed by chelation-assisted copper-catalyzed azide-alkyne cycloaddition (CuAAC). In the PRIME step, Escherichia coli lipoic acid ligase site-specifically attaches a picolyl azide derivative to a 13-amino acid recognition sequence that has been genetically fused onto the protein of interest. Proteins bearing picolyl azide are chemoselectively derivatized with an alkyne-probe conjugate by chelation-assisted CuAAC in the second step. We describe herein the optimized protocols to synthesize picolyl azide, perform PRIME labeling, and achieve CuAAC derivatization of picolyl azide on live cells, fixed cells, and purified proteins. Reagent preparations, including synthesis of picolyl azide probes and expression of lipoic acid ligase, take 12 d, while the procedure to perform site-specific picolyl azide ligation and CuAAC on cells or on purified proteins takes 40 min-3 h.
Introduction
PRobe Incorporation Mediated by Enzymes (PRIME) is a versatile tool for site-specific tagging of proteins with chemical probes that possess useful biophysical properties such as fluorescence or photocrosslinking ability1-3, enabling functional studies of a given protein in vitro or in cells. The central component of the PRIME method is an engineered mutant of E. coli lipoic acid ligase (LplA) that catalyzes the covalent tagging of the desired probe onto a specific lysine residue within a 13-amino acid recognition sequence called the LplA acceptor peptide (LAP), which is genetically fused to the protein of interest (POI). LplA's high specificity for the single lysine in LAP ensures that there is no labeling on other proteins that are present in the labeling environment (such as other cellular proteins), nor on other sites within the LAP fusion protein. Probe targeting can be accomplished in a single step if the probe is small enough to fit into the engineered small-molecule binding pocket of LplA. The fluorescent probes coumarin2, Pacific Blue4, aminocoumarin5, and resorufin (Liu et al, unpublished results), as well as the photocrosslinker aryl azide6 have been targeted in this way. Larger fluorophores and probes that cannot fit into the LplA active site – including most green and red organic fluorophores and quantum dots (QDs) - must be targeted in two steps: first by using LplA to ligate a functional group handle to LAP, and second by using bioorthogonal ligation chemistry1,3,7 or a protein-ligand binding interaction8 to target the probe to the functional group handle.
Since the labeling efficiency of two-step PRIME depends not only on LplA ligation kinetics but also on the kinetics of the derivatization chemistry, the best schemes utilize fast derivatization reactions such as the reverse-electron-demand Diels-Alder cycloaddition3, chelation-assisted copper-catalyzed azide-alkyne cycloaddition (chelation-assisted CuAAC1), and HaloTag-mediated labeling8. Among our two-step labeling schemes, PRIME with Halotag is most suited for QD targeting to cell-surface proteins because of HaloTag's superior labeling kinetics over all bioorthogonal chemistries (QDs can only be practically supplied for cell labeling at low nM concentrations), while PRIME with Diels-Alder is our best protocol for labeling of intracellular proteins with diverse organic probes. For site-specific labeling of cell-surface proteins or purified proteins with small molecules, which is the focus of this protocol, PRIME with chelation-assisted CuAAC is preferred over PRIME with HaloTag or Diels-Alder chemistry. This is because HaloTag adds undesired steric bulk (35 kDa) to the POI, which could disrupt its function, and trans-cyclooctene and tetrazine reagents for Diels-Alder are not as synthetically accessible as reagents for CuAAC.
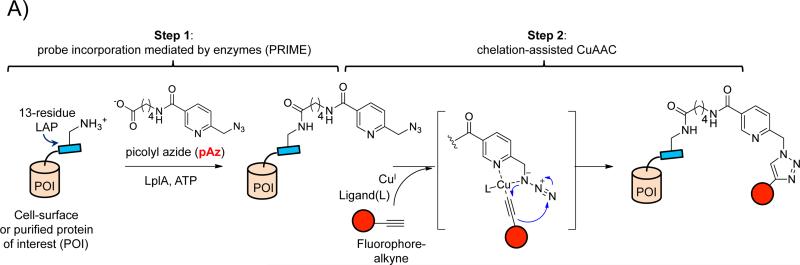
In two-step labeling with PRIME and chelation-assisted CuAAC, a copper-chelating picolyl azide (pAz) molecule is used as the LplA substrate (Figure 1A). pAz ligation onto LAP-tagged cell-surface proteins can be accomplished in two distinct ways (Figure 1B). The first option is to add purified LplA mutant (W37VLplA) to the cell culture media, along with the pAz probe and ATP1. This protocol tags the cell-surface pool of a LAP fusion protein selectively, not labeling intracellular subpopulations in the endoplasmic reticulum (ER) or Golgi. The second option is to co-express with the LAP fusion construct an LplA mutant targeted to the cell's endoplasmic reticulum. For this option, we utilize a quadruple mutant of LplA shown to have higher activity in the secretory pathway than the simple W37VLplA mutant. This construct is called W37A,T57I,F147L,H267RLplA, or AILRLplA9.
Figure 1.
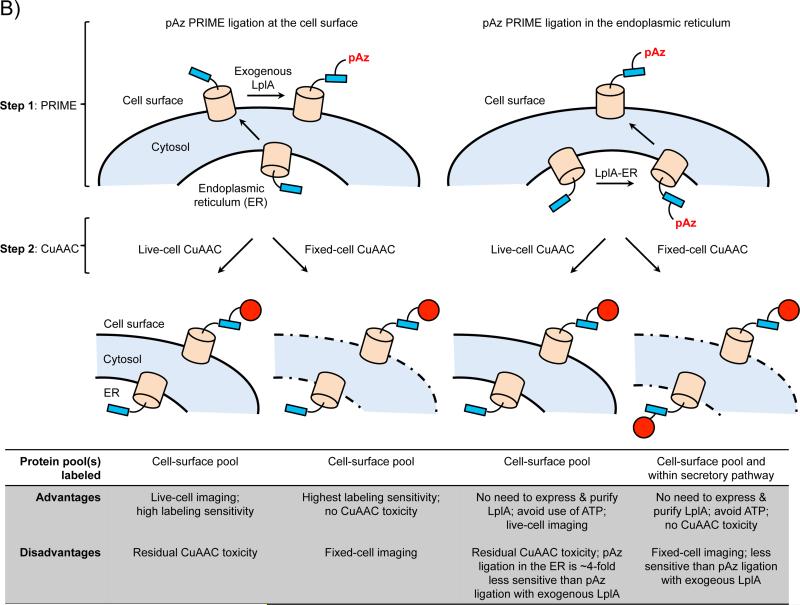
Site-specific protein labeling via PRIME and chelation-assisted CuAAC1. A) The two-step labeling scheme. In the first step, an engineered PRIME ligase (Trp37→Val mutant of lipoic acid ligase, or LplA) covalently attaches a copper-chelating picolyl azide derivative (pAz) onto LplA's acceptor peptide (LAP), which is genetically fused to a cell-surface or purified protein of interest (POI). In the second step, picolyl azide-modified proteins are chemoselectively derivatized with a terminal alkyne-probe conjugate (red circle) by chelation-assisted CuAAC. CuI is generated in-situ from 10-100 μM CuIISO4 and 2.5 mM sodium ascorbate. Ligand (L) represents CuI-stabilizing ligands, such as THPTA10, BTTAA27, or TBTA29. The LAP sequence is GFEIDKVWYDLDA24 (lysine labeling site underlined). B) Four different configurations for PRIME/CuAAC labeling. PRIME ligation of pAz can be performed at the cell surface (left), with application of exogenous LplA enzyme to the cell media. Alternatively, pAz ligation can be performed in the cell's secretory pathway (right), using ligase expressed in the endoplasmic reticulum (ER). Thereafter, CuAAC derivatization of picolyl azide-modified proteins can be performed on live cells, or after cell fixation. Key features of each labeling configuration are listed in the table below.
Once pAz is ligated to LAP, it is chemoselectively derivatized with alkyne-probe conjugates via chelation-assisted CuAAC. This variant of CuAAC is faster (due to increased local copper concentration induced by the picolyl moiety) and more cell-compatible (due to the lower concentration requirement for toxic copper) than conventional CuAAC using alkyl azides1.
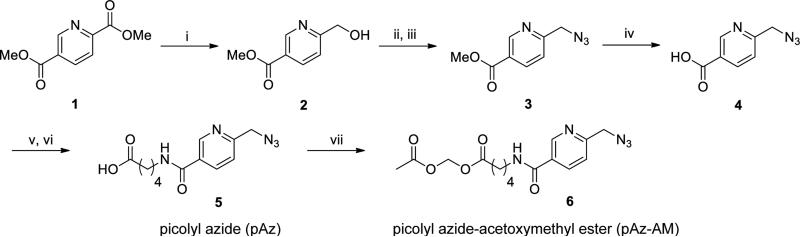
Because pAz is charged, it does not efficiently cross the plasma membrane of living cells. Consequently, for the PRIME labeling option using ER-expressed AILRLplA (Figure 1B), it is necessary to protect pAz as an acetoxymethyl (AM) ester (Figure 2) so that it can access LplA in the ER. Once inside the cell, the AM ester is cleaved by endogenous cellular esterases, releasing the parent pAz. Prior to unmasking, pAz-AM itself is not an LplA substrate since a free carboxylate is required for conjugation of the probe to LAP.
Figure 2.
Synthesis of picolyl azide (pAz) and pAz-acetoxymethyl ester (pAz-AM) reagents for PRIME labeling. The synthesis begins with commercially-available dimethyl 2,5-pyridine dicarboxylate and proceeds through six steps to give pAz. pAz-AM is made from pAz in one additional step. i) NaBH4, CaCl2, THF/MeOH; ii) TsCl (p-toluenesulfonyl chloride), TEA (triethylamine), CH2Cl2; iii) NaN3, THF; iv) LiOH, MeOH/H2O; v) disuccinimidyl carbonate, TEA, DMF; vi) 5-aminovaleric acid, TEA, DMF; vii) bromomethyl acetate, TEA, DMF.
