Highlight
The barley GTPase RACB is a disease susceptibility factor. However, RACB is not a key regulator of plant immune responses but acts in polar cell development.
Key words: Blumeria graminis, disease susceptibility, epidermis, MAP kinase, nucleus, oxidative burst, polarity, ROP GTPase.
Abstract
RHO GTPases are regulators of cell polarity and immunity in eukaryotes. In plants, RHO-like RAC/ROP GTPases are regulators of cell shaping, hormone responses, and responses to microbial pathogens. The barley (Hordeum vulgare L.) RAC/ROP protein RACB is required for full susceptibility to penetration by Blumeria graminis f.sp. hordei (Bgh), the barley powdery mildew fungus. Disease susceptibility factors often control host immune responses. Here we show that RACB does not interfere with early microbe-associated molecular pattern-triggered immune responses such as the oxidative burst or activation of mitogen-activated protein kinases. RACB also supports rather than restricts expression of defence-related genes in barley. Instead, silencing of RACB expression by RNAi leads to defects in cell polarity. In particular, initiation and maintenance of root hair growth and development of stomatal subsidiary cells by asymmetric cell division is affected by silencing expression of RACB. Nucleus migration is a common factor of developmental cell polarity and cell-autonomous interaction with Bgh. RACB is required for positioning of the nucleus near the site of attack from Bgh. We therefore suggest that Bgh profits from RACB’s function in cell polarity rather than from immunity-regulating functions of RACB.
Introduction
Plants possess an innate immunity, which constantly monitors the cell surface and cytoplasm for the presence or activity of pathogenic organisms. Plant immune receptors can detect conserved molecular patterns that derive from microbes (MAMPs, microbe-associated molecular patterns) or from host cell damage (damage-associated molecular patterns). Such receptors are called pattern recognition receptors (PRRs). They are localized in the host plasma membrane and function in basal resistance to non-adapted and virulent pathogens (Macho and Zipfel, 2014). Additionally, a second class of plant immune receptors co-evolved with specific, largely polymorphic virulence effectors (Jones and Dangl, 2006). Most of these so-called resistance (R) proteins are localized in the cytoplasm and nucleoplasm. For triggering immunity, R proteins directly interact with effector proteins, monitor functionality of effector targets, or mimic effector targets (Dodds and Rathjen, 2010). Plant immunity is robust in most environments. Nevertheless, microbes adapt to plant hosts by evolution of virulence effectors that suppress or circumvent host immunity.
Plant disease resistance can be observed as a result of MAMP- or effector-triggered immunity but also as a consequence of mutations of susceptibility genes. Susceptibility genes encode host factors that are required for pathogenesis in interactions of susceptible hosts with adapted virulent pathogens. Mechanistically, loss of susceptibility can result from de-regulated or primed immunity if susceptibility genes code for negative regulators of plant defence. Alternatively, loss of susceptibility can be explained by lack of or mutation of effector targets that serve demands of the pathogen other than suppressing immunity. Furthermore, susceptibility factors might provide developmental or metabolic prerequisites for attraction, accommodation, or feeding of the pathogen (Hückelhoven et al., 2013; Lapin and Van den Ackerveken, 2013). Loss of susceptibility is usually recessively inherited after loss of gene function and often accompanied by pleiotropic effects that limit applicability in plant breeding (Pavan et al., 2010; Hückelhoven et al., 2013; van Schie and Takken, 2014). Thus a deeper understanding of susceptibility is required to inform plant breeding.
