Abstract
The advent of next-generation sequencing has allowed huge amounts of DNA sequence data to be produced, advancing the capabilities of microbial ecosystem studies. The current challenge is to identify from which microorganisms and genes the DNA originated. Several tools and databases are available for annotating DNA sequences. The tools, databases and parameters used can have a significant impact on the results: naïve choice of these factors can result in a false representation of community composition and function. We use a simulated metagenome to show how different parameters affect annotation accuracy by evaluating the sequence annotation performances of MEGAN, MG-RAST, One Codex and Megablast. This simulated metagenome allowed the recovery of known organism and function abundances to be quantitatively evaluated, which is not possible for environmental metagenomes. The performance of each program and database varied, e.g. One Codex correctly annotated many sequences at the genus level, whereas MG-RAST RefSeq produced many false positive annotations. This effect decreased as the taxonomic level investigated increased. Selecting more stringent parameters decreases the annotation sensitivity, but increases precision. Ultimately, there is a trade-off between taxonomic resolution and annotation accuracy. These results should be considered when annotating metagenomes and interpreting results from previous studies.
Keywords: DNA sequencing, metagenomics, metagenome analysis, microbial ecology, sequence annotation
The sequence annotation accuracies of metagenomic analysis tools were evaluated using a simulated metagenome.
INTRODUCTION
The advent of next-generation sequencing and metagenomics has resulted in increasing numbers of ever-larger datasets describing the community structure and function of a variety of different environments, from the human gut (Arumugam et al. 2011; David et al. 2014) to arctic peat soils (Lipson et al. 2013) and deep-sea vents (Xie et al. 2011; Anderson, Sogin and Baross 2015), to name a few. Next-generation sequencing technologies have greatly reduced sequencing costs and speed, and researchers can now affordably study whole microbial communities and functions. Prior to this, the focus was on community species composition, studied using 16S rRNA targeted amplicon sequencing. Amplicon sequencing does not require the DNA coverage that metagenomic studies require and can accurately identify which species are present in a sample (Woese and Fox 1977; Lane et al. 1985; Hugenholtz 2002), but it does not provide the depth of information, such as gene function, that full metagenome sequencing and annotation provides. Cost is no longer the primary limiting factor for undertaking metagenomic studies, rather it is now bioinformatics and processing power required to process the data produced. Illumina's HiSeq platform, for example, can affordably sequence the most complex of microbial communities, and the challenge now is to interpret the data produced.
Henry et al. (2014) provide an extensive directory of tools available for different tasks involved in a metagenomic project pipeline, related to a range of ‘omics’ studies. These may include bespoke bioinformatics pipelines, downloadable programs and web-based services. MEGAN (Huson et al. 2007) is a popular Graphical User Interface program for analyzing and visualizing BLAST results to study the taxonomy of microbial communities. While MEGAN typically analyses BLAST results in a few minutes, running BLAST searches against reference sequences in a database is computationally intensive and slow for metagenomes (Desai et al. 2012; Hunter et al. 2012; Thomas, Gilbert and Meyer 2012). Web-based servers are increasingly popular for processing large amounts of data. With an intuitive web interface and a variety of analytical tools to choose from, MG-RAST (Meyer et al. 2008) is increasingly cited. MG-RAST allows users to upload raw sequence files that are processed through quality filters and annotated using a selection of user-defined parameters, such as reference databases, minimum identity cut-off values, maximum E-values or expect-values, and minimum alignment lengths. Details of the processing procedure can be found in the MG-RAST Technical Report (Wilke et al. 2013).
In response to the growing size of data sequenced, faster alignment methods are being produced. RAPSearch2 (Zhao, Tang and Ye 2012) translates nucleotide sequences and aligns them with annotated protein sequences, reporting to be c. 100-fold faster than BLASTX with only a 1.3%–3.2% reduction in sensitivity (the proportion of sequences annotated). With ‘accelerate’ mode, the speed increase is up to 1000-fold. PAUDA (Huson and Xie 2014) uses a similar approach and claims to be 10 000-fold faster than BLASTX, although with a significant reduction in sensitivity. DIAMOND (Buchfink, Xie and Huson 2015) purports to be both fast and accurate, with a 20 000-fold increase in processing speed compared to BLASTX. In sensitive mode, 99% of sequences are aligned, with a speed increase of 2000-fold compared to BLASTX. Like BLAST with Megablast, RAPSearch2 and DIAMOND offer fast and sensitive modes, each coming at the cost of the other. The outputs from both programs can be viewed and analyzed using MEGAN.
One Codex is a web-based program that uses a different technique to BLAST and MG-RAST to classify sequences (https://onecodex.com/). The program designers report that it runs 900 times faster than BLAST while maintaining similar genus-level sensitivity and precision (the proportion of annotated sequences that are correctly identified), taking hours rather than days to classify most metagenomes. One Codex works by comparing k-mers (sequences of a set length) from a sequence to a reference database of k-mers; the greatest number of 100% k-mer matches determines the classification. BLAST and MG-RAST classify sequences by matching them with the most similar sequences in a database. Unlike MG-RAST, One Codex does not annotate genes for function.
The choice of database, minimum identity cut-off value (i.e. sequence match stringency), minimum alignment length cut-off value and minimum E-value limit (the probability a match has occurred by chance) all influence sequence annotation accuracy, which, in turn, affect the reproducibility and interpretation of the data. An inherent issue with metagenomic studies is that establishing the accuracy of sequence annotation for environmental samples is practically impossible, given that the quantities of organisms and genes are unknown. Therefore, determining the most effective annotation method is fundamental to investigating environmental communities with confidence.
Databases
There are a variety of different reference nucleotide and amino acid databases available for annotating gene or protein sequences (Table S1, Supporting Information). The M5NR database (Wilke et al. 2012) incorporates information from a selection of different databases (see Table S1, Supporting Information), increasing the amount of reference data available for annotation. Using a single reference database may be the best option in some cases, for example, where 16S rRNA amplicons are used as a method to identify taxa, rather than other genes.
Whereas taxonomic nomenclature is universal, governed by international conventions, there are multiple approaches for functional classification. Two popular methods include Clusters of Orthologous Groups (COGs) (Tatusov, Koonin and Lipman 1997) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000). COGs comprise orthologous functions that allow for functional description of poorly characterized genomes based on protein orthologs. KEGG provides a reference database of sequences with functional pathway annotations. Both methods include a hierarchy of functional descriptions. At the highest level, COG descriptions are characterized under cellular processes, information storage and processing, metabolism and poorly characterized. KEGG descriptions are characterized under cellular processes, environmental information processing, genetic information processing, human diseases and metabolism. Due to the differences in characterization approaches, COG and KEGG annotations cannot be compared directly. COG is currently freely available. KEGG operates on a subscription basis, and MG-RAST uses the latest freely available version (updated in 2008).
Parameters
Selecting a minimum identity cut-off value for metagenome analysis is challenging because interspecific sequence identity varies among genes. Too high a value will accurately identify genes with highly conserved regions, such as 16S rRNA or highly conserved coding genes with little synonymous substitution, but may fail to identify genes or non-coding regions that are highly variable. Conversely, a value too low will allow for highly variable genes to be identified, but may also incorrectly identify an organism/function, thus providing false community/function profiles.