Both PRIME and CuAAC labeling steps are efficient. PRIME ligation at the cell surface with pAz goes to ~80% completion after 20 minutes using purified W37VLplA1. For chelation-assisted CuAAC performed in vitro, we achieved complete conversion to product in less than 5 min in the presence of 10-40 μM copper and a CuI-stabilizing ligand THPTA10, in contrast to < 40% conversion with conventional CuAAC under the same conditions. In cells, chelation-assisted CuAAC increased labeling yields by 2.7- to 25-fold compared to conventional CuAAC1. We estimate the overall two-step labeling yields for both cell surface proteins and purified proteins to be >70% using 30-60 minute labeling protocols (vide infra). Since much less cytotoxic copper (10-100 μM, compared to low mM amounts in traditional CuAAC) can be used to achieve similar or better reaction rates, chelation-assisted CuAAC is inherently less toxic than conventional CuAAC1.
Applications of the method
Two-step PRIME/CuAAC labeling has been applied to fluorophore tagging of a variety of cell-surface and purified proteins, including neurexin, neuroligin, and kinesin. The labeling is highly specific in all eukaryotic cell types we have tested, including yeast, human cell lines, and rat neurons. Versatile chemical probe targeting to cell surface proteins can be useful for diverse applications, such as single-molecule tracking of receptor motion,11 super-resolution imaging of cell-surface adhesion proteins and receptors12,13, and imaging assays that probe the fate and properties of the surface subpopulation of proteins14 (as opposed to the total or intracellular protein pool).
PRIME/CuAAC can also be adapted not just for imaging specific cellular proteins, but to monitoring protein-protein interactions. In a separate study, we have used interaction-dependent PRIME/CuAAC tagging to visualize trans-cellular neurexin-neuroligin interactions in cultured HEK cells and rat neurons9.
Apart from its pairing with PRIME, chelation-assisted CuAAC is by itself useful for a variety of detection assays. For example, we have shown that it can be used to detect proteins and RNA metabolically labeled in live cells with azide analogs1.
Comparison with other protein labeling methods
Two-step PRIME/CuAAC labeling offers several advantages over existing site-specific protein labeling methods. First, the total label size is small (13 amino acids for LAP plus the fluorophore of interest joined via a stable triazole linker as in Figure 1A), ensuring minimal interference with the protein of interest, unlike the much larger GFP15, SNAP16, and HaloTag17 labels (which range between 20 kDa and 35 kDa). Second, the label is monomeric and will not induce artificial crosslinking like wild-type streptavidin18 and antibodies19. Third, both labeling steps are kinetically efficient, and therefore can be performed quickly and with low concentrations of precious reagents without sacrificing yield. This contrasts with strain-promoted azide-alkyne cycloaddition-based labeling7 and the aliphatic aldehyde tag20,21, for example, whose much slower second order rate constants result in lower labeling yields even when much higher probe concentrations are used. Fourth, labeling is highly specific, not only because of the inherent specificity of LplA-mediated ligation and CuAAC, but also because the reaction partners (LAP, picolyl azide, alkyne) are biologically inert until reaction catalysts (LplA, then copper) are supplied. This contrasts with components in many protein labeling methods (e.g., FlAsH22) and bio-orthogonal ligation chemistries3,23 that have side reactions with endogenous biomolecules. Lastly, even though PRIME/CuAAC labeling requires tailored small-molecule reagents, the reagents are straightforward to synthesize or commercially available, and are stable to long-term storage.
Limitations
At 13 amino acids, the LAP tag can still interfere with the function or trafficking of proteins to which it is fused. If possible, one should perform control experiments to ensure proper function of the tagged recombinant protein. For example, we confirmed that LAP-tagged postsynaptic adhesion protein neuroligin-1 traffics correctly to the postsynapse, via co-localization analysis with a postsynaptic protein marker fused to a fluorescent protein1,9 and that its binding to its adhesion partner neurexin is not impaired by an introduction of a labeling tag.
One should also ensure that overexpression of the tagged protein does not alter its function. If needed, expression levels of the protein can be tuned via the use of tunable promoters, promoters of different strengths, or by generating stable cell lines.
Even at 10-40 μM copper concentration, CuAAC can still be somewhat toxic to cells and damaging to proteins. We have included measures to ensure maximal cell viability when performing CuAAC, including use of the best CuI-stabilizing ligands available, immediate sequestration of copper ions after labeling (via use of a cell-compatible copper chelator bathocuproin sulfonate), and addition of a radical scavenger TEMPOL to quench reactive oxygen species. Using the optimized protocol presented here, we show that chelation-assisted CuAAC is as non-toxic as copper-free strain-promoted azide-alkyne cycloaddition, and can be performed on delicate cultured rat neurons1. As CuAAC toxicity directly correlates with the amount of copper used and the labeling duration, one should finely tune these two parameters to obtain a balance between maximal signal and minimal disruption of cellular and protein function.
In its current form, the two-step PRIME/CuAAC is not applicable to labeling of intracellular proteins. While pAz ligation can occur effectively inside the cytosol, it is currently not possible to perform CuAAC inside living cells for many reasons. First, CuI/II would need to be delivered across the cell membrane. Second, once inside, glutathione in the cytosol would likely reduce any CuII species to the CuAAC-competent CuI species, but a major problem would be sequestration of CuI by the same thiol. Third, cell-protective CuI-ligands such as THPTA and BTTAA would also have to be delivered across the cell membrane.
Designing LAP-tagged cell-surface proteins
LAP sequence
the most kinetically efficient LplA acceptor peptide LAP2 (GFEIDKVWYDLDA)24 generally works best and should be tested first as a fusion tag to the protein of choice. An alternative LAP sequence, called LAP4.2 (GFEIDKVWHDFPA)24 has also performed well in our hands, especially when fused to cell-surface proteins. In the case that expression, surface trafficking, or labeling of a LAP2-tagged protein is problematic, one should test LAP4.2 as a possible remedy, along with other strategies such as changing the LAP fusion site or inserting a linker to increase flexibility around the LAP sequence. To increase labeling signal, tandem LAPs (such as 2x or 3xLAP) can also be used, though one caveat might be a decrease in surface trafficking of the fusion protein due to the larger, more disordered tag.
LAP fusion site
LplA can catalyze probe ligation onto LAP at the N terminus, C terminus, or in an internal loop of a protein. For tagging of cell-surface proteins as described in this protocol, the LAP sequence must be on the extracellular/luminal face of the protein. If needed, include a few Gly or Ser on either side of LAP sequence to separate LAP from the folded protein domain, and to make LAP more sterically accessible to LplA.
Probe synthesis
A synthetic route to access the picolyl azide substrate for LplA (structure 5) is shown in Figure 2. To access other picolyl azide derivatives such as picolyl azide-fluorophore conjugates, one can use the succinimidyl ester of 6-azidomethylnicotinic acid 4 to react with an amine of choice. Many alkyne-fluorophore conjugates can be purchased (for example, from Life Technologies or Sigma-Aldrich), or synthesized in a single step via amide coupling between a commercially available fluorophore-succinimidyl ester and an amino-alkyne such as propargylamine1.
Design of labeling experiments
In initial protein labeling tests (whether at the cell surface or in vitro), it is best to include negative controls (omit LplA, omit picolyl azide, omit copper, or introduce a Lys→Ala point mutation in LAP; see Figure 3A) to ensure that the labeling signal is specific and results from the activity of LplA and the chemistry of CuAAC. When designing the protein construct, it is very helpful to insert a short epitope tag (HA, FLAG, c-Myc, or V5 for example) in addition to the LAP tag, so that one can verify expression of the fusion protein by immunofluorescence staining or western blotting, independent of PRIME/CuAAC labeling.
Figure 3.
Cell-surface protein labeling via PRIME and chelation-assisted CuAAC. A) Demonstration of labeling specificity. Rat hippocampal neurons (at 14 days in vitro) expressing LAP-neuroligin-1 (LAP on the extracellular N-terminus) and Homer1b-GFP were labeled live with 10 μM W37VLplA, 200 μM picolyl azide, 1 mM ATP, and 5 mM Mg(OAc)2 for 20 min at 37 °C. After two rounds of washing, cells were fixed using formaldehyde, and blocked with 0.5% casein. CuAAC was performed using 1 mM CuSO4, 100 μM TBTA, 2.5 mM sodium ascorbate, and 5 μM AF647-alkyne for 1 hour at room temperature. Negative controls are shown with picolyl azide omitted during the PRIME step, CuSO4 omitted during CuAAC, or AP-tagged neuroligin-1 instead of LAP-neuroligin-1 (AP = acceptor peptide for biotin ligase30 rather than LplA). B) Comparison of PRIME ligation of pAz at the cell surface versus within the ER. HEK cells expressing LAP-neurexin-1β (LAP on the extracellular N-terminus), AILRLplA-ER (= W37A,T57I,F147L,H267RLplA with a KDEL sequence for ER-retention) and histone 2B-YFP (a transfection marker) were labeled with exogenous W37VLplA as in A) (left), or with 100 μM picolyl azide-acetoxymethyl ester (pAz-AM) for 1 hour at 37 °C (right). Thereafter, CuAAC was performed on the live HEK cells using 50 μM CuSO4, 300 μM BTTAA, 2.5 mM sodium ascorbate, and 20 μM Alexa Fluor 647-alkyne for 5 min at room temperature. Quantitation shows that the AF647 signal is ~4.6-fold higher in the case of exogenous labeling. Scale bars, 10 μm.
To selectively label the cell-surface pool of a protein with PRIME/CuAAC, at least one of the labeling steps must be performed on live cells with membrane-impermeant reagents, so that the intracellular protein pool will not be labeled. If PRIME ligation with pAz is performed on live cells with exogenously supplied W37VLplA, the CuAAC derivatization step is flexible: it can be performed on living cells to enable real-time tracking of surface proteins; or it can be performed after cells are fixed and permeabilized. For the purpose of detecting protein localization, the fixed-cell CuAAC protocol should be considered, as it provides more sensitive detection than live-cell CuAAC (because the CuAAC conditions can be more forcing), and any residual toxicity from CuAAC becomes irrelevant. If PRIME ligation of pAz is performed with AILRLplA-ER, intracellular pools of the protein will also be tagged with pAz in addition to the surface pool. It is thus necessary to use live-cell CuAAC (copper ions are not membrane-permeant) following ER-pAz ligation if one wants to detect only the surface pool of the protein.