Plant monomeric RHO GTPases (rat sarcoma homologues, also called RAC for rat sarcoma-related C3 botulinum toxin substrate or ROP for RHO of plants) are involved in immunity and susceptibility to plant diseases. Type I RAC/ROPs possess a typical motif for post-translational prenylation at their C-terminus, and can be additionally palmitoylated after activation. In contrast, type II RAC/ROPs are often constitutively S-acylated (Yalovsky, 2015). In rice, the type II RAC/ROP protein RAC1 is a central regulator of immune response mediated by either PRRs or R proteins (Kawano et al., 2014). The rice chitin elicitor PRR CERK1 can activate RAC1 via a plant-specific guanine nucleotide exchange factor RACGEF1. RAC1 orchestrates the elicitor-activated production of reactive oxygen species (ROS), mitogen-activated protein (MAP) kinase (MAPK) activation, transcriptional responses, and changes of the host proteome (Kawano et al., 2014). Additionally, RAC1 is activated downstream of the R protein Pit, a nucleotide-binding leucine-rich repeat protein, which confers effector-triggered immunity to the rice blast fungus Magnaporthe oryzae (Kawano et al., 2010). However, in rice and barley, there are also other type I and type II RAC/ROP GTPases that limit basal resistance or support susceptibility to fungal diseases (Chen et al., 2010; Schultheiss et al., 2002, 2003). Barley RACB, a type I RAC/ROP, is required for full susceptibility to barley powdery mildew caused by the biotrophic ascomycete Blumeria graminis f.sp. hordei (Bgh). Transient or stable gene silencing of RACB by RNAi limits fungal entry and formation of fungal haustoria in barley epidermal cells (Schultheiss et al., 2002; Hoefle et al., 2011). Molecular cell biology suggests a role for RACB in organization of the cytoskeleton (Opalski et al., 2005; Hoefle et al., 2011; Huesmann et al., 2012). Transgenic expression of constitutively activated (CA) RACB (RACB G15V) enhances powdery mildew susceptibility, supports establishment of haustoria in barley epidermal cells, but has little effect on cellular defence reactions. When overexpressed in single epidermal cells or transgenic plants, barley type II CA RAC/ROPs (RAC1, RAC3, and ROP6) can support susceptibility to powdery mildew too, whereas RACD, another type I RAC/ROP, appears not to influence the outcome of interaction with Bgh (Schultheiss et al., 2003; Pathuri et al., 2008). Little is known about the function of barley RAC/ROPs in interaction with other microbes. However, when ectopically expressed, barley CA RACB and CA RAC3 can support susceptibility to Pseudomonas syringae pv. tabaci in tobacco, and barley CA RAC1 can support penetration resistance to hemibiotrophic M. oryzae in transgenic barley (Pathuri et al., 2008, 2009).
RAC/ROPs function in plant cell polarity. This is well established for root hair tip growth, pollen tube tip growth, and epidermal pavement cell interdigitation (Yang, 2008). In particular, type I RAC/ROP GTPases of dicots function in cell polarity. Little is known about RAC/ROP functions in polar cell growth in monocots. It has been described that barley CA RACB enhances epidermal cell size in leaves. Root hair phenotypes of transgenic CA RACB barley were reported as root hair swelling on solid medium, which is typical in dicots expressing CA ROP genes (Jones et al., 2002; Pathuri et al., 2008, 2009). In contrast, silencing of RACB by RNAi in transgenic barley led to a defect in the ability to form root hairs (Hoefle et al., 2011). In maize, the development of stomatal complexes was reported to depend partially on ROP2 and ROP9, two type I RAC/ROP proteins very similar to barley RACB (Humphries et al., 2011).
It has not been studied whether RACB interferes with pattern-triggered immunity or defence gene expression in response to Bgh. Here, we show that RACB does not limit early MAMP-triggered immune responses and supports rather than limits expression of defence genes. However, knock down of RACB strongly affects polar cell growth and positioning of the nucleus in barley epidermal cells. Bgh may hence profit from functions of RACB in cell polarity during invasion of host cells.
Materials and methods
Plant material and growth conditions
For all experiments, the barley (Hordeum vulgare) cultivar Golden Promise and transgenic RACB plants with the genetic background of Golden Promise were used. The overexpressor line of CA RACB 17/1-11 and RACB RNAi 16/2-4B and 15/1-16 have been described previously (Schultheiss et al., 2005; Hoefle et al., 2011). Kernels were surface-sterilized in 20ml of sterilization solution (4% NaOCl, Tween-20) for 1.5h with shaking. After washing with H2O for 30min, husks were carefully removed without damaging the embryo to guarantee equal germination of seeds. Seeds were pre-germinated on wet filter paper for 2 d in the dark before being sown into soil (Typ ED73, Einheitserde- und Humuswerke, Gebr. Patzer GmbH & Co KG, Sinntal-Jossa, Germany). Plants were grown in a growth chamber (Conviron, Winnipeg, Canada) at 18 °C with relative humidity of 65% and a photoperiod of 16h. Both transgenic genotypes do not produce homozygous offspring. Offspring of transgenic T3 donor plants were genotyped according to previous studies to separate transgenic offspring carrying the T-DNA from azygous offspring that lost the T-DNA due to segregation. Azygous sister plants are similar to the wild type (WT; Schultheiss et al., 2005; Hoefle et al., 2011) and thus served as ideal controls. Arabidopsis thaliana ecotype Columbia 0 (Col-0) seeds were purchased from Lehle Seeds (Round Rock, USA) and stratified for 2^d at 4 °C before placing into a growth chamber. Plants were grown at 22 °C with a photoperiod of 10h and a relative humidity of 65%.