The optimum identity cut-off point for species identification using the 16S rRNA gene is widely accepted as 97% (Stackebrandt and Goebel 1994; Rosselló-Mora and Amann 2001; Chun et al. 2007; Richter and Rosselló-Móra 2009; Mende et al. 2013; Větrovský and Baldrian 2013), although this value has its limitations. Some species, such as certain Rickettsia spp., have a 16S rRNA gene similarity greater than 97%, thus a cut-off value at this level would not differentiate between the species (Fournier et al. 2003). Stackebrandt and Goebel (1994) suggest that a higher value may be more appropriate, but fewer sequences would be annotated due to sequencing errors and sequence mutations. Typically, lower cut-off values are suitable for metagenomic studies as the multitude of genes that contain varying degrees of conservation are sequenced. The default value used by MG-RAST, and used in many metagenomic studies (e.g. Tatusov, Koonin and Lipman 1997; Lipson et al. 2013), is 60%, as this allows for identification using less conserved genes and non-coding regions.
Minimum alignment lengths set the minimum length of sequence considered for annotation. A lower value allows shorter sequences to be annotated, although the chance of incorrectly annotating a shorter sequence is higher. A higher value will reduce this chance, but may also reduce the number of annotations overall. Combining a low minimum alignment length with a strict minimum identity cut-off value allows shorter sequences to be annotated but with a high match criteria.
Setting maximum E-values and minimum alignment lengths allows stringency of annotations to be controlled. E-values denote the maximum probability that a sequence annotation has occurred by chance. Lower maximum E-values will reduce the number of possible incorrect annotations, although this also reduces number of annotations retained for analysis. The default maximum E-value used by MG-RAST is 1-e−5.
Aims
The aim of this study is to evaluate the accuracy of MEGAN, MG-RAST and One Codex annotation methods while investigating how using different databases and parameters impact the annotation of metagenomes. To do this, a novel simulated metagenome was generated using the NCBI whole bacterial genome database and annotated using each pipeline and, for MG-RAST, with different reference databases, minimum identity cut-off values, minimum alignment lengths and maximum E-values.
Using a simulated metagenome comprising known genome abundances allows the accuracy of annotation to be quantified. The simulated metagenome was also annotated using Megablast, a faster variation of BLAST, to provide a control and so that MEGAN, MG-RAST and One Codex could be compared to a standard in sequence annotation. Comparing the MEGAN, MG-RAST and One Codex annotations to the Megablast annotations will quantify the accuracy of these programs for annotating sequences from organisms whose genomes are stored in the NCBI databases.
METHODOLOGY
Metagenome simulation
A simulated metagenome, hereafter Simmet, was created using NeSSM (Jia et al. 2013), comprising the complete NCBI bacterial genome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/all.fna.tar.gz, May 2013 collection, accessed on 29/04/14). NeSSM creates synthetic metagenomes from input genomes based on user-defined parameters (e.g. sequence count, length and abundance distributions) that aim to simulate real sequencing data, including expected sequencing errors (i.e. substitutions, insertions and deletions) based on the chosen sequencing technology simulated (see Step II: error models and sequencing coverage bias estimation in Jia et al. 2013). A total of 2400 000 sequences with a read length of 450 base pairs were designated for simulation, based on 454 pyrosequencing.
One strain for each of the 1505 species in the NCBI bacterial genome database was randomly selected to be included in the simulation because certain species, e.g. model organisms and human pathogens such as Escherichia coli, Salmonella enterica, Mycobacterium tuberculosis, Bacillus cereus and Staphylococcus aureus, have been extensively studied and are overrepresented in the databases. The resulting genus richness was 688. The species abundance distribution used for simulation was derived from the abundance distribution of a pasture soil metagenome (sequence count: 2378 586, MG-RAST ID: 4554767.3) (See Equation 1).
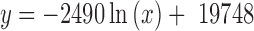 |
(1) |
where x is the randomly selected species rank.
The sequences were processed with Sickle (Joshi and Fass 2011) to trim low-quality ends, with the average threshold phred score set at 20 (a base call error rate of 1%).
Analysis
The Simmet metagenome file was annotated with Megablast (available from: http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE = BlastDocs&DOC_TYPE = Download) as a control, using a reference database of the genomes used to create Simmet. This quantifies the effect that the simulated sequencing errors have on the annotations. The NCBI nucleotide database (updated 17/11/2014) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) was also used to assess the annotation performance of Megablast. The maximum E-value selected was 1-e−5 and the minimum alignment length, 15 bases. Megablast annotations using the Simmet database will be referred to as ‘control’ and those using the NCBI nucleotide database will be referred to as ‘Megablast’. The BLAST results were uploaded to MEGAN (version 5.2.3) and analyzed using the same parameters used in the BLAST.
Simmet was uploaded to MG-RAST and One Codex. The databases investigated within MG-RAST were GenBank, GreenGenes, RDP, RefSeq, SEED, SwissProt and TrEMBL. The M5NR and M5RNA databases were excluded from individual sequence analysis, as individual sequence annotations were not available for download from MG-RAST for these databases. RefSeq was used for One Codex. For both the Megablast and MG-RAST annotations, which use a minimum sequence alignment match to annotated sequences, the minimum identity cut-off values tested were as follows: 40%, 50%, 60%, 70%, 80%, 90%, 95% and 97%. The minimum alignment lengths tested were as follows: 10, 15, 20, 25, 30, 25, 40, 45, 50, 55 and 60 bp. The maximum E-values tested were as follows: 1-e−1, 1-e−5, 1-e−10 and 1-e−15. Aside from testing, default parameters were used: 60%, 15 bp and 1-e−5, respectively, for minimum identify cut-off, minimum alignment length and maximum E-value.
The sequence IDs and annotations were extracted from the Megablast results (https://github.com/sandyjmacdonald/blast_parser), and full taxonomic lineages were generated for each sequence using the NCBI taxonomy database (available from: ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/; NCBI database version generated 13/05/2013). Species level was excluded from analysis due to the high variation in annotated species nomenclature and the accepted caveats associated with microbial species classification (Gevers et al. 2005; Achtman and Wagner 2008), e.g. horizontal gene transfer (Gogarten and Townsend 2005; Bapteste and Boucher 2009). Discrepancies identified between databases for organism names were corrected for, such as NCBI using the old name Chloroflexia (as of 17/11/14) and MG-RAST using the new name Chloroflexi for the same class. Those that were not annotated were named ‘Unidentified’ and those that were annotated but were either ambiguously annotated or not annotated at all taxonomic levels had the corresponding levels in the lineage replaced with ‘Unclassified’. For MEGAN and One Codex, NCBI taxa IDs were used to generate the lineages.