For labeling of the surface pool of a protein, we generally recommend using purified W37VLplA over AILRLplA-ER as the former is ~4-5-fold more sensitive than the latter (Figure 3B). AILRLplA-ER is technically simpler and might be advantageous in certain biological applications—such as labeling and imaging in tissue slices—where protein delivery may be a concern. Performing pAz ligation in the ER also eliminates the need to add ATP during labeling, which is a preferred practice for neuron cultures as ATP can activate purinergic receptors in neurons and cause excitotoxicity25.
When performing live-cell CuAAC on hardy cell lines (for example, HEK 293T, HeLa, and COS-7 cells), one can omit the cell-protective reagents TEMPOL and bathocuproin sulfonate without affecting labeling efficiency. One should not omit these reagents when working with more delicate cells such as neurons.
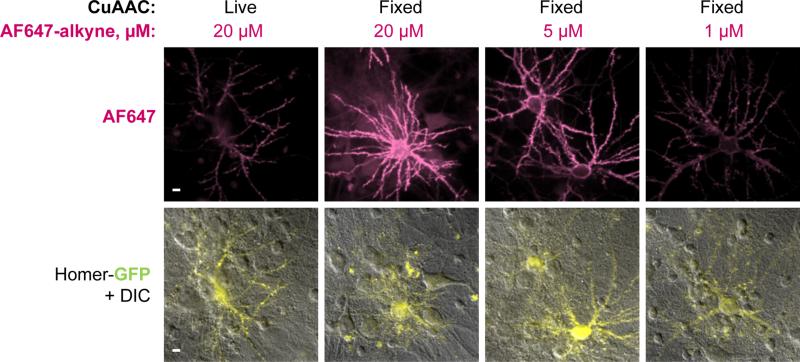
The choice of fluorophore-alkyne and its labeling concentration matters for both live-cell and fixed-cell CuAAC. For live-cell CuAAC, choose fluorophores that are not membrane-permeable (such as Alexa Fluor dyes, almost all of which are polysulfonated) to avoid intracellular uptake. For fixed-cell CuAAC, it is also best to choose polysulfonated, hydrophilic fluorophores in order to minimize non-specific sticking to fixed cells. Higher fluorophore-alkyne concentration results in faster CuAAC labeling, but also greater non-specific sticking. Ideally, one should perform a titration of dye concentrations to identify conditions that give maximal signal-to-noise ratio. An example of cell-surface protein labeling with both the live-cell and fixed-cell CuAAC protocol, at different fluorophore-alkyne concentrations, is shown in Figure 4. In this example, 5 μM Alexa Fluor 647-alkyne (AF647-alkyne) for fixed-cell CuAAC provides the highest signal-to-noise ratio.
Figure 4.
Comparison of live-cell and fixed-cell CuAAC labeling protocols. Rat hippocampal neurons were transfected with LAP-neuroligin-1 and Homer1b-GFP, and labeled live with picolyl azide as in Figure 3A. Then, cells were either labeled live by CuAAC using 50 μM CuSO4, 250 μM BTTAA, 2.5 mM sodium ascorbate, 100 μM TEMPOL, and 20 μM AF647-alkyne for 5 min, or fixed. For fixed cells, CuAAC was performed as described in Figure 3A, except that the AF647-alkyne concentration was varied (1, 5, or 20 μM). All AF647 images are shown with the same intensity thresholds. For fixed-cell CuAAC, higher labeling signals were obtained, as well as higher background due to AF647-alkyne non-specific sticking to the fixed cells. Scale bars, 10 μm.
For labeling of purified proteins (Figure 5), we describe the expression and purification of two LplA variants: hexahistidine-tagged W37VLplA (His6-W37VLplA) and tagless W37VLplA. The choice of which enzyme to use depends on how one wishes to purify proteins after PRIME labeling. For example, it is helpful to use tagless LplA to label a His6-tagged protein, so that the latter can be easily separated from LplA following labeling. On the other hand, His6-tagged LplA can be used to label a protein with an orthogonal purification tag (such as FLAG or GST).
Figure 5.
In vitro protein labeling with PRIME and chelation-assisted CuAAC. LAP-tagged kinesin K56031 (at 10 μM) was labeled with 5 μM W37VLplA, 100 μM picolyl azide, 500 μM ATP, and 2.5 mM Mg(OAc)2 for 1 hour at room temperature. Excess small-molecule reagents were then removed by centrifugation through a dialysis membrane. CuAAC labeling was performed with 200 μM CuSO4, 1 mM BTTAA ligand, 2.5 mM sodium ascorbate, and 20 μM AF647-alkyne for 30 min at room temperature. The reaction was quenched with EDTA (final concentration 20 mM), then analyzed on 12% SDS-PAGE. Negative controls are shown with picolyl azide omitted during the PRIME step (lane 2), CuSO4 omitted during CuAAC (lane 3), or kinesin K420 (no LAP) replacing LAP-kinesin K560 (lane 4). The left gel shows total protein (silver stain), and the right gel shows AF647 fluorescence. Kinesin K560 and kinesin K420 are the 560-amino acid and 420-amino acid N-terminal fragments of human kinesin respectively.
Materials
Reagents for chemical synthesis
Dimethyl 2,5-pyridine dicarboxylate (Alfa Aesar, cat. No. A17250)
Sodium borohydride (Sigma-Aldrich, cat. No. 241-004-4)
Calcium chloride, anhydrous (Alfa Aesar, cat. No. 12316)
Sodium azide (Alfa Aesar, cat. No. 14314)
- Caution: sodium azide is toxic and potentially explosive.
p-Toluenesulfonyl chloride (Sigma-Aldrich, cat. No. 89730)
Triethylamine (Sigma-Aldrich, cat. No. T0886)
Lithium hydroxide monohydrate (Sigma-Aldrich, cat. No. 43171)
N,N′-Disuccinimidyl carbonate (Sigma-Aldrich, cat. No. 43720) 5-Aminovaleric acid (Alfa Aesar, cat. No. B24788)
Bromomethyl acetate (Sigma-Aldrich, cat. No. 303208)
Sodium sulfate (Sigma-Aldrich, cat. No. 239313)
Silica gel 150 Å (200-425 mesh) (Sigma-Aldrich, cat. No. 236810)
Methanol, puriss, absolute, over molecular sieve (Sigma-Aldrich, cat. No. 66542)
Hexanes, ACS reagent grade (Sigma-Aldrich, cat. No. 178918)
Ethyl acetate, ACS reagent grade (Sigma-Aldrich, cat. No. 319902)
Dichloromethane, puriss, absolute, over molecular sieve (Sigma-Aldrich, cat. No. 66749)
Chloroform, ACS reagent grade (Sigma-Aldrich, cat. No. 437581)
N,N-Dimethylformamide, puriss, absolute, over molecular sieve (Sigma-Aldrich, cat. No. 40228)
Tetrahydrofuran, ACS reagent grade (Sigma-Aldrich, cat. No.676764)
Reagents for LplA expression in E. coli
pYFJ16-His6-W37VLplA plasmid, available from Addgene (ID 34838)
pYFJ16-His6-ENLYFQG-W37VLplA plasmid, available on request from Ting lab. ENLYFQG is a 7-amino-acid recognition sequence for the Tobacco Etch Virus (TEV) protease26.
E. coli BL21 (DE3) pLysS chemically competent cells (Novagen)
Luria-Broth (LB) medium and LB-Ampicillin plates (ampicillin 100 μg ml−1)
Ampicillin, sodium salt (Amresco). Dissolve at 100 mg ml−1 in water and filter sterilize.
Isopropyl-β-D-thiogalactopyranoside (Calbiochem), 100 mg ml−1 in water
- Critical: store at −20 °C at 1000x stock
B-PER for bacterial lysis (Pierce)
Phenylmethylsulfonylfluoride (Amresco). Make 100 mM solution in isopropanol (100x stock).
- Caution: harmful. Store at −20 °C
Protease inhibitor cocktail (Sigma-Aldrich)
- Critical: the protease inhibitor must be EDTA-free, as EDTA can remove nickel from the nickel-affinity column. Store at −20 °C
Nickel-nitrilotriacetic (NTA) acid agarose resin (Qiagen)
Nickel-NTA binding buffer (recipe given in the reagent setup section)
Nickel-NTA wash buffer (recipe given in the reagent setup section)
Nicket-NTA elution buffer (recipe given in the reagent setup section)
1 M imidazole
Dithiothreitol (DTT)
LplA dialysis buffer (recipe given in the reagent setup section)
AcTEV protease (Life Technologies, cat. No. 12575)
20X TEV protease cleavage buffer (Life Technologies)
Standard reagents for protein gel electrophoresis
Reagents for mammalian cell labeling experiments
Human Embryonic Kidney 293T (HEK) cells (ATCC) or mammalian cell type of interest. The protocol has been shown to work in HEK cells and in dissociated rat neuron cultures.
Human fibronectin (Millipore), for promotion of adherence of HEK cells to glass coverslips
pDisplay-LAP2-CFP-TM plasmid, available from Addgene (ID 34842)
pcDNA4-AILRLplA-ER, available on request from the Ting Lab
Standard media for mammalian cell culture
Standard reagents for DNA cloning
Lipofectamine 2000 (Life Technologies, cat. No. 11668) or other transfection reagent of choice
W37VLplA (expression and purification protocols given in Box 2)
Picolyl azide (pAz, no commercial vendor, synthetic procedure given in Box 1)
Picolyl azide-acetoxymethyl ester (pAz-AM, no commercial vendor, synthetic procedure given in Box 1)
Adenosine 5’-triphosphate disodium salt hydrate (ATP, Sigma)
Magnesium acetate heptahydrate (Mg(OAc)2, Alfa Aesar, cat. No. 11596)
Dulbecco's phosphate-buffered saline (DPBS), no calcium, no magnesium (Life Technologies)
Bovine serum albumin (BSA, Amresco, cat. No. 0332)
BSA blocking buffer (recipe given in the reagent setup section)
Casein, vitamin free (MP biomedicals, cat. No. 904520)
Casein blocking buffer (recipe given in the reagent setup section)
Tyrode's buffer (for use with cultured neurons; recipe given in the reagent setup section)
Copper(II) sulfate pentahydrate (CuSO4, Mallinckrodt chemicals, cat. No. 4752-10)
Sodium ascorbate (Spectrum, cat. No. S1349)
Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, Sigma, cat. No. 678937).