Elicitors
The flagellin elicitor flg22 (Felix et al., 1999) was synthesized as described before (Ranf et al., 2011). Chitin from shrimp shells (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) was ground to a fine powder and suspended in H2O (20mg ml–1). Insoluble chitin fragments were removed by centrifugation (1900 g, 10min) and the supernatant was used for experiments.
Immunoblot analysis
For detection of activated MAPKs, we used 10 leaf discs of 5mm diameter from second leaves of 14-day-old barley plants or from 6-week-old Arabidopsis plants per time. Leaf discs were incubated in 2ml of H2O/well for 16h in 24-well plates, transferred to fresh H2O for 30min, and subsequently elicited with 1 µM flg22 or 100 µg ml–1 chitin. Detection of activated MAPKs with anti-pTEpY (α-phospho-p44/42-ERK, Cell Signaling Technology, Boston, USA) was performed as previously described (Saijo et al., 2009; Ranf et al., 2011).
Detection of ROS production of barley leaves
ROS production was assayed by H2O2-mediated oxidation and luminescence of the luminol derivative L-012 (Wako Chemicals GmbH, Neuss, Germany). Leaf discs (5mm diameter) of 7-day-old barley plants were floated in 200 µl of H2O/well overnight in a 96-well plate. After removal of H2O, leaf discs were incubated for 30min in 2 µg ml–1 horseradish peroxidase (HRP) and 10 µM L012. Subsequently, leaf discs were elicited with 100nM flg22 or 100 µg ml–1 chitin. Luminescence was measured at 1min intervals with a Tecan Reader (infinite M200, Tecan, Männedorf, Switzerland) for 30min. We calculated relative luminescence units (RLU) by subtraction of leaf disc-specific background (recorded for 5min before elicitation) and of mock treatment-associated blanks.
Quantitative reverse transcription PCR
Gene expression analysis was carried out by reverse transcription quantitative real-time PCR (RT–qPCR) in a Mx3005P cycler (Agilent Technologies, Santa Clara, CA, USA) using the Maxima SYBR Green qPCR master mix (2×) (Thermo Fisher Scientific, St. Leon-Rot, Germany). Reactions were performed in duplicate with 10ng of cDNA and 330nM forward and reverse primer each in a final volume of 10 µl. Expression values of defence genes and RACB were normalized to a barley housekeeping ubiquitin (HvUBI) (Ovesna et al., 2012) using primer efficiency correction as suggested by Pfaffl (2001). The program consisted of an initial step at 95 °C for 10min and 95 °C for 30s, followed by 40 cycles at 55 °C for 30s and at 72 °C for 1min. The melting curve analysis was performed at 55–95 °C. All primers (Table 1) were designed using Primer3 software (Untergasser et al., 2012) and were checked for specificity using the Basic Local Alignment Search Tool (BLAST) and therein with nucleotide blast against the H. vulgare database (http://blast.ncbi.nlm.nih.gov), and amplicon size assessment in agarose gels before running RT–qPCR.
Table 1.