The taxonomic lineage for each annotated sequence was compared to the lineage for the corresponding source sequence in Simmet to determine the annotation sensitivity and precision at each taxonomic level. The effect that minimum identity cut-off values, minimum alignment lengths and maximum E-values had on annotation sensitivity and precision were established using Megablast and MG-RAST. The correlations between the relative abundances for each taxon in Simmet and in the annotations were calculated using Pearson's product moment correlation coefficient. Domain was excluded due to the small number of taxa. The natural logarithms of the relative abundance values were calculated for plotting, as the original distributions would not visually convey the variations in low abundance taxa. The taxa richness values for each taxonomic level were calculated.
Unlike investigating taxa, correct functional annotations cannot be ascertained with 100% confidence. To investigate functional annotation performance, protein sequences associated with the sequences in Simmet were extracted from GenBank records and annotated using the KEGG Automatic Annotation Server (KAAS) (Moriya et al. 2007) and WebMGA (Wu et al. 2011). Both are web-based functional annotation tools independent of those investigated in this study. They did not contain sequencing errors and thus provided the best possible indication of the functional annotation accuracy, although the caveats associated with sequence annotation (e.g. possibly incorrectly assigning a function) are present.
KEGG Orthology and COG IDs were extracted from the KASS and WebMGA results, respectively, for each sequence annotated and compared with the IDs assigned by MG-RAST. The parameters set for the taxonomic investigation were used, and the minimum identity cut-off values investigated were as follows: 40%, 50%, 60%, 70%, 80%, 90% and 95%. The minimum alignment lengths tested were as follows: 10, 15, 20, 25, 30, 25, 40, 45, 50, 55 and 60 base pairs. The maximum E-values tested were as follows: 1-e−1, 1-e−5, 1-e−10 and 1-e−15.
RESULTS
Simulation and annotation
NeSSM produced 2399 077 sequences (length range: 195–459 bp, median length 377 bp). The average phred quality scores remained above 20 until beyond 400 bases (Fig. S1, Supporting Information) and 98.7% of sequences are between 300 and 400 base pairs long (Fig. S2, Supporting Information). Of the 2399 077 sequences, KASS annotated 1 341 362 (55.9%) sequences and WebMGA annotated 1945 674 sequences (81.1%).
Parameters (Blast and MG-RAST)
More stringent parameter values resulted in fewer sequence annotations but had a greater precision; lower values resulted in more annotations being made, but these comprised increases in both correct and incorrect annotations. For example, with cut-off values of 95% and 40%, MG-RAST RefSeq annotated 40.2% and 90.3% sequences, respectively, with incorrect annotation rates of 2.9% and 34.5% at the genus level. This was observed for all parameters tested and for both taxonomic and functional annotations (Figs 1–6). As the taxonomic level moved up the taxonomic hierarchy, more sequences were correctly annotated (e.g. 0.5% and 10.2% for MG-RAST RefSeq with cut-off values of 95% and 40%, respectively, at the class level). Note that sensitivity is unaffected by the taxonomic level investigated.
Figure 1.
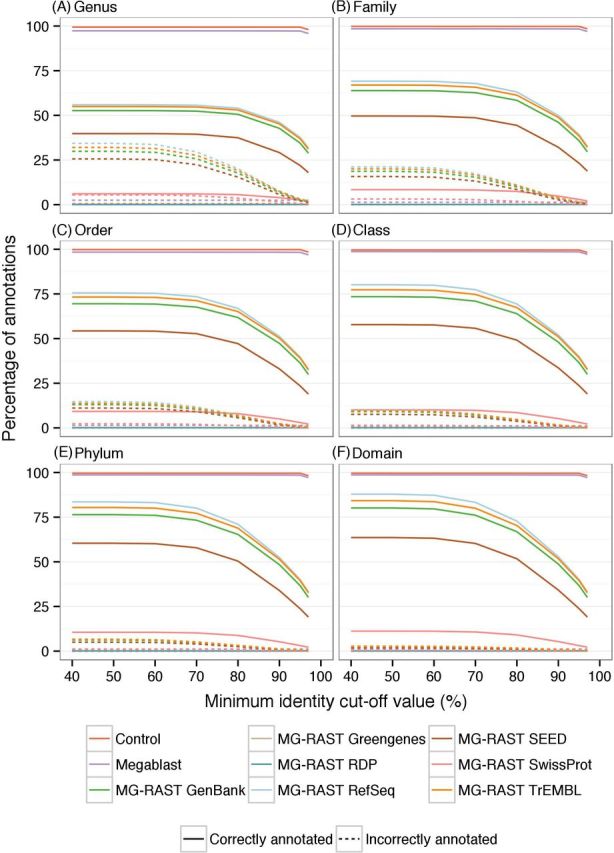
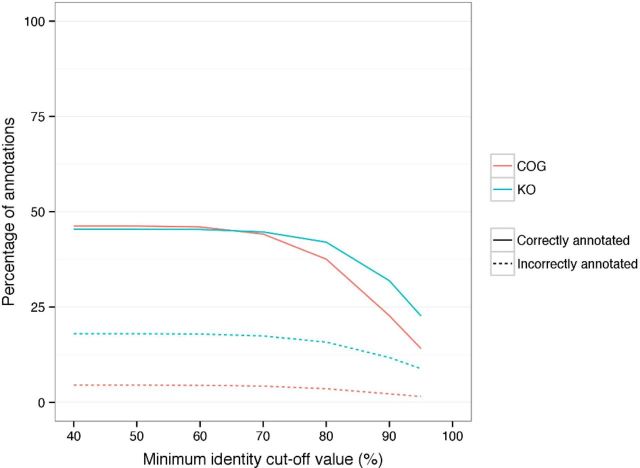
Effect of minimum identity cut-off values on taxonomic annotation. The effect of changing minimum identity cut-off value on the number of sequences correctly and incorrectly annotated across the taxonomic levels.
Figure 6.
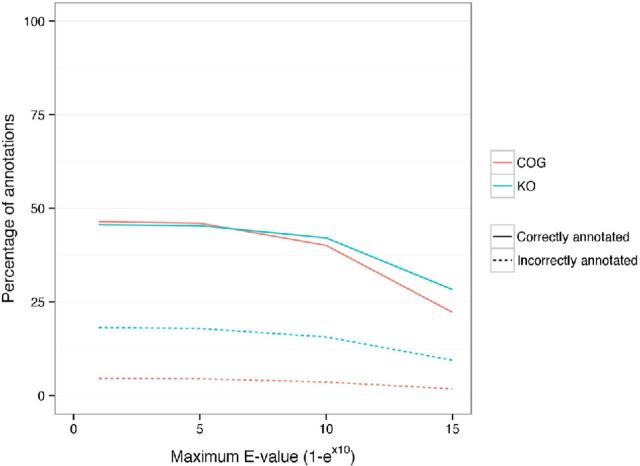
Effect of maximum E-value on functional annotation. The effect of changing maximum E-value value on the number of sequences correctly and incorrectly annotated for functions.
Figure 2.
Effect of minimum identity cut-off values on functional annotation. The effect of changing minimum identity cut-off value on the number of sequences correctly and incorrectly annotated for functions.
Figure 3.