Tris-(hydroxypropyltriazolylmethyl)amine (THPTA, no commercial vendor, synthesize according to Hong et al10).
Bis[(tertbutyltriazoyl)methyl]-[(2-carboxymethyltriazoyl)methyl]-amine (BTTAA, no commercial vendor, synthesize according to Besanceney-Webler et al27.).
TEMPOL (Calbiochem, cat. No. 581500). Store at −20 °C.
Alexa Fluor (AF) 647-alkyne, triethylammonium salt (Life Technologies, cat. No. A10278).
Alkyne conjugates to AF488, AF594, and AF555 are also commercially available from Life Technologies.
Bathocuproin sulfonate (Alfa Aesar, cat. No. B22550-MD)
10% formaldehyde solution in water, methanol-free (Polysciences, cat. No. 04018)
- Caution: formaldehyde is carcinogenic, toxic, irritant, and lachrymatory. Handle in a well-ventilated fume hood while wearing protective gear. Wash hands after use.
Equipment for chemical syntheses
Weighing balance
Spatulas
Weighing paper
Syringes and needles
Fume hood
Magnetic stir plate
N2 gas and an silicone oil-filled bubbler
Round-bottomed flasks
Teflon-coated magnetic stir bars
Graduated cylinders
Erlenmeyer flasks
Separatory funnels
Pasteur Pipettes
SiO2 TLC plate and spotter
TLC developing chamber
Tweezers
UV handheld lamp
Heat gun
Columns for flash chromatography
Rotary evaporator
Vacuum pump
Electric oven capable of heating up to 130 °C
Access to 1H- and 13C-NMR spectrometers
Equipment for LplA expression in E. coli
Standard equipment for bacterial cell culture
Standard equipment for protein gel electrophoresis
Spectrophotometer to measure Abs600 to monitor bacterial growth
Floor centrifuge, capable of spinning down 1-liter of bacterial culture at 8,000 g
Centrifuge tubes for spinning liter cultures
Motorized pipettor (Drummond)
Poly-prep protein chromatography column (Bio-Rad)
Snakeskin dialysis tubing, 10-kDa cutoff (Thermo Scientific, cat. No. 68100)
Stir plate and large stir bar
Access to a temperature-controlled (4 °C) room or cabinet
Equipment for mammalian cell labeling experiments
Standard equipment for mammalian cell culture
0.2 μm syringe and bottletop filters for sterile filtration of reagents
Sterile 48-well cell culture plates
22 × 22 mm2 no. 1 or no. 1.5 thickness glass coverslips
Tweezer
Fluorescence microscope, with filter sets appropriate to the relevant fluorophore
REAGENT SETUP
W37VLplA. W37VLplA is expressed from pYFJ16- His6-W37VLplA or pYFJ16-His6-ENLYFQG-W37VLplA in E. coli as described in Box 2. Store LplA protein aliquots at −80 °C for > 6 months without loss of activity.
Nickel-NTA binding buffer. 50 mM Tris base, 300 mM NaCl, adjusted to pH 7.8 with 1 M HCl. Store at 4 °C.
1 M imidazole in nickel-NTA binding buffer, adjusted with 5 M HCl to pH 7.8. Dilute the imidazole stock solution in nickel-NTA binding buffer to make:
- Nickel-NTA wash buffer. Imidazole concentration 10 mM
- Nickel-NTA elution buffer. Imidazole concentration 100 mM
LplA dialysis buffer. 20 mM Tris base, 1 mM DTT, and 10% v/v glycerol, adjusted to pH 7.5 with 1 M HCl.
- Critical: add DTT fresh before dialysis
20X TEV protease cleavage buffer (minus DTT). 1 M Tris, 10 mM EDTA, adjust to pH 8.0 with 1 M HCl.
TEV protease cleavage buffer. Dilute 20X TEV buffer and add DTT to make a solution of 50 mM Tris pH 8.0, 0.5 mM EDTA, and 1 mM DTT.
- Critical: add DTT fresh before dialysis
Reagent setup for labeling experiments
pAz. Make 20 mM stock solution in water. Store at −80 °C for > 1 year without evidence of decomposition.
pAz-AM. Make 100 mM stock solution in dimethyl sulfoxide (DMSO). Store at −80 °C for > 6 months without evidence of decomposition.
ATP. Make single-use 100 mM aliquots (100x stock), adjusted to pH 7.5 with 1 M NaOH, store at −80 °C.
- Critical: avoid freeze-thawing. Adjust pH carefully to ensure pH does not overshoot over 8, where ATP will be rapidly hydrolyzed
Mg(OAc)2. Make 500 mM stock solution in water. Store at 4 °C.
BSA blocking buffer. Make 3% w/v BSA solution in DPBS and filter-sterilize. Store at 4 °C.
Casein blocking buffer. Make approximately 0.5% w/v casein solution in DPBS. As casein is not very soluble in DPBS, we first microwave DPBS, add casein, and stir at 40-50 °C for 30 min. Insoluble casein is then removed via centrifugation (at 10,000 g for 10 min). The supernatant is collected via decanting without disturbing the pellet, and filter-sterilized. Store at −80 °C.
Tyrode's buffer. 145 mM NaCl, 1.25 mM CaCl2, 3 mM KCl, 1.25 mM MgCl2, 0.5 mM NaH2PO4·H2O, 10 mM glucose, 10 mM HEPES, adjusted to pH 7.4 with 1 M HCl and filter-sterilized, store at 4 °C.
CuSO4. Make 20 mM stock solution in water. Store at 4 °C.
Sodium ascorbate. Make 100 mM stock solution in water.
- Critical: make stock solution fresh right before use. Discard unused portions.
TBTA. Make 1.7 mM stock solution in 20% v/v DMSO in tert-butanol. Store at −20 °C for > 6 months without evidence of decomposition.
THPTA. Make 50 mM stock solution in water. Store at −80 °C for > 1 year without evidence of decomposition.
BTTAA. No commercial supplier. Make 50 mM stock solution in water. Store at −80 °C for > 1 year without evidence of decomposition.
TEMPOL. Make 100 mM stock solution in water.
- Critical: make stock solution fresh right before use. Discard unused portions.
AF647-alkyne. Make 20-100 mM stock solution in DMSO. In general, aim to make a DMSO probe stock that is at least 1000x more concentrated than the final probe concentration used for labeling. Store at −80 °C for > 1 year without evidence of decomposition.
Bathocuproin sulfonate. Make 500 mM stock solution in water. Store at −20 °C for > 6 months without evidence of decomposition.
Formaldehyde fixative solution. Make 4% v/v formaldehyde solution in DPBS. Store at 4 °C.
Formaldehyde fixative solution for cultured neurons. Assemble the fixative solution as described below, adjust the final volume with water and the final pH to 7.4. Freeze-store in aliquots at −20 °C. Right before use, thaw appropriate portions at 37 °C. Discard unused portions.
| Component | Stock concentration | Final concentration |
|---|---|---|
| Formaldehyde | 10% | 4% |
| PIPES pH 7.0 | 500 mM | 60 mM |
| HEPES pH 7.0 | 500 mM | 25 mM |
| EGTA pH 8.0 | 500 mM | 10 mM |
| MgCl2 | 1 M | 2 mM |
| Sucrose | 1.2 M | 120 mM |
- Caution: formaldehyde is carcinogenic, toxic, irritant, and lachrymatory. Handle in a well-ventilated fume hood while wearing protective gear. Wash hands after use.
Equipment setup
Glassware and stir bars used for tosylation, succinimidyl ester formation, acetoxymethyl ester formation, and amide coupling reactions should be dried before use in an oven or by flame-drying.
- Critical: these reactions are conducted using anhydrous solvents under inert atmosphere. The presence of moisture in the reaction vessels due to improper drying can cause unwanted hydrolysis of the reagents or desired products.
Glass coverslips for cell plating. Cut 22 × 22 mm2 no.1 or no. 1.5 coverslips by hand into 7 × 7 mm2 using a diamond glass-cutter. The cut coverslips are sterilized by irradiation under a UV lamp in the cell culture hood for at least 1 hour. Mammalian cells are then grown on these 7 × 7 mm2 coverslips placed in a 48-well plate.
- Critical: we often perform cell labeling experiments in a 48-well plate format to conserve precious labeling reagents. In each 48-well plate well, only ~150 μL of the labeling solution is needed. For imaging, we transfer the coverslip to an imaging dish with a no. 1 glass bottom.
Procedure
1. Picolyl azide ligation onto cell-surface proteins with LplA
Picolyl azide ligation can be performed exogenously with purified W37VLplA (Module A), or endogenously with AILRLplA-ER (Module B). Exogenous pAz ligation with W37VLplA is our recommended protocol for most cell-surface protein labeling experiments since it is more sensitive than pAz ligation in the ER. Users should only consider using AILRLplA-ER if it is known that: 1) the 39 kDa LplA might have problems accessing the labeling site on the protein of interest at the cell surface, whereas such hindrance is not present if the labeling is performed within the ER; or 2) reagents required for exogenous labeling such as ATP are toxic to the cell type of interest. Relating to the toxicity concern, note that pAz-AM needed for ER labeling is not water-soluble and has to be supplied as DMSO stock, which increases the toxicity of the ER labeling procedure. pAz needed for exogenous LplA labeling, on the other hand, is water-soluble.
This step is not required for labeling of purified proteins; users wanting to do this should go immediately to Step 4.
- pAz ligation of cell-surface proteins using exogenous W37VLplA
- Plate cells onto 7 × 7 mm2 coverslips in a 48-well plate. For HEK cells, we coat coverslips with human fibronectin (50 μg/mL) according to the manufacturer's instructions, and generally plate ~8,000 cells in 250 μL media per well. This cell density should result in cells that are 80% confluent ~24 hours after plating, which is optimal for lipofectamine transfection.
- Transfect cells with an expression plasmid for the LAP-tagged cell-surface protein. For transient transfection using Lipofectamine 2000, we typically use 200-600 ng of DNA, and maximal expression is often reached at ~24 hours after transfection. We recommend co-expressing a fluorescent protein marker along with the LAP-tagged protein, for easy identification of transfected cells during imaging.