Oligonucleotides for RT–qPCR
| Gene | Accession number | Forward primer (5'→3')/reverse primer (5'→3') | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|---|
| HvUBI | AK252410, M60175 | TCTCGTCCCTGAGATTGCCCACAT/TTTCTCGGGACAGCAACACAATCTTCT | 58 | 263 |
| HvPR1 | Z26333 | AAGCTGCAAGCGTTCGCC/AGGTGTTGGAGCCGTAGTC | 60 | 184 |
| HvPR3 | AK364132 | CTACACGTACGACGCCTTCAT/GTGGCCTTGCTTATCTCTTCC | 60 | 194 |
| HvPR5 | AK371265 AJ001268 | CACGGACATCACCAAGGATT/TTGCCCTTGAAGAACATTGAG | 60 | 152 |
| HvPR10 | AK360974 | AGGGCGACAAGGTAAGTGG/CATCTTGAGCAGGTCGAGGTA | 60 | 181 |
| HvJIP23 | AB251339 | TGTTGCAGACTATGCCATGAA/TGCCAATCGTTGTACTTAGCC | 60 | 167 |
| HvJIP60 | AK372562 | TTCTTCTTCCGGGCTGTAAAT/GTACGCTGAGCTACCCAGACA | 60 | 150 |
| HvRACB | AJ344223 | TGCACCAGGTGTGCCTATTATC/CTTCGCCCTTGTTCTTTGTC | 60 | 309 |
Scanning electron microscopy
For scanning electron microscopy (SEM), root and leaf material was harvested and fixed in 4% paraformaldehyde (4% PFA) in 1× phosphate-buffered saline buffer (1× PBS), pH 7.4 as described (Sauer and Friml, 2010). Fixed material was washed in 1× PBS, pH 7.4 three times for 10min, followed by three washing steps in distilled water for 10min. Dehydration occurred in an increasing ethanol series of 25% (v/v), 50% (v/v), and 75% (v/v) in distilled water and pure ethanol three times each for at least 10min. For critical point drying, the EM CPD300 Automated Critical Point Dryer (Leica, Vienna, Austria) was used and drying was done following the ‘Rice Root’ protocol for root tissue and the ‘Tobacco Leaf’ protocol for barley leaf material as described in the manufacturer′s manual. The imaging was done using the TM300 Tabletop Microscope (Hitachi, Tokyo, Japan). The image editing program GIMP 2.8 was used to merge single root pictures to generate root overviews and to colour subsidiary cells in the leaf images.
Fluorescence microscopy of root tissue
Seedling roots were harvested, fixed, and washed as described above. For staining, propidium iodide (PI; Applichem, Darmstadt, Germany) was dissolved in distilled water to a final concentration of 100 µg ml–1 for RACB RNAi root material and 40 µg ml–1 for the azygous control plants. The root material was incubated in the staining solution for 1h in the dark. Subsequently, stained roots were transferred to a clearing solution, prepared by mixing chloral hydrate, glycerol, and water in the ratio 4:1:2 (w/v/v) and kept there for 15h. After clearing, the roots were directly mounted in Hoyer’s solution consisting of 1g of glycerol, 10g of chloral hydrate, and 1.5g of gum arabic dissolved in 2.5ml of distilled water. Visualization followed immediately using a Leica TCS SP5 Confocal Microscope and the Leica LAS AF software (Leica Microsystems, Mannheim, Germany). PI was excited by a 561nm laser line and emission was detected from 560nm to 675nm.
Measurement of the nucleus attraction index
At 8h after inoculation, the leaf material was harvested and halved along the longitudinal axis using a razor blade. One half of the leaf blade was used for relative quantification of RACB expression. Leaf pieces were fixed, de-waxed, and destained (Sauer and Friml, 2010). For the last rehydration step, 1× PBS, pH 7.4 was used. To remove RNA from the tissue, RNase A (DNase free, Applichem, Darmstadt, Germany) was dissolved in 10mM Tris–HCl, pH 7.5 to a final concentration of 10mg ml–1. The stock solution was subsequently diluted in 1× PBS, pH 7.4 to 100 µg ml–1 to achieve the RNase A solution in which leaf material was incubated for 1h for RNA digestion. Subsequently the leaves were placed in the staining solution (100 µg ml–1 PI in distilled water) for at least 5min. To determine the nucleus attraction index (NAI), epidermal B cells (Koga et al., 1990), which were attacked by a single fungal appressorium, were imaged. A z-stack was recorded starting from the brightest fluorescence of the fungal appressorium to the brightest fluorescence of the plant nucleus. The picture number and increments were adjusted for each cell and z-stack, depending on the vertical distance between the appressorium and plant nucleus. The NAI was calculated as follows: where a reflects the depth of the z-stack and b the planar distance between the appressorium and the nucleus. Both represent the legs of a right-angled triangle. The diagonal of the B cell is represented by d. Cell size measurement was performed using the software ImageJ (Schneider et al., 2012).