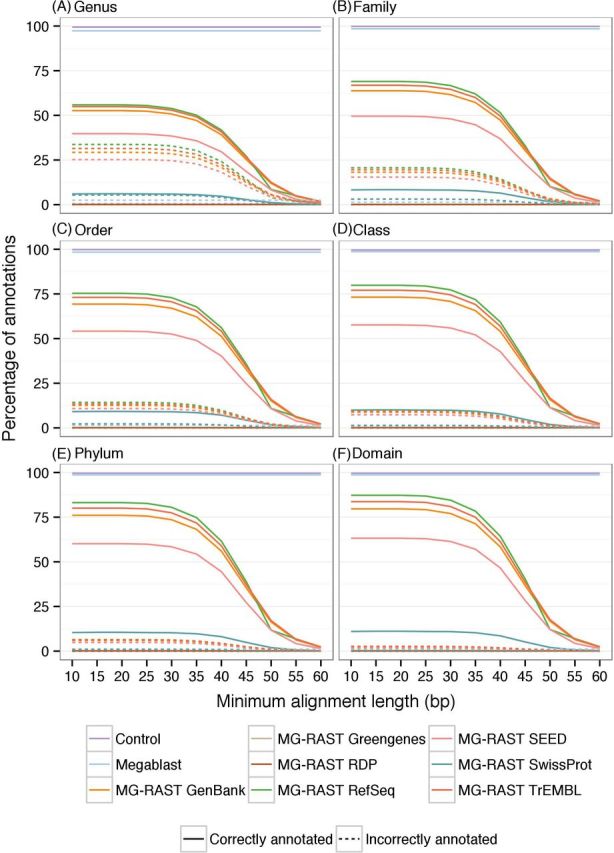
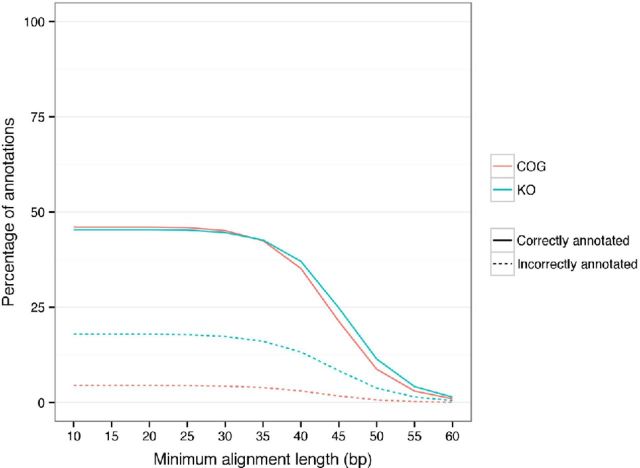
Effect of minimum alignment length on taxonomic annotation. The effect of changing minimum alignment length on the number of sequences correctly and incorrectly annotated across the taxonomic levels.
Figure 4.
Effect of minimum alignment length on functional annotation. The effect of changing minimum alignment length on the number of sequences correctly and incorrectly annotated for functions.
Figure 5.
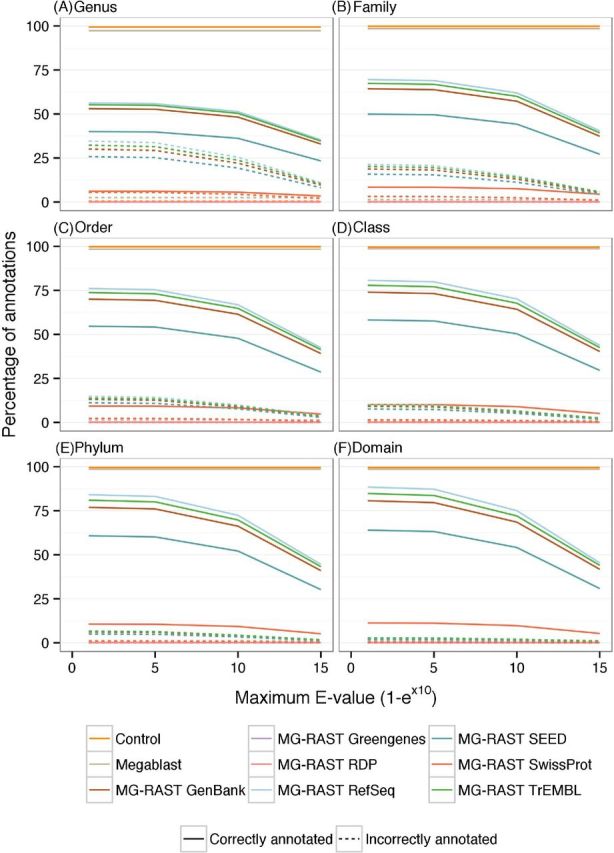
Effect of maximum E-value on taxonomic annotation. The effect of changing maximum E-value on the number of sequences correctly and incorrectly annotated across the taxonomic levels.
The correlation coefficients between the taxa relative abundances in Simmet and in the annotations decreased as parameter stringency increased (Fig. 7, associated scatter plots in Figs S3–S5, Supporting Information). Most databases achieved maximum correlations with a minimum identity cut-off value of 50%, a minimum alignment length of 30 bp and a maximum E-value of 1-e−1. Greater decreases in correlation coefficients occurred with a minimum identity cut-off value above 70% and a minimum alignment length greater than 40 bp.
Figure 7.
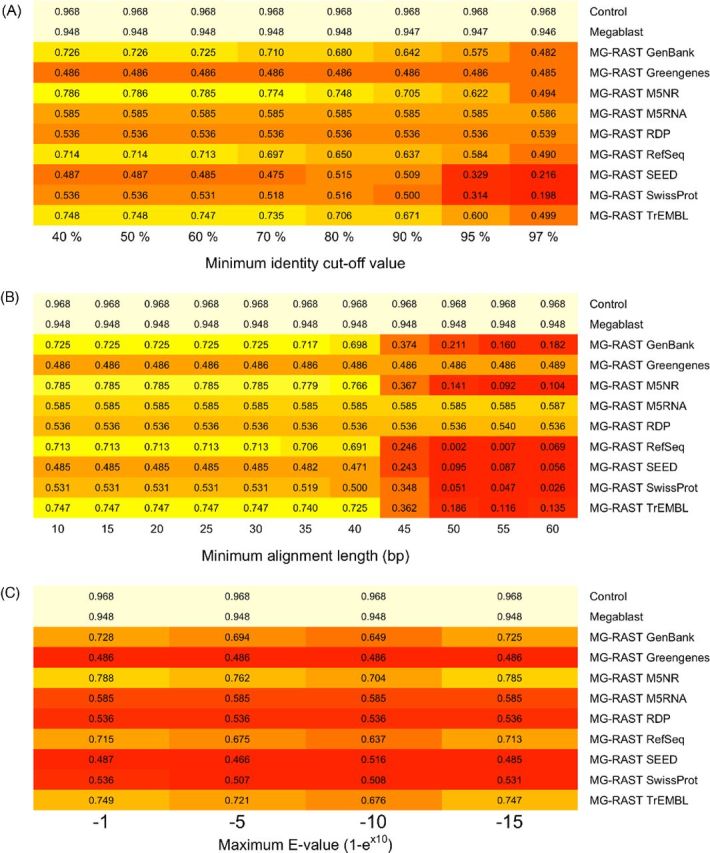
Abundance correlations for different parameter values. The Pearson's product-moment correlation coefficients for the correlations between the genus relative abundances from Simmet and those from various annotation methods using different (A) minimum identity cut-off values, (B) minimum alignment lengths and (C) maximum E-values.