- To perform pAz ligation, add 10 μM W37VLplA, 200 μM pAz, 1 mM ATP, and 5 mM Mg(OAc)2 in regular cell growth media for 15-60 min at room temperature or 37 °C. If other labeling medium such as PBS or calcium-free DPBS is preferred, add 3% w/v bovine serum albumin (BSA) to the solution to prevent non-specific sticking of labeling reagents to the cell surface.
- - Critical: do not use any buffer that contains high Ca2+ (such as Ca2+-containing DPBS or Tyrode's buffer) during labeling, as Ca2+ inhibits LplA. If serum-containing media is used as a labeling buffer, check that lipoic acid content in the serum is not high (not more than 5 μM). Lipoic acid is a natural, more efficient substrate for LplA than pAz and will reduce the labeling yield of pAz.
- Wash cells at least twice with cell growth media to remove labeling agents before proceeding to the CuAAC derivatization step.
- - Critical: for labeling on cultured neurons, ensure that neurons are not exposed to air for longer than 2-3 seconds during reagent addition or wash steps. You can use two pipettes to simultaneously add and withdraw solution from the wells.
- pAz ligation of cell-surface proteins using AILRLplA-ER
- After cell plating, co-transfect cells with expression plasmids for the LAP-tagged cell-surface protein and AILRLplA-ER (pcDNA4-AILRLplA-ER). For transfection using lipofectamine 2000, we typically use 200-600 ng of each plasmid DNA. Use a fluorescent co-transfection marker if desired. Wait 18-24 hours before performing labeling.
- To perform pAz ligation in the ER, add 25-100 μM pAz-AM in serum-free DMEM for 1-4 hour at 37 °C.
- - Critical: do not use serum-containing media, as trace amount of esterases present in the serum can hydrolyze pAz-AM and prevent its cell entry.
- To remove excess pAz-AM from cells, wash cells with cell growth media three times over 15 minutes at 37 °C.
2. CuAAC derivatization of picolyl azide-tagged cell-surface proteins
There are two ways to detect picolyl azide on LAP-tagged cell-surface proteins: live-cell CuAAC (Module A) and fixed-cell CuAAC (Module B). The fixed-cell CuAAC protocol can be used when one wants to completely avoid residual toxicity of live-cell CuAAC. As illustrated in Figure 1B, pAz-labeled cells from either Step 1A or 1B can be used for either CuAAC procedure.
- pAz derivatization on live-cells using CuAAC
- Mix CuSO4 solution with either THPTA or BTTAA solution at stock concentrations in an eppendorf tube. Use required volume of each such that the final concentration of CuSO4 is 40-100 μM and THPTA or BTTAA is 200-500 μM (recommended concentrations) after dilution (step iii below). THPTA and BTTAA are used at 5 molar equivalents with respect to CuSO4.
- - Critical: use lowest CuSO4 concentration needed to obtain adequate signal, as cytotoxicity is proportional to CuSO4 concentration. It is possible to lower CuSO4 concentration below 40 μM if the protein target is overexpressed. We do not recommend using more than 100 μM CuSO4.
- - Critical: THPTA and BTTAA are interchangeable in most cases of cell-surface protein labeling, but we observed lower toxicity when CuAAC is performed on cultured neurons with BTTAA. BTTAA also yields slightly higher labeling signal for cell-surface protein labeling1.
- - Critical: do not use water-insoluble ligands such as TBTA in a live-cell labeling experiment.
- Add sodium ascorbate, then TEMPOL at stock concentrations such that the final concentration of sodium ascorbate is 2.5 mM and TEMPOL is 100 μM after dilution (step iii below). Incubate the mixture at room temperature for 5-10 min. Keep the tube cap closed during incubation.
- - Critical: the 5-10-min incubation step is crucial, as it allows THPTA/BTTAA and Tempol to quench reactive oxygen species that are generated as by-products of the CuAAC reaction.
- Dilute the mixture in DPBS. For labeling of cultured neurons, use Tyrode's buffer
- - Critical: do not use MEM, DMEM, serum-supplemented media, BSA-containing buffer, or other protein-containing buffer as a labeling buffer. High amine or protein content in these buffers can sequester copper ions and reduce labeling yields.
- Add 5-20 μM fluorophore-alkyne to the labeling solution. Higher dye-alkyne concentration than 20 μM (up to 100 μM) can be used to increase labeling sensitivity and decrease labeling time. Ensure that the fluorophore-alkyne stock solution is sufficiently concentrated such that no more than 0.1% (v/v) DMSO is added to cells.
- Add the complete labeling solution to cells for 1-10 min at room temperature. 5 min is a good starting point for labeling time optimization. Labeling can be performed at lower temperature to reduce endocytosis, but labeling yield will decrease. Keep the labeled cells from air and light exposure by doing the reaction in a foil-wrapped, lidded well plate.
- After labeling, add regular cell growth media containing 500 μM bathocuproin sulfonate to cells for 30 seconds to remove copper from cells.
- - Critical: This bathocuproin sulfonate wash step helps reduce toxicity as copper is immediately removed after labeling.
- Wash cells two more times with regular cell growth media.
- pAz derivatization in fixed cells using CuAAC
- Fix cells following the advice in Box 3. Fixed cells should be blocked with casein blocking buffer for at least 1 hour at room temperature. Alternatively, block with casein at 4 °C overnight.
- Mix CuSO4 solution with TBTA solution at stock concentrations in an Eppendorf tube. We recommend 1 mM CuSO4, and 100 μM TBTA final concentration after dilution (step iv below).
- - Critical: do not use THPTA or BTTAA for fixed-cell CuAAC labeling. Fixed-cell CuAAC requires the use of highly concentrated protein blocking reagent such as casein, which is incompatible with THPTA- and BTTAA-based CuAAC.
- Add 2.5 mM sodium ascorbate. Incubate the mixture at room temperature for 5-10 min. Keep the tube cap closed during incubation.
- Dilute the mixture in casein blocking buffer. If casein is not available, BSA blocking buffer can be used as an inferior substitute.
- Add 1-5 μM fluorophore-alkyne to the labeling solution.
- - Critical: using higher dye-alkyne concentration can lead to high non-specific sticking background. It is better to use lower dye-alkyne concentration with longer labeling time to increase labeling sensitivity.
- Add the complete labeling solution to cells for 30-120 min at room temperature. 60 min is a good starting point for optimizing labeling time. Reduce air and light exposure to labeled cells by carrying out the reaction in a foil-wrapped, lidded plate.
- After labeling, wash cells three times with casein blocking buffer over 15 minutes.
3. Labeling of purified proteins with picolyl azide and CuAAC
LAP-tagged proteins can be recombinantly expressed and purified using standard techniques appropriate for a given protein. We have successfully performed both pAz ligation and CuAAC labeling steps on ~10 μM purified protein solution using the protocol provided below. For protein samples more concentrated than 50 μM, use more pAz such that pAz concentration is at least twice the concentration of the protein. It is also permissible to add more LplA in the labeling solution; up to 20 mol% with respect to the amount of the LAP-tagged protein should be more than adequate. ATP and Mg(OAc)2 are already provided in large excess in the protocol below and need no adjustment for labeling of proteins of up to 100 μM.
- Critical: if the target LAP-tagged protein is His6-tagged, it is best to use tagless W37VLplA here so the target protein can be purified after labeling using nickel affinity column. If the target protein is not His6-tagged, you can use His6-W37VLplA, and elute out the desired target protein from the nickel-NTA resin while His6-LplA remains bound to the resin.
A. pAz ligation
- To label a 10 μM purified protein solution, add 1 μM W37VLplA, 50 μM pAz, 500 μM ATP, and 2.5 mM Mg(OAc)2 to the protein solution for 15-60 min at room temperature.
- - Critical: pAz ligation is compatible with standard buffers such as phosphate and HEPES, but avoid using high Ca2+ or high lipoic acid-containing buffer during labeling. LplA has pI of 5.8 and will precipitate near that pH.
To purify away small molecule reagents (pAz, ATP), perform more than five rounds of centrifugal washing using a centricon centrifugal filter unit with appropriate molecular weight cutoff. Add more than 10x of target protein-friendly buffer during each wash step. Gel filtration, such as with NAP resins, can also be used to remove small molecule reagents. It might not be necessary to perform this small-molecule purification step if you plan to purify your target protein using affinity capture.
To remove LplA, use nickel affinity column. For labeling with a His6-tagged target protein and a non-tagged LplA, use nickel-NTA resin to bind to the target protein and wash away LplA. For labeling with a non-His6-tagged protein and a His6-tagged LplA, use nickel-NTA resin to bind and remove LplA.
B. Reaction with CuAAC
Refer to Presolski et al.28, which reported a detailed protocol on how to perform CuAAC on biomolecules, including proteins, in vitro. Pay attention to many of their recommendations on how to perform optimal CuAAC, such as which buffers are compatible with CuAAC. We have used 200 μM CuSO4, 1 mM BTTAA (THPTA can likewise be used), 20 μM AF647-alkyne, 2.5 mM sodium ascorbate, and 100 μM TEMPOL to label ~10 μM LAP-kinesin for 30 min at room temperature, and saw that labeling signal already saturated after 30 min of CuAAC.
C. Estimation of in vitro protein labeling yield
After excess fluorophore is removed from the protein sample via gel filtration, dialysis, or centrifugal washing, one can estimate the overall labeling yield on the protein by measuring the dye-to-protein molar ratio. Measure absorbance at 280 nm (A280) and maximal absorbance at a particular wavelength for each fluorophore (for example, absorbance at 650 nm for Alexa Fluor 647) to determine the molar concentration of the protein and the fluorophore in a given sample respectively. To calculate molar concentrations, find out the extinction coefficients (ε) of the protein and the fluorophore. If ε of a protein is not known in literature or cannot be determined experimentally, one can roughly estimate ε from the protein's primary sequence using computational means (for example, ProtParam tool from ExPASy, which estimates ε based on numbers of Cys, Trp and Tyr present in a given protein). As fluorophores also absorb at 280 nm, one must apply correction factors for A280 that is contributed by the fluorophore. Information about fluorophores’ extinction coefficients and A280 correction factors as well as more details on dye-to-protein ratio calculation can be obtained from websites of common fluorophore manufacturers and vendors.