Rhodamine 123 staining and trichoblast quantification
Rhodamine 123 (R123) selectively stains mitochondria in living cells (Wu, 1987). A stock solution was prepared by dissolving R123 (Sigma-Aldrich, St Louis, MO, USA) in DMSO to a final concentration of 10mg ml–1 For the staining solution, the stock solution was diluted in 0.5× Murashige and Skoog medium with modified vitamins (0.5× MS; Duchefa Biochemie, Harleem, The Netherlands) mixed with sucrose to 1% (w/v) and 2-(N-morpholino)ethanesulphonic acid (MES; Carl Roth, Karlsruhe, Germany) to 0.05% (w/v) final concentration, pH 5.6 to 1 µg ml–1. Intact seedlings were incubated in the staining solution for 10min in the dark. After staining, seedlings were briefly rinsed in an excess of 0.5× MS, pH 5.6 and immediately visualized by confocal microscopy. R123 was excited by a 488nm laser line and the emission was detected from 515nm to 575nm. In an early developmental state, trichoblasts were counted before root hair initiation each in an area of 0.024mm2.
Results
RACB does not control early MAMP responses in barley
Specific RAC/ROP GTPases modulate immune responses and NADPH oxidase-dependent ROS production in plants. Therefore, we tested barley WT Golden Promise and corresponding transgenic barley plants silenced for RACB by RNAi or overexpressing CA RACB for their ability to respond to MAMPs by production of ROS. Transgenic lines used have been validated before as being silenced or overexpressors, respectively, and are representatives of several (CA RACB) or two (RACB RNAi) independent lines with consistent transgene-associated phenotypes (Schultheiss et al., 2005; Hoefle et al., 2011). The barley type I RAC/ROP gene RACD is co-silenced in the RACB RNAi line, whereas other RAC/ROPs show WT-like expression (Hoefle et al., 2011).
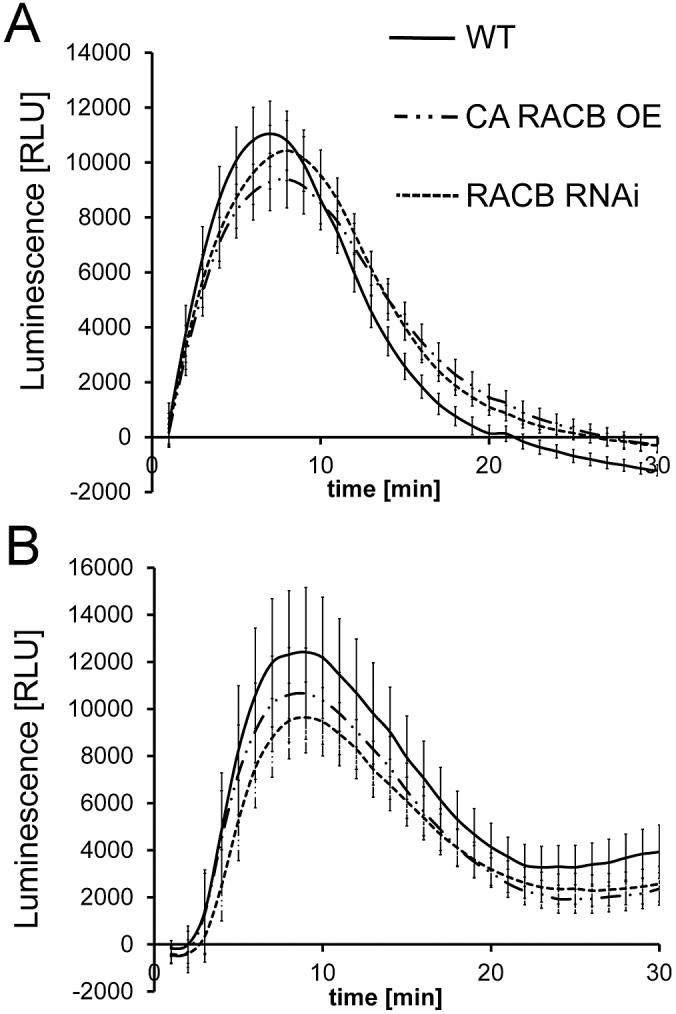
WT barley plants showed a typical MAMP-triggered oxidative burst when challenged with a chitin elicitor preparation. No ROS burst was recordable in mock-treated plants. After elicitor treatment, barley leaf discs rapidly produced ROS. The kinetics and amount of ROS produced appeared similar in WT, CA RACB, and RACB RNAi barley (Fig. 1A). To test whether RACB might influence the oxidative burst elicited by a fungus-unrelated MAMP, we included the bacterial flagellin-derived elicitor flg22 in our experiments (Felix et al., 1999). The flg22 peptide elicited an oxidative burst that was, when taking variance of biological repetitions into account, indistinguishable between WT, CA RACB, and RACB-RNAi barley (Fig. 1B).