Annotation sensitivity and precision
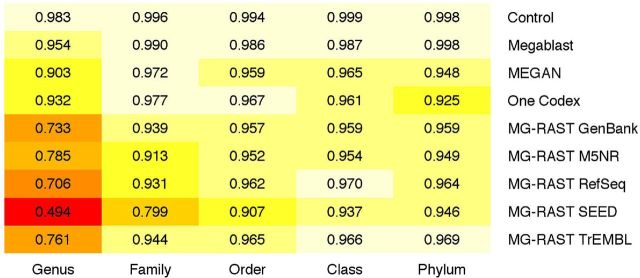
The control taxonomically annotated 99.9% of sequences and had a genus precision of 99.5%. This produced the greatest number of correct annotations (99.4%) (Table 1, Fig. 8, Table S2, Supporting Information, for all taxonomic levels). Megablast annotated 99.8% of sequences and had a genus precision of 97.5%. One Codex annotated all of the sequences, but incorrectly annotated more sequences (5.8%) than MEGAN (2.9%), Megablast (2.5%) and the control (0.5%). Megablast, MEGAN and One Codex correctly annotated 97.3%, 95.7% and 94.2% sequences respectively, significantly more than the next most successful methods: MG-RAST RefSeq (55.9%), MG-RAST TrEMBL (54.9%) and MG-RAST GenBank (52.7%). MG-RAST RDP and Mg-RAST Greengenes, both rRNA databases, annotated less than 1% of the sequences. This is consistent with the expected frequency of rRNA genes within bacterial genomes (Větrovský and Baldrian 2013). As the taxonomic level increases, precision increases and becomes more similar across the different databases.
Table 1.
Taxonomic annotation statistics. The Simmet taxonomic annotation statistics for each method and database at the genus level using default parameters.
| Method | Database | Sensitivity (%) | Correctly annotated (%) | Incorrectly annotated (%) |
|---|---|---|---|---|
| Megablast | Control | 99.88 | 99.39 | 0.49 |
| Megablast | NCBI | 99.81 | 97.32 | 2.49 |
| MEGAN | MEGAN | 98.56 | 95.65 | 2.91 |
| MG-RAST | GenBank | 81.94 | 52.65 | 29.30 |
| MG-RAST | Greengenes | 0.11 | 0.08 | 0.03 |
| MG-RAST | RDP | 0.13 | 0.10 | 0.03 |
| MG-RAST | RefSeq | 89.58 | 55.90 | 33.68 |
| MG-RAST | SEED | 64.97 | 39.75 | 25.22 |
| MG-RAST | SwissProt | 11.49 | 6.08 | 5.42 |
| MG-RAST | TrEMBL | 86.37 | 54.93 | 31.44 |
| One Codex | One Codex | 100.00 | 94.18 | 5.82 |
Figure 8.
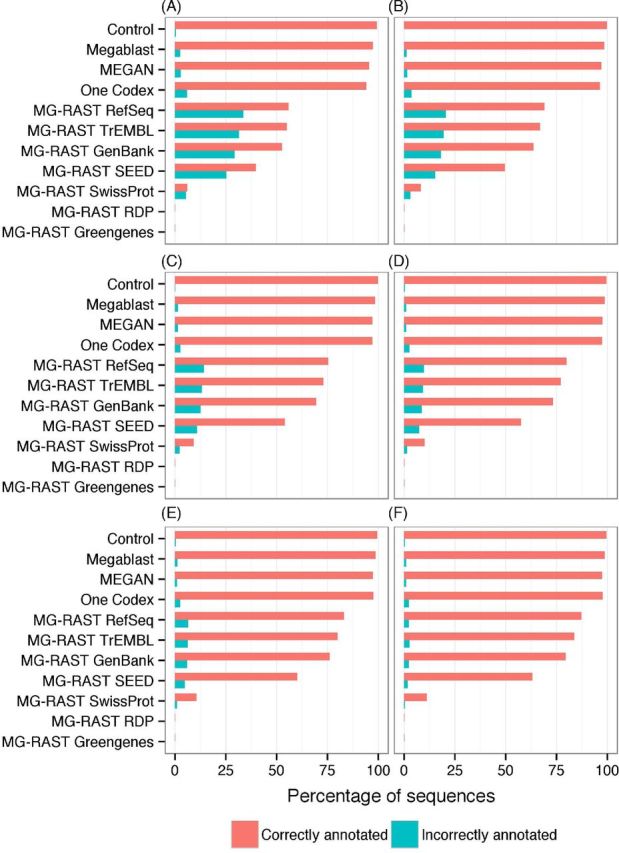
Annotation performance. The annotation sensitivity and number of sequences correctly annotated from a variety of methods and databases across the taxonomic levels investigated.
MG-RAST KEGG annotated 63.3% of the sequences and had a precision of 71.7%, with 45.4% of sequences correctly assigned a function and 17.9% incorrectly assigned a function. MG-RAST COG annotated 50.5% of the sequences and had a precision of 91.1%, resulting in 46.0% of sequence being correctly assigned a function and 4.5% being incorrectly assigned a function (Tables S3–S5, Supporting Information). The portions of sequences correctly annotated by both methods were 81.5% for MG-RAST KEGG and 55.4% for MG-RAST COG.
Taxa abundance correlations
The control showed the greatest genus-level correlation (r2 = 0.98). Megablast had the greatest genus-level correlation with Simmet after the control (r2 = 0.95), while MG-RAST SEED had the weakest (r2 = 0.49). MEGAN and One Codex had genus-level correlations of r2 = 0.90 and r2 = 0.93, respectively. The greatest correlation achieved, aside from the control, was by Megablast at the phylum level (r2 > 0.99) (Fig. 9, Fig. S6, Supporting Information).
Figure 9.
Abundance correlations for different taxonomic levels. The Pearson's product-moment correlation coefficients for the correlations between the relative abundances from Simmet and those from the annotation methods.
MG-RAST M5NR and MG-RAST RefSeq generated 87 and 56 false positive class identifications, respectively (Table 2). MEGAN had only two false positive class identifications (‘Unidentified’ and Insecta) and one false negative identification (Solibacteres). One Codex also had a low abundance of false positive class identifications (eight) and no false negative class identifications. Classes with many false positive identifications include eukaryotes, particularly fungi and bacteria such as Spartobacteria. The greatest fold differences for classes can be found in Table S6 (Supporting Information).
Table 2.