Timing
Box 1. With all reagents available, the syntheses, purifications and characterizations of picolyl azide and picolyl azide-acetoxymethyl ester are anticipated to require a week. Synthesis of 6-hydroxymethyl-nicotinic acid methyl ester 2: module A, 6 h. Synthesis of methyl 5-(azidomethyl)nicotinate, intermediate 3: module B, 32 h. Synthesis of 6-Azidomethylnicotinic acid 4: module C, 4 h. Synthesis of picolyl azide 5: module D, 12 h. Synthesis of picolyl azideacetoxymethyl ester 6, module E, 5 h.
Box 2. Expression of W37VLplA: module A, 2.5 days. Purification of W37VLplA: module B, 18 h. TEV protease treatment to make tagless W37VLplA: module C ii-xi, 2.5 days.
Step 1. Picolyl azide ligation using exogenous W37VLplA: module A, 2 days for i-ii, 30 min for iii-iv. Picolyl azide ligation using AILRLplA-ER: module B, 2 days for i, 90 min for ii-iii.
Step 2. Live-cell CuAAC: module A, 10-20 min. Fixed-cell CuAAC: module B, 1 h for i, 1.5 h for ii-vii.
Step 3. In vitro pAz ligation: module A, 1 h for i-ii, 1 h for iii, 1 h for iv. CuAAC: module B, 30 min for labeling.
Troubleshooting
See Table 1 for troubleshooting guidelines.
Anticipated results
Analytical data
6-Hydroxymethyl-nicotinic acid methyl ester, intermediate 2
1H NMR (500 MHz, CDCl3): 9.16, (d, 1H, J = 2.0 Hz), 8.29 (dd, 1H, J = 2.0, 8.5 Hz), 7.36 (d, 1H, J = 8.5 Hz), 4.83 (s, 2H), 3.96 (s, 3H). 13C NMR (75 MHz, CDCl3): 165.7, 163.7, 149.9, 138.3, 125.2, 120.4, 64.4, 52.7. HR-ESI-MS: [M+H]+ m/z 168.0655 calculated, 168.0655 observed.
Methyl 5-(azidomethyl)nicotinate, intermediate 3
1H NMR (500 MHz, CDCl3): 9.18 (d, 1H, J = 2.0 Hz), 8.32 (dd, 1H, J = 8.5, 2.0 Hz), 7.44 (d,1H, J = 8.5 Hz), 4.56 (s, 2H), 3.95 (s, 3H). 13C NMR (125 MHz, CDCl3): 165.7, 160.3, 151.6, 138.4, 125.5, 121.6, 55.7, 52.7. HR-ESI-MS: [M+H]+ m/z 193.0726 calculated, 193.0733 observed.
6-Azidomethylnicotinic acid, intermediate 4
1H NMR (400 MHz, CD3OD): 9.10 (dd, J = 2.1, 0.8 Hz), 8.39 (dd, 1H, J = 8.1, 2.1 Hz), 7.57 (dd, 1H, J = 8.1, 0.8 Hz), 4.59 (s, 2H). 13C NMR (100 MHz, CD3OD): 167.7, 161.3, 151.5, 139.9, 127.6, 123.3, 56.0. HR-ESI-MS: [M+H]+ m/z 179.0569 calculated, 179.0563 observed.
Picolyl azide 5, (5-(6-(Azidomethyl)nicotinamido)pentanoic acid)
1H NMR (500 MHz, D2O): 8.83 (s, 1H), 8.18 (d, 1H, J = 8.5 Hz), 7.59 (d, 1H, J = 8 Hz), 4.62 (s, 2H), 3.42 (m, 2H), 2.32 (m, 2H), 1.65 (m, 4H). 13C NMR (100 MHz, CD3OD): 177.7, 167.6, 160.0, 149.2, 137.7, 131.1, 123.3, 55.9, 40.7, 34.7, 29.8, 23.5. HR-ESI-MS: [M+H]+ m/z 278.1248 calculated, 278.1264 observed.
Picolyl azide-acetoxymethyl ester 6, (acetoxymethyl 5-(6-(azidomethyl)pyridine-3-carboxamido)pentanoate)
1H NMR (400 MHz, CDCl3) 8.95 (d, 1H, J = 1.7 Hz), 8.14 (dd, 1H, J = 2.3, 8.1 Hz), 7.43 (d, 1H, J = 8.4), 5.72 (s, 2H), 4.54 (s, 2H), 3.47 (m, 2H), 2.43 (t, 2H, J = 6.8), 2.09 (s, 3H), 1.70 (m, 4H). 13C NMR (100 MHz, CDCl3): 172.5, 169.9, 165.4, 158.8, 147.9, 136.4, 129.7, 121.8, 79.5, 55.5, 39.8, 33.5, 28.9, 21.7, 21.0. HR-ESI-MS: [M+H]+ m/z 350.1459 calculated, 350.1449 observed.
LplA expression
The typical yield of W37VLplA from a 500-ml culture of bacteria is 20-40 mg protein.
Cell-surface protein labeling
Typical labeling results with negative controls for picolyl azide ligation and CuAAC derivatization of cell-surface proteins in neurons are shown in Figure 3A. A comparison of labeling signal obtained from pAz ligation using exogenous LplA versus ER-pAz ligation is shown in Figure 3B. A comparison of labeling signal obtained from live-cell CuAAC versus fixed-cell CuAAC is shown in Figure 4.
In vitro protein labeling
Labeling specificity of picolyl azide and CuAAC labeling on LAP-tagged kinesin is shown in Figure 5. Using labeling protocols provided in Step 5, we have obtained the dye-to-protein ratio of ~0.7 on LAP-kinesin with AF647-alkyne.
The overall cell-surface labeling yield from picolyl azide ligation with exogenous W37VLplA followed by live-cell CuAAC is shown in Figure S1.
Supplementary Material
Table 1.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 2 | Low/no labeling | Poor transfection | Check for expression of a co-transfection fluorescent marker. Compare transfection efficiency obtained to literature reports; obtain new batch of transfection reagents if they do not perform well. |
| 2 | Low/no labeling | LAP-tagged protein not trafficked properly to the cell surface | Check for surface expression of the protein using antibody against an extracellular epitope. Performing immunofluorescence staining to visualize the total pool of protein, and see what percentage is trafficked to the surface. Check literature for precedence of successful surface trafficking of the target protein upon fusion to an epitope tag or a fluorescent protein. |
| 2 | Low/no labeling | LAP not accessible to LplA | Use more forcing labeling conditions (more enzyme, more pAz, longer labeling time) to see if LAP can be labeled at all. Perform lipoic acid ligation, followed by immunostaining with anti-lipoic acid antibody24, as a positive control that LAP is recognized by LplA. Change the LAP location within the protein or introduce flexible linkers if needed. |
| 2 | Low/no labeling | Purified W37VLplA not active | Check to see that there is no enzyme precipitate; re-express and purify if necessary. Use a positive control LAP-construct (available on Ting Lab's Addgene site) and see if labeling works on the positive control. |
| 2 | Low/no labeling | Degraded reagents | Do not use re-thawed ATP and W37VLplA. We have observed that ligands such as THPTA perform worse if left at room temperature for a long time, likely due to degradation. Make fresh sodium ascorbate solution. |
| 2 | High labeling background with live-cell CuAAC | Poor cell health | Use milder CuAAC conditions: less copper, shorter labeling time. Ensure that the supplements that preserve cell health (sodium ascorbate, TEMPOL, bathocuproin sulfonate) are good. Sodium ascorbate and TEMPOL solutions should be made fresh. |
| 2 | High labeling background with fixed-cell CuAAC | Non-specific sticking of fluorophore-alkynes | Poor blocking reagent is often the main reason for probe sticking. Test different batches of the blocking reagents to compare efficiency. Use casein over BSA when possible. |
| 1 | Cell toxicity | Toxicity from pAz ligation step | Reduce ATP concentration, which can activate purinoreceptors in neurons and cause excitotoxicity. Perform labeling at 37 °C instead of room temperature. Use CO2-independent medium for labeling at room temperature outside of an incubator. Especially for neurons, ensure that they are never left dry throughout labeling. |
| 2 | Cell toxicity | Toxicity from CuAAC step | Reduce copper concentration and labeling time, compensate for loss in labeling yield by increasing dye-alkyne concentration. Ensure that the supplements that preserve cell health (Ascorbate, TEMPOL, bathocuproin sulfonate) are good. Ascorbate and TEMPOL solutions should be made fresh. |
Box 1. Syntheses of picolyl azide (pAz) and picolyl azide-acetoxymethy ester (pAz-AM).
- Critical: all reactions are carried out under slightly positive pressure of dry nitrogen, and stirred with a Teflon-coated magnetic stir bar.
Depending on whether you are planning to label the cell-surface protein exogenously (using exogenous W37VLplA) or endogenously (using AILRLplA-ER), you should prepare either picolyl azide (pAz) or picolyl azide-acetoxymethy ester (pAz-AM). To label purified proteins, use pAz. To prepare pAz, perform Modules A-D; to prepare pAz-AM, perform Modules A-E.
- Synthesis of methyl 6-(hydroxymethyl)nicotinate 2
- Weigh 10.0 g of dimethyl 2,5-pyridine dicarboxylate (51.2 mmol) and 22.8 g of anhydrous CaCl2 (205.4 mmol), and transfer both to a 1-liter round-bottom flask equipped with an ice bath. Transfer 100 ml of anhydrous tetrahydrofuran and 200 ml of anhydrous methanol to the flask, and stir the slurry mixture. Allow the mixture to cool to 0 °C for 5-10 min.
- Weigh 3.9 g of sodium borohydride (NaBH4, 103.1 mmol), and transfer in portions to the reaction flask. Stir the reaction at 0 °C for ~2 hours.
- Quench excess NaBH4 by adding 100 ml of ice-cold water to the reaction mixture.
- Optional: concentrate the reaction mixture using rotary evaporation until the total volume is reduced down to ~150 ml. This makes the subsequent extraction step easier to handle.