Fig. 1.
MAMP-triggered ROS burst is unaffected by RACB transgenes. (A) Chitin- (100 µg ml−1) triggered ROS in barley is unaffected by overexpression of CA RACB (RACB OE) or by silencing RACB (RACB RNAi). (B) Flagellin- (100nM flg22) triggered ROS in barley is unaffected by CA RACB (RACB OE) or by silencing RACB (RACB RNAi). Elicitors were added to leaf discs at 0min and ROS-dependent luminol luminescence recorded over 30min. Data show relative luminescence units (RLU) that have been corrected by subtraction of leaf disc-specific background (recorded for 5min before elicitation) and mock treatment-associated blanks (average of eight leaf discs). Error bars show the SE over the mean of four (A) or three (B) experiments each with eight elicited leaf discs per genotype.
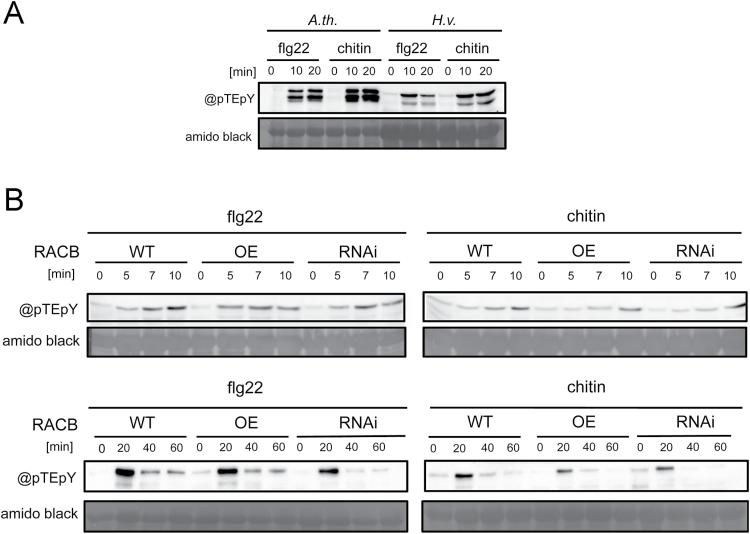
MAPK activation is another typical early MAMP response and potentially modified by plant RAC/ROPs. Activation of MAPKs can be detected by immunodetection of a phosphorylated MAPK-typical TEY motif (pTEpY), such as that present in Arabidopsis MPK3 and MKP6 (Ranf et al., 2011). We detected phosphorylated MAPKs (MAPK-P) in Arabidopsis and barley leaf discs treated in parallel either with chitin or with flg22 (Fig. 2A). In Arabidopsis, typical phosphorylation of MAPKs was detected after elicitation with chitin or flg22. Both elicitors induced a similar pattern of activated MAPK-P in protein extracts from barley leaf discs. However, one or two bands appeared predominant in most experiments, whereas in Arabidopsis extracts two to three bands were detected. This suggested that barley reacts to MAMPs with activation of MAPKs. We then compared patterns of MAPK-P in WT barley, CA RACB barley, and RACB RNAi barley (Fig. 2B). This revealed, at the given level of detection, similar MAPK activation in all three genotypes, with a slight increase visible after 5min and a decline between 20min and 40min after elicitation. This suggests that activity and abundance of RACB do not strongly influence barley competence to react to MAMPs with typical early MAMP responses.