False positive and negative class abundances. The false positive and negative classes from the MG-RAST M5NR, RefSeq, One Codex and MEGAN annotations. The values displayed are the relative abundances.
| Class | Simmet | Annotation |
|---|---|---|
| A.1 The top 10 false positive classes for MG-RAST M5NR | ||
| Erysipelotrichi | 0.0 | 0.00109 |
| Dehalococcoidetes | 0.0 | 0.00100 |
| Ktedonobacteria | 0.0 | 0.00014 |
| Spartobacteria | 0.0 | 0.00013 |
| Mammalia | 0.0 | 0.00012 |
| Insecta | 0.0 | 0.00011 |
| Eurotiomycetes | 0.0 | 0.00009 |
| Sordariomycetes | 0.0 | 0.00009 |
| Saccharomycetes | 0.0 | 0.00008 |
| Liliopsida | 0.0 | 0.00007 |
| A.2 The top 10 false positive classes for MG-RAST RefSeq | ||
| Spartobacteria | 0.0 | 0.00011 |
| Ktedonobacteria | 0.0 | 0.00010 |
| Insecta | 0.0 | 0.00009 |
| Eurotiomycetes | 0.0 | 0.00008 |
| Mammalia | 0.0 | 0.00007 |
| Saccharomycetes | 0.0 | 0.00007 |
| Lentisphaeria | 0.0 | 0.00007 |
| Anthozoa | 0.0 | 0.00005 |
| Amphibia | 0.0 | 0.00005 |
| Zetaproteobacteria | 0.0 | 0.00005 |
| A.3 The false positive classes for One Codex | ||
| Sordariomycetes | 0.0 | 0.00001 |
| Holophagae | 0.0 | <0.00000 |
| Ktedonobacteria | 0.0 | <0.00000 |
| Eurotiomycetes | 0.0 | <0.00000 |
| Leotiomycetes | 0.0 | <0.00000 |
| Dothideomycetes | 0.0 | <0.00000 |
| Nitrospinia | 0.0 | <0.00000 |
| Saccharomycetes | 0.0 | <0.00000 |
| A.4 The false positive classes for MEGAN | ||
| Insecta | 0.0 | 0.00042 |
| B.1 The false negative classes for MG-RAST M5NR | ||
| Dehalococcoidia | 0.00122 | 0.0 |
| Ignavibacteria | 0.00059 | 0.0 |
| Erysipelotrichia | 0.00057 | 0.0 |
| Chthonomonadetes | 0.00055 | 0.0 |
| Phycisphaerae | 0.00049 | 0.0 |
| Caldilineae | 0.00045 | 0.0 |
| Caldisericia | 0.00019 | 0.0 |
| B.2 The false negative classes for MG-RAST RefSeq | ||
| Anaerolineae | 0.00127 | 0.00000 |
| Ignavibacteria | 0.00059 | 0.00000 |
| Chthonomonadetes | 0.00055 | 0.00000 |
| Phycisphaerae | 0.00049 | 0.00000 |
| Caldilineae | 0.00045 | 0.00000 |
| Thermodesulfobacteria | 0.00040 | 0.00000 |
| Caldisericia | 0.00019 | 0.00000 |
| B.3 The false negative classes for One Codex | ||
| NA | ||
| B.4 The false negative classes for MEGAN | ||
| Solibacteres | 0.00111 | 0.0 |
Taxa richness
Six of the annotation methods underestimated the genus richness and six overestimated it (Table 3, Fig. 10). The control perfectly estimated the genus richness. The next closest estimate was achieved by MEGAN (97.7%), followed by MG-RAST SwissProt (95.5%), MG-RAST M5RNA (95.2%), Megablast (110.2%) and MG-RAST RefSeq (118.2%). MG-RAST M5NR produced the most incorrect richness value at 1244 genera (180.8%). One Codex overstated the genus richness by 26.7%. The methods were inconsistent in response to the taxonomic level. With increasing taxonomic level, some estimates increased in accuracy while others decreased (Fig. 10, Table S7, Supporting Information). Excluding the control and the domain level, where the number of taxa is low, MEGAN achieved the most accurate richness value (101.2%) at the family level. MG-RAST M5NR achieved the most inaccurate richness value (253.3%) at the order level. Megablast and One Codex achieved accurate results relative to other methods, but they still overstated taxa richness at every taxonomic level.
Table 3.
Genus richness. The genus richness estimates and the differences from Simmet for each annotation method. Due to the low numbers, domain is excluded from comparisons. Richness values at all taxonomic levels can be found in Table S7 (Supporting Information).
| Method | Database | Richness | Difference (%) |
|---|---|---|---|
| Simmet | N/A | 688 | N/A |
| Megablast | Control | 688 | 100.00 |
| Megablast | Megablast | 758 | 110.17 |
| MEGAN | MEGAN | 672 | 97.67 |
| MG-RAST | GenBank | 1090 | 158.43 |
| MG-RAST | Greengenes | 404 | 58.72 |
| MG-RAST | M5NR | 1245 | 180.96 |
| MG-RAST | M5RNA | 655 | 95.20 |
| MG-RAST | RDP | 469 | 68.17 |
| MG-RAST | RefSeq | 813 | 118.17 |
| MG-RAST | SEED | 445 | 64.68 |
| MG-RAST | SwissProt | 657 | 95.49 |
| MG-RAST | TrEMBL | 1094 | 159.01 |
| One Codex | One Codex | 872 | 126.74 |
Figure 10.
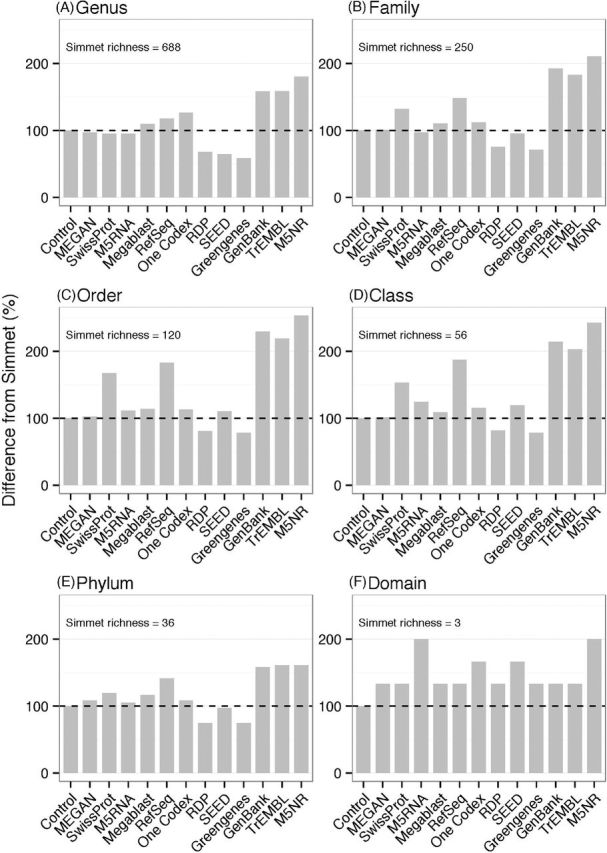
Taxa richness. The differences between annotated richness values and the actual richness value (dashed line) for each taxonomic level.