- Extract the reaction mixture four times with chloroform (70 ml for each extraction). Combined organic extract is washed twice with water, dried with sodium sulfate, and concentrated using rotary evaporation to obtain methyl 6-(hydroxymethyl)nicotinate 2 as white solid (4.5 g, 52% yield). Rf = 0.27 (1:1 hexanes/ethyl acetate).
- Critical: the aqueous wash step is critical, as it removes a polar by-product (caused by over-reduction of two esters instead of one) without the need for a subsequent chromatographic purification step.
- - Pause point: Store methyl 6-(hydroxymethyl)nicotinate at −20 °C for > 1 year without evidence of decomposition.
- Synthesis of methyl 5-(azidomethyl)nicotinate 3
- Weigh 4.0 g of 6-(hydroxymethyl)nicotinate 2 (23.9 mmol), transfer to a 500-ml round-bottom flask, and dissolve it in 240 ml anhydrous dichloromethane.
- Add 13.4 ml of anhydrous triethylamine (95.7 mmol) and 6.8 g of p-toluenesulfonyl chloride (35.9 mmol) consecutively to the reaction flask. Stir the reaction at room temperature for 3 hours.
- Remove dichloromethane from the reaction mixture using rotary evaporation.
- Caution: ensure that dichloromethane is completely removed, as it can react with sodium azide used in the subsequent step to form potentially explosive diazidomethane.
- Dissolve the resulting residue in 240 ml of tetrahydrofuran.
- Add 15 g of sodium azide (230.9 mmol). Stir the reaction at room temperature for 24 hours.
- Critical: note that the Rf of methyl 5-(azidomethyl)nicotinate 3 is almost identical to that of the tosylate intermediate, making reaction monitoring by TLC difficult. If needed, use 1H-NMR to monitor the reaction progress instead.
- Reduce the volume of the reaction mixture to ~60 ml through rotary evaporation.
- Caution: do not use heat during rotary evaporation. While sodium azide is not explosive except when heated near its decomposition temperature (300 °C), heating should generally be avoided when working with it.
- Dilute the resulting residue with ethyl acetate and water. Further extract the aqueous layer three times with ethyl acetate (70 ml each time). Wash the combined organic extract with brine solution, then dry it over Na2SO4 and concentrate using rotary evaporation.
- Purify the resulting residue by flash chromatography on silica, using isocratic 4:1 hexanes:ethyl acetate to afford methyl 5-(azidomethy)nicotinate 3 as a light yellow solid (3.26 g, 71% yield). Rf = 0.79 (1:1 hexanes/ethyl acetate).
- - Pause point: Store methyl 5-(azidomethyl)nicotinate at −20 °C for > 1 year without evidence of decomposition.
- Synthesis of 6-azidomethylnicotinic acid 4
- Transfer 1.32 g of methyl 5-(azidomethyl)nicotinate 3 to a 500-ml round-bottom flask, and dissolve it in 68 ml of methanol.
- Add 20.5 ml of 1.0 M solution of LiOH in water (20.5 mmol). Stir the mixture at room temperature for 25 min.
- Critical: be careful not to prolong the LiOH hydrolysis step as the azido group can be labile under aqueous basic conditions.
- Add ~700 μl of acetic acid to the reaction mixture, and concentrate the crude mixture using rotary evaporation.
- Purify the resulting residue by flash chromatography on silica, using isocratic ethyl acetate + 1% (v/v) acetic acid to provide 6-azidomethylnicotinic acid 4 as a yellow solid (1.12 g, 92% yield). Rf = 0.35 (ethyl acetate + 1% acetic acid).
- - Pause point: Store 6-azidomethylnicotinic acid at −20 °C for > 1 year without evidence of decomposition.
- Synthesis of 5-(6-(azidomethyl)nicotinamido)pentanoic acid 5, or picolyl azide (pAz)
- Transfer 300 mg of 6-azidomethylnicotinic acid 4 (1.68 mmol) to a 100-ml round-bottom flask, and dissolve it in 5 ml of anhydrous N,N-dimethylformamide (DMF).
- Add 353 μl of anhydrous triethylamine (2.53 mmol) and 650 mg of disuccinimidyl carbonate (2.53 mmol). Stir the reaction at room temperature for 3 hours.
- Dilute the reaction mixture with chloroform and water. Further extract the aqueous layer three times with chloroform (20 ml each time). Wash the combined organic extract with brine solution, dry it over Na2SO4, and concentrate using rotary evaporation.
- Purify the resulting residue by flash chromatography on silica, using isocratic 1:1 hexanes:ethyl acetate to afford the succinimidyl ester of 6-azidomethylnicotinic acid (166 mg, 36% yield). Rf = 0.67 in 9:1 chloroform:methanol.
- Dissolve 166 mg of the succinimidyl ester of 6-azidomethylnicotinic acid (0.36 mmol) in 1.75 ml of anhydrous DMF.
- Add 150 μl of anhydrous triethylamine (1.08 mmol) and 127 mg of 5-aminovaleric acid (1.08 mmol). Stir the reaction at room temperature for 3 hours.
- Concentrate the reaction mixture using rotary evaporation.
- Purify picolyl azide by either normal-phase purification on a hand-packed silica column (Step ix) or reverse-phase purification using HPLC (Step x). Purification on a hand-packed silica column is suitable for large-scale purifications, while HPLC purification affords the product with higher purity.
- Dissolve the crude reaction mixture in a minimum volume of 90:5:5 ethyl acetate:methanol:acetic acid. Load the resulting mixture directly on to a silica gel column equilibrated with ethyl acetate + 1% (v/v) acetic acid. Elute the product using isocratic ethyl acetate + 1% acetic acid to afford the product as light yellow oil (131 mg, 93% yield). Critical: 1H-NMR analysis shows that picolyl azide purified according to procedure a coelutes with N-hydroxysuccinimide (δ 2.69 in CD3OD, singlet).
- For an alternative HPLC purification, use a Varian Prostar 210 HPLC equipped with Agilent 325 UV/Vis dual-wavelength detector, Agilent 440-LC fraction collector, and a Microsorb C18 column (Varian, 5 μm particle size, 21mm × 250 mm dimension). We use a 0-10% v/v acetonitrile in water gradient over 30 min, at a 10 ml/min flow rate. Picolyl azide elutes at 29-30 minutes. Collect the desired fractions, remove acetonitrile using rotary evaporation, then flash-freeze and lyophilize the resulting aqueous solution. Picolyl azide purified this way appears as a pale yellow solid, and does not contain the N-hydroxysuccinimide contaminant.
- - Pause point: Store picolyl azide at −20 °C for > 1 year without evidence of decomposition.
- Synthesis of acetoxymethyl 5-(6-(azidomethyl_nicotinamido)pentanoate 6 or picolyl azide-acetoxymethyl ester (pAz-AM)
- Transfer 60 mg of picolyl azide 5 (0.216 mmol) to a 5-ml round-bottom flask, and dissolve it in 1.2 ml of anhydrous DMF.
- Add 60 μl of anhydrous triethylamine (0.432 mmol) and 42 μl of bromomethyl acetate (0.432 mmol). Stir the reaction at room temperature for 1 hour.
- Dilute the reaction mixture with chloroform (10 ml) and water (10 mL). Further extract the aqueous layer three times with chloroform (10 ml each time). Wash the combined organic extract with brine solution, dry it over Na2SO4, and concentrate using rotary evaporation.
- Purify the resulting residue by flash chromatography on silica, using isocratic 1:2 hexanes:ethyl acetate to afford the acetoxymethyl ester of pycolyl azide 6 as pale yellow solid (35 mg, 46% yield). Rf = 0.37 (1:1 hexanes/ethyl acetate).
- - Pause point: Store methyl 5-(azidomethyl)nicotinate at −80 °C for > 6 months without evidence of decomposition.
Box 2. W37VLplA expression and purification.
If you are performing exogenous cellular labeling with pAz (Step 3A) or you are intending to label a purified protein, then you need to express and purify W37VLplA. If pAz ligation is going to be performed endogenously with ER-targeted AILRLplA, this step is not required; go directly to Step 3B. Modules A and B describe the expression and purification of His6-W37VLplA. These same steps could also be performed to prepare His6-ENLYFQG-W37VLplA. Module C describes how to remove the His-tag from His6-ENLYFQG-W37VLplA and purify the resulting W37VLplA. TEV protease-mediated cleavage of the ENLYFQG sequence removes His6-ENLYFQ (cleavage occurs between Gln and Gly) from W37VLplA. Thereafter, the reaction mixture is run through nickel-NTA resin, which selectively retains His6-ENLYFQ peptide and His6-AcTEV protease, and allows tagless W37VLplA to flow through.
- Expression of His6-W37VLplA in E. coli
- Transform chemically competent BL21 (DE3) pLysS E. coli with pYFJ16-W37VLplA, and plate on an LB-Amp agar plate. Incubate at 37 °C overnight.
- Pick one colony from the plate and inoculate into 5 ml LB-Amp (LB with 100 μg ml−1 ampicillin). Shake at 220 rpm at 37 °C overnight.
- To make an expression culture, dilute the 5 ml overnight culture into 500 ml LB-Amp in a 2-liter culture flask. Shake at 37 °C until A600 reaches 0.5.
- - Critical: for maximal LplA protein yield, it is better to induce expression when A600 is below 0.7. Once A600 reaches 0.1, the cell density should double at every 20 minutes.
- Move the culture flask to a room-temperature shaker, allow the flask to cool down to room temperature for ~5 min.
- Add isopropyl-β-D-thiogalactopyranoside to a final concentration of 100 μg ml−1.
- Shake the culture flask at room temperature overnight (~8 hours).
- - Critical: the lower culture temperature during induction greatly improves the folding efficiency of W37VLplA
- Harvest bacteria by centrifuging the culture at 5,000 g for 10 min at 4 °C. Discard the supernatant via decanting. Keep the pellet on ice.
- - Pause point: the bacterial cell pellet can be stored at −80 °C for at least three months.
- Purification of His6-W37VLplA on a nickel-affinity column
- - Critical: always keep the cell pellet, cell lysate, and any LplA-containing solutions on ice to minimize LplA aggregation and precipitation.