Fig. 2.
MAMP-triggered MAPK phosphorylation is unaffected by RACB transgenes. (A) Elicitation of Arabidopsis (A.th.) or barley (H.v.) leaf discs induced phosphorylation of similar MAPKs as detected by an antibody (@pTEpY) that detects the phosphorylated TEY motif in MAPKs (Ranf et al., 2011) (B) Chitin- (100 µg ml−1) or flagellin- (100nM flg22) triggered MAPK phosphorylation in barley is unaffected by overexpression of CA RACB (OE) or by silencing RACB (RNAi). Elicitors were added to leaf discs at 0min and proteins extracted at 5, 7, and 10min or at 20, 40, and 60min.
We further studied pathogenesis-related (PR) PR1, PR3, PR5, and PR10 gene expression after high density inoculation with Bgh, because the strength of PR gene expression has been linked to penetration resistance in barley (Peterhänsel et al., 1997; Molitor et al., 2011). We further wanted to test whether RACB possibly acts as a negative regulator of PR or jasmonate-associated gene expression (jasmonate-induced genes JIP23 and JIP60) (Kogel et al., 1995). We compared gene expression after mock inoculation and at 12h and 32h after inoculation (HAI), because these times represent stages of fungal penetration attempts and haustorium expansion, and both processes are influenced by RACB (Hoefle et al., 2011). PR genes showed an enhanced expression level in supersusceptible CA RACB barley 12h after mock inoculation. Conversely, PR genes were underexpressed 12h after mock inoculation in less susceptible RACB RNAi barley when compared with the WT (Fig. 3A). At 32h, similar and partially stronger de-regulation of PR genes was observed in RACB-transgenic barley. However, this was not statistically significant for each individual PR gene (Fig. 3A). All four PR genes were up-regulated after inoculation with Bgh in the WT. At 12 and 32 HAI, CA RACB barley and RACB RNAi barley reacted similarly to the WT when inoculated with Bgh (Fig. 3B). Quantitative differences in the strength of the PR gene expression post-inoculation are explained by differences in constitutive gene expression. CA RACB barley reacted to Bgh with a less strong PR gene expression response because genes were already expressed at a higher level without inoculation. JIP gene expression was not strongly deregulated in RACB-transgenic barley. However, there was slightly enhanced expression of JIP23 and JIP60 in CA RACB barley (Fig. 3A, C). Other effects of genotype or inoculation on JIP gene expression were less consistent between the two sampling times. At 12 and 32 HAI with Bgh, the differences in gene expression between WT and RACB-transgenic genotypes were less pronounced when compared with the situation without inoculation (compare Fig. 3A and C). However, inoculated CA RACB barley still expressed single PR genes and JIP23 on an up to a 2.7-fold higher level than the WT, whereas RACB RNAi barley expressed the same genes at the level of the WT or below. Together, the RACB transgenes influenced defence gene expression but this did not reflect altered susceptibility of RACB-transgenic barley.
Fig. 3.
Marker gene expression does not reflect the susceptibility status of RACB-transgenic barley. Wild-type (WT), CA RACB-overexpressing, and RACB RNAi plants were either mock-treated or inoculated with Bgh (100 spores mm−2) and collected at 12 or 32 HAI for RT–qPCR (A) Genotype-dependent expression of defence-related genes [pathogenesis-related (PR) genes and jasmonate-induced protein (JIP) genes]. Genes are constitutively overexpressed in mock-treated CA RACB plants versus WT plants and partially underexpressed in RACB RNAi plants. (B) Bgh-triggered expression of PR genes and JIP genes is only weakly affected by the transgenes. Less strong PR gene expression in CA RACB plants is explained because genes are already constitutively expressed on a higher level (see A). (C) Genotype-dependent expression of PR genes and JIP genes in Bgh-inoculated plants. Columns show the average fold change of three biological repetitions of gene expression relative to that of constitutively expressed HvUBI. Error bars show the SE of three fully independent experiments. * indicate significant changes at P<0.05 according to a two-sided one-sample t-test.
RACB operates in root trichoblast polarity of barley