DISCUSSION
In the study, we evaluated the performances of MEGAN, MG-RAST, One Codex and Megablast by determining their sequence annotation accuracies. All common taxonomic levels above species are studied, building on the work by Lindgreen, Adair and Gardner (2016) who study several tools at the genus and phylum levels. By studying a range of taxonomic levels, we provide a guideline for researchers to establish the annotation accuracy costs of investigating lower taxonomic levels, allowing them to optimize their investigations depending on their requirements for taxonomic resolution. MG-RAST and Megablast use a selection of parameters to determine the stringency of matching a sequence with a reference sequence in a database. Less stringent parameters (i.e. lower minimum identity cut-off values, lower minimum alignment lengths and higher maximum E-values) annotate more sequences, but more incorrect annotations are made, thus producing an incorrect community profile. More stringent parameters reduce the number of incorrect annotations, but many fewer annotations are made, resulting in much of the data being rejected. Shakya et al. (2013) drew similar conclusions for varying minimum identity cut-off values. Decreases in sensitivity generally occur from minimum identify cut-off values above 60%, a minimum alignment lengths greater than 30 bp or a maximum E-value below 1-e−5; therefore, the default values used by MG-RAST maximize sensitivity. According to Carr and Borenstein (2014), the impact of parameters such as E-value will vary depending on read length, something that should be considered in future evaluations as newer sequencing technologies produce longer reads (e.g. nanopore sequencing; Branton et al. 2008). The sensitivities and the number of sequences correctly annotated are relatively low for MG-RAST at the genus and family levels. At the order level, the values are higher, suggesting that this would be the optimum taxonomic level to study, which maximizes the amount of data used without producing too many incorrect annotations. Ultimately, there is a trade-off between taxonomic resolution and annotation accuracy, and this must be considered when determining methods for metagenomic studies.
A marginal number of sequences were not annotated by the control and an even smaller number were incorrectly annotated. These discrepancies are due to the sequencing errors inserted into the simulation. We can therefore conclude that 0.5% of intersample difference at the genus level may be attributed to sequencing error, an important consideration when interpreting data obtained from environmental samples using these methods. This is supported by Hoff (2009) and Carr and Borenstein (2014), who found that increasing error rates decrease gene prediction accuracy. As the error rates of next-generation sequencing technologies improve, this effect will reduce.
One Codex had the greatest annotation sensitivity and the fourth highest annotation precision. This is likely to be due to a combination of the kmer-based annotation method that it uses and that the simulated metagenome was created using the NCBI genome database, the primary reference source for One Codex. Other than the control, Megablast correctly annotated the most sequences at the genus level (97.3%), although the sensitivity of this method was 0.2% less than One Codex. MEGAN had the second highest precision, annotating 98.6% of sequences, with 95.7% correct annotations. This suggests that Megablast is the most reliable method for annotating sequences, and indicates that it is more conservative than One Codex when assigning a sequence hit but also less likely to misidentify a sequence. MEGAN's performance was similar to Megablast, which is expected as MEGAN processed the Megablast output. Discrepancies between the two are therefore derived from MEGAN's processing.
MG-RAST RefSeq had the fifth greatest annotation sensitivity and the greatest of the MG-RAST annotations (excluding MG-RAST M5NR, for which sequence-specific annotation data were unavailable), although it also achieved the greatest number of misidentifications. At the genus level, 33.7% of sequences were misidentified and 55.9% were correctly identified, leaving the remainder unassigned despite the fact that all taxa in Simmet are fully sequenced. This would suggest that investigating metagenomes at the genus level would be unreliable, generating many false positives and implying an incorrect community structure and composition. This supports Garcia-Etxebarria, Garcia-Garcerà and Calafell (2014), who found that more annotations are made at higher taxonomic levels and that discrepancies between known frequencies and annotations increase at lower taxonomic levels, and Lindgreen, Adair and Gardner (2016), who report decreases in community annotation accuracy at the genus level compared to phylum. At the class level, the proportion of incorrect annotations is reduced to fewer than 10% for MG-RAST RefSeq, with 80% being annotated correctly. While taxonomic resolution is reduced, it ensures that the confidence in the annotations remains high.
MG-RAST KEGG correctly annotated a similar number of sequences to MG-RAST COG, but incorrectly annotated many more. KEGG offers a more descriptive annotation as it comprises specific gene and pathway annotations, whereas COG provides descriptions based on orthologous sequences. However, the specificity of KEGG classifications may be the cause of the incorrect annotations as there are more annotations to be selected from and there may be more closely related functions, increasing the chance of misidentification. Because KEGG is now subscription based, and MG-RAST uses the last free version (2008), it will not contain information added after that date. Our results are in line with those produced by Lindgreen, Adair and Gardner (2016), who also conclude that MG-RAST's functional annotation was accurate.
The control, One Codex, Megablast and MEGAN achieved the greatest correlation coefficients between Simmet and annotation abundances at the genus level, all above 0.9. For all MG-RAST annotations, the correlation coefficients were less than 0.8. For MG-RAST, the greatest correlation of all abundances was achieved at the order level by the M5NR database, closely followed by TrEMBL and RefSeq. These correlations inform us about community-wide analyses, but they are not as sequence sensitivity and precision as correlating abundances values may occur from coincidental incorrect annotations.
MG-RAST overannotated many more classes than MEGAN and One Codex, for which the most abundant feature was the unidentified group. This supports the sensitivity and precision data in suggesting that One Codex is more likely to categorize unknown sequences as unidentified, rather than incorrectly identifying them.
The genus richness estimated by MG-RAST M5NR was 81.0% greater that Simmet's actual richness, the highest overstatement, while MEGAN achieved the most accurate genus richness value (2.3% lower) after the control (100.2%). This overstatement could be due to the greater number of sequences present in MG-RAST M5NR. MG-RAST M5RNA produced a relatively accurate estimate of genus richness (95.2%); as M5RNA is a 16S rRNA database, it is unlikely to annotate non-16S rRNA sequences, reducing the number of incorrect identifications. However, the taxa abundance correlations show that MG-RAST M5RNA achieved the second lowest correlation with Simmet at the genus level, and the lowest at all other taxonomic levels. MG-RAST RefSeq generated the fifth most accurate richness value, greater than One Codex, although not as accurate as Megablast and MEGAN. Combined with its high abundance correlation with Simmet, this suggests that MG-RAST RefSeq provides a relatively accurate representation of both the richness of a community and the abundance of organisms present. MEGAN and One Codex achieve more accurate taxa richness values and taxa abundance correlations than MG-RAST RefSeq at the family level and above, suggesting they would be a viable alternative to MG-RAST RefSeq.