- Lyse bacteria using Bacterial Protein Extraction Reagent (B-PER). First, mix 10 ml of B-PER with 50 μl of protease inhibitor cocktail and 50 μl of 100 mM phenylmethylsulfonylfluoride (PMSF). Transfer the mixture to a pellet-containing culture bottle. Completely resuspend the pellet in B-PER using pipetting or vortexing. Add an additional 10 ml of B-PER (also with protease inhibitor cocktail and PMSF).
- Gently agitate the homogenous cell suspension for 10 min at 4 °C on a motorized shaker.
- Centrifuge the cell suspension at 10,000 g for 5 min at 4 °C. Collect the supernatant in a 50-ml conical tube as the cell lysate.
- Prepare the nickel-NTA resin. Load a poly-prep column with ~1 ml (packed volume) of nickel-NTA agarose resin, and wash the resin with five column volumes of ice-cold nickel-binding buffer by allowing the buffer to flow through the resin.
- - Critical: only use gravity-driven flow. Do not apply extra pressure.
- Add ~20 ml of ice-cold nickel-binding buffer to the cell lysate, then transfer the washed nickel-NTA resin to the same tube. Gently agitate via shaking for 20 min at 4 °C to ensure His-tagged protein binding to the resin.
- Add the cell lysate/resin mixture to a poly-prep column and let the lysate flow through. Avoid letting the column run dry.
- Wash the resin with 10 column volumes of nickel-NTA wash buffer to remove non-specifically bound proteins.
- Elute LplA from the column using 500-μl aliquots of nickel-NTA elution buffer. Collect each 500-μl fraction, and check for the presence of protein in the fractions by measuring A280. Pool the positive fractions.
- - Critical: W37VLplA can precipitate if its eluted concentration is too high (>5 mg/mL). It is best not to use imidazole concentration higher than 100 mM during elution.
- Dialyze W37VLplA two times, each for > 6 hours, against at least 1000-fold ice-cold LplA dialysis buffer at 4 °C.
- Measure LplA concentration using standard protein concentration measurement assays, such as the bicinchoninic acid (BCA) assay.
- - Critical: ensure that the protein concentration measurement assay of choice is not affected by the presence of 1 mM DTT in the dialysis buffer. BCA assay is compatible with 1 mM DTT.
- Confirm purity of W37VLplA using protein gel electrophoretic analysis. LplA molecular weight is 39 kDa.
- Aliquot LplA, and store at −80 °C.
- - Pause point: W37VLplA is stable at −80 °C for > 2 years.
- Preparation of tagless W37VLplA from His6-ENLYFQG-W37VLplA
- Express and purify His His6-ENLYFQG-W37VLplA as described in Steps 2A-B.
- After nickel-NTA purification, dialyze the protein aliquots in LplA dialysis buffer two times, each for > 6 hours, against at least 1000-fold ice-cold buffer at 4 °C.
- Further dialyze LplA two times in TEV protease cleavage buffer, each time for > 6 hours, against at least 1000-fold ice-cold buffer at 4 °C. After dialysis, measure LplA concentration to make sure that it does not exceed 5 mg/mL; make dilutions if necessary. We typically work with 2-3 mg/mL solution during the protease cleavage step.
- - Critical: do not directly dialyze LplA into TEV protease cleavage buffer, which does not contain glycerol and can cause LplA to precipitate
- To perform protease-mediated cleavage, add AcTEV protease to the LplA solution. We generally use ~10 units of the protease per 60 μg of LplA (~3-times less protease than recommended by Life Technologies).
- Incubate the reaction at 4 °C for 36-48 h.
- Prepare nickel-NTA resin by loading the resin onto a column, and wash the resin with five column volumes of ice-cold nickel-binding buffer. We use ~1 mL (packed volume) of resin per ~5 mg of protein
- Add the LplA/protease reaction directly to the column, and collect the flow-through. viii. Wash the resin once using one column volume of ice-cold nickel-binding buffer. Collect the flow-through.
- Check for the presence of protein in the flow-through fractions using A280. Pool the positive fractions.
- Dialyze W37VLplA two times, each for > 6 hours, against at least 1000-fold LplA dialysis buffer at 4 °C.
- Measure concentration and confirm purity of LplA as described in Step 2B. Aliquot LplA and store at −80 °C.
- - Pause point: W37VLplA is stable at −80 °C for > 2 years.
Box 3. Cell fixation (optional).
Cells can be fixed after pAz ligation step (after Step 3) if one wants to perform CuAAC post-cell fixation to maximize labeling sensitivity, or after both labeling steps are performed (after Step 4) to preserve cells for subsequent imaging experiments.
A. Cell fixation after pAz ligation step.
If you want to detect the surface pool of the cell-surface protein, you can fix the cells using formaldehyde without additional membrane permeabilization or protein precipitation step. Glutaraldehyde can also be used, but do not use a reducing agent such as sodium borohydride to quench glutaraldehyde as picolyl azide will also be reduced. If you want to access the intracellular pool of the cell-surface protein, fix the cells and use methanol to precipitate proteins and permeabilize the membrane. It is best to avoid detergents as they can extract cell-surface proteins from the cell membrane.
B. Cell fixation after CuAAC step.
Use standard fixation methods with formaldehyde or methanol. Glutaraldehyde can also be used, but do not use a reducing agent such as sodium borohydride to quench glutaraldehyde as many fluorophores can be reduced and destroyed.
Acknowledgments
We thank Ting lab members who have contributed to PRIME-related protocols, particularly M. Fernández-Suárez and K.A. White. We also thank A. Karunakaran (UCSF) and R. D. Vale (UCSF) for LAP-kinesin K560 and kinesin K420 proteins, C. Garner (Stanford) for neuron fixative recipe, and M.G. Finn (Scripps) for helpful advice on CuAAC. Funding was provided by NIH (R01 GM072670) and the Dreyfus Foundation.
Footnotes
Author contribution statements
C.U., M.I.S., D.S.L., J.Z.Y., K.A.W., S.G., S.C., K.R.G., and A.Y.T. developed protocols. C.U. contributed all the data. C.U., M.I.S. and A.Y.T. wrote the paper.
Competing financial interests
The authors declare competing financial interests: details are available in the online version of the paper.
Contributor Information
Chayasith Uttamapinant, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA..
Mateo I. Sanchez, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Daniel S. Liu, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Jennifer Z. Yao, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Katharine A. White, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Scott Grecian, Life Technologies, Eugene, Oregon, USA..
Scott Clarke, Life Technologies, Eugene, Oregon, USA..
Kyle R. Gee, Life Technologies, Eugene, Oregon, USA.
Alice Y. Ting, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
References
- 1.Uttamapinant C, et al. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed Engl. 2012;51(24):5852. doi: 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107(24):10914. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu DS, et al. Diels-Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J. Am. Chem. Soc. 2012;134(2):792. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen JD, Thompson S, Ting AY. Structure-guided engineering of a Pacific Blue fluorophore ligase for specific protein imaging in living cells. Biochemistry. 2011;50(38):8221. doi: 10.1021/bi201037r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin X, Uttamapinant C, Ting AY. Synthesis of 7-aminocoumarin by Buchwald-Hartwig cross coupling for specific protein labeling in living cells. Chembiochem. 2011;12(1):65. doi: 10.1002/cbic.201000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baruah H, et al. An engineered aryl azide ligase for site-specific mapping of protein- protein interactions through photo-cross-linking. Angew. Chem. Int. Ed Engl. 2008;47(37):7018. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao JZ, et al. Fluorophore targeting to cellular proteins via enzyme-mediated azide ligation and strain-promoted cycloaddition. J. Am. Chem. Soc. 2012;134(8):3720. doi: 10.1021/ja208090p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu DS, et al. Quantum Dot Targeting with Lipoic Acid Ligase and HaloTag for Single- Molecule Imaging on Living Cells. ACS Nano. 2012;6(12):11080. doi: 10.1021/nn304793z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu DS, et al. Imaging trans-cellular neurexin-neuroligin interactions by enzymatic probe ligation. PLoS One. 2013;8(2):e52823. doi: 10.1371/journal.pone.0052823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong V, et al. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem. Int. Ed Engl. 2009;48(52):9879. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2(3):168. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 12.Dani A, et al. Superresolution Imaging of Chemical Synapses in the Brain. Neuron. 2010;68(5):843. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellner RR, et al. Nanoscale organization of nicotinic acetylcholine receptors revealed by stimulated emission depletion microscopy. Neuroscience. 2007;144(1):135. doi: 10.1016/j.neuroscience.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 14.Zou P, Ting AY. Imaging LDL receptor oligomerization during endocytosis using a co-internalization assay. ACS Chem. Biol. 2011;6(4):308. doi: 10.1021/cb100361k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh DR, et al. Distribution of an NMDA receptor:GFP fusion protein in sensory neurons is altered by a C-terminal construct. J. Neurochem. 2001;77(1):23. doi: 10.1046/j.1471-4159.2001.t01-1-00182.x. [DOI] [PubMed] [Google Scholar]
- 16.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15(2):128. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Los GV, et al. HatoTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3(6):373. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 18.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods. 2006;3(4):267. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 1998;16:569. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 20.Shi X, et al. Quantitative fluorescence labeling of aldehyde-tagged proteins for single- molecule imaging. Nat. Methods. 2012;9(5):499. doi: 10.1038/nmeth.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dirksen A, et al. Nucleophilic catalysis of hydrazone formation and transimination: implications for dynamic covalent chemistry. J. Am. Chem. Soc. 2006;128(49):15602. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 22.Martin BR, et al. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23(10):1308. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 23.Beatty KE, et al. Live-cell imaging of cellular proteins by a strain-promoted azide alkyne cycloaddition. Chembiochem. 2010;11(15):2092. doi: 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puthenveetil S, et al. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J. Am. Chem. Soc. 2009;131(45):16430. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods. 2008;5(5):397. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrington JC, Dougherty WG. A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc. Natl. Acad. Sci. U. S. A. 1988;85(10):3391. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besanceney-Webler C, et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem. Int. Ed Engl. 2011;50(35):8051. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presolski SI, Hong VP, Finn MG. Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011;3(4):153. doi: 10.1002/9780470559277.ch110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan TR, et al. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 2004;6(17):2853. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 30.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 2008;3(3):534. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woehlke G, et al. Microtubule interaction site of the kinesin motor. Cell. 1997;90(2):207. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.