One limitation with evaluating annotations using organism nomenclature, rather than taxon IDs (which were unavailable for MG-RAST sequence-specific annotation data), is the lack of taxonomic metadata curation in some databases. Some genomes in the NCBI database are stored with the abbreviated species name rather than complete name, thus Amycolatopsis mediterranei would not automatically be identified as an Amycolatopsis species. Furthermore, as names are updated, disparities can form between different databases. For example, the class Chloroflexia has been renamed to Chloroflexi, and is called this by MG-RAST. However, NCBI is using the old name Chloroflexia (as of 17/11/14), thus sequences identified as Chloroflexi would not be correctly matched in Simmet. These issues were corrected for during data processing; however, there may be other cases of disparities in the plethora of organisms present in the analysis. A solution to this would be to use the taxon IDs instead; however, these were not available for sequence-specific annotations downloaded from MG-RAST.
In conclusion, we found that One Codex, Megablast and MEGAN are suitable methods for annotating DNA sequences that are located in the reference databases that they use for annotation, with One Codex offering fast, web-based analyses and MEGAN providing a user-friendly Graphical User Interface to analyze BLAST results. Results appear to vary significantly depending on the program and parameters used, a conclusion also drawn by Lindgreen, Adair and Gardner (2016). While MG-RAST appears to have a greater rate of incorrect assignments, this is reduced when investigating higher taxonomic levels (e.g. with RefSeq: over 33% at the genus level compared to less than 15% and 10% at the order and class levels). The correlations between the annotated taxa abundances are greatest for MG-RAST at the order level, using M5NR, TrEMBL or RefSeq. In many of the tests, MG-RAST M5NR proved to be a reliable database, but the diversity indices suggest that it is less reliable than MG-RAST RefSeq; at the class, order and family levels, MG-RAST M5NR estimates more the double the actual richness values. Therefore, we hypothesize that MG-RAST M5NR would generate more false positive sequence annotations than MG-RAST RefSeq.
A simulated metagenome allows for the quantification of annotation errors. This study compliments the work by Mavromatis et al. (2007), who evaluated different metagenomic processing methods using simulated metagenome developed from 113 isolated genomes, and by Pignatelli and Moya (2011), who used simulated data to study the performances of de novo short-read assembly programs. It should be noted that the performances of the methods discussed in this study are likely to differ from the reported results when annotating environmental sequence data; a greater number of sequences are likely to be unidentified due to the multitude of uncultured microorganisms (Streit and Schmitz 2004) and non-sequenced microbial genomes (Tringe et al. 2005) that are currently absent from the NCBI whole bacterial genome database. While this research focused on a selection of annotation methods, the overall conclusions drawn should be considered for any pipeline.
In this study, we highlight and quantify the annotation errors for a selection of parameters and databases. We show that analysis pipelines are not equivalent and certain parameters can significantly reduce the confidence in results. These findings should be used as a guideline when determining methods for annotating metagenomic sequences and considered when interpreting metagenomic results. Ultimately, the most appropriate balance between taxonomic resolution, annotation sensitivity and annotation precision needs to be identified for each study conducted.
Supplementary Material
Acknowledgments
We sincerely thank Sandy Macdonald and Tobias Hodges for their technical support and assistance with data analysis techniques. Sandy in particular provided advice and guidance throughout the study that ultimately contributed to the work produced.
SUPPLEMENTARY DATA
FUNDING
This work was supported by the and joint seedcorn Ph.D. studentship.
Conflict of interest. None declared.
REFERENCES
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–40. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Anderson RE, Sogin ML, Baross JA. Biogeography and ecology of the rare and abundant microbial lineages in deep-sea hydrothermal vents. FEMS Microbiol Ecol. 2015;91:1–11. doi: 10.1093/femsec/fiu016. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, Boucher Y. Epistemological Impacts of Horizontal Gene Transfer on Classification in Microbiology. In: Gogarten DMB, Gogarten DJP, Olendzenski DLC, editors. Horizontal Gene Transfer. New York City, USA: Humana Press; 2009. pp. 55–72. [DOI] [PubMed] [Google Scholar]
- Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Carr R, Borenstein E. Comparative analysis of functional metagenomic annotation and the mappability of short reads. PLoS One. 2014;9:e105776. doi: 10.1371/journal.pone.0105776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Lee J-H, Jung Y, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Micr. 2007;57:2259–61. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Antonopoulos D, Gilbert JA, et al. From genomics to metagenomics. Curr Opin Biotech. 2012;23:72–6. doi: 10.1016/j.copbio.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Fournier P-E, Dumler JS, Greub G, et al. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Etxebarria K, Garcia-Garcerà M, Calafell F. Consistency of metagenomic assignment programs in simulated and real data. BMC Bioinformatics. 2014;15:90. doi: 10.1186/1471-2105-15-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–9. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–87. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Henry VJ, Bandrowski AE, Pepin A-S, et al. OMICtools: an informative directory for multi-omic data analysis. Database. 2014;2014:bau069. doi: 10.1093/database/bau069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ. The effect of sequencing errors on metagenomic gene prediction. BMC Genomics. 2009;10:520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:reviews0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CI, Mitchell A, Jones P, et al. Metagenomic analysis: the challenge of the data bonanza. Brief Bioinform. 2012;13:743–6. doi: 10.1093/bib/bbs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–86. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Xie C. A poor man's BLASTX–high-throughput metagenomic protein database search using PAUDA. Bioinformatics. 2014;30:38–9. doi: 10.1093/bioinformatics/btt254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Xuan L, Cai K, et al. NeSSM: a next-generation sequencing simulator for metagenomics. PLoS One. 2013;8:e75448. doi: 10.1371/journal.pone.0075448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NA, Fass JN. 2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software] https://github.com/najoshi/sickle (18 May 2016, date last accessed) [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–9. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep. 2016;6:19233. doi: 10.1038/srep19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson DA, Haggerty JM, Srinivas A, et al. Metagenomic insights into anaerobic metabolism along an Arctic peat soil profile. PLoS One. 2013;8:e64659. doi: 10.1371/journal.pone.0064659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova N, Barry K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Mende DR, Sunagawa S, Zeller G, et al. Accurate and universal delineation of prokaryotic species. Nat Methods. 2013;10:881–4. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, et al. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M, Moya A. Evaluating the fidelity of de novo short read metagenomic assembly using simulated data. PLoS One. 2011;6:e19984. doi: 10.1371/journal.pone.0019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. P Natl Acad Sci USA. 2009;106:19126–31. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Shakya M, Quince C, Campbell JH, et al. Comparative metagenomic and rRNA microbial diversity characterization using Archaeal and Bacterial synthetic communities. Environ Microbiol. 2013;15:1882–99. doi: 10.1111/1462-2920.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–9. [Google Scholar]
- Streit WR, Schmitz RA. Metagenomics–the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492–8. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. Microb Inform Exp. 2012;2:3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke A, Glass EM, Bischof J, et al. MG-RAST Technical Report and Manual v3.3.6 r1. Argonne National Laboratory: University of Chicago; 2013. [Google Scholar]
- Wilke A, Harrison T, Wilkening J, et al. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformatics. 2012;13:141. doi: 10.1186/1471-2105-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. P Natl Acad Sci USA. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, et al. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Wang F, Guo L, et al. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 2011;5:414–26. doi: 10.1038/ismej.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–6. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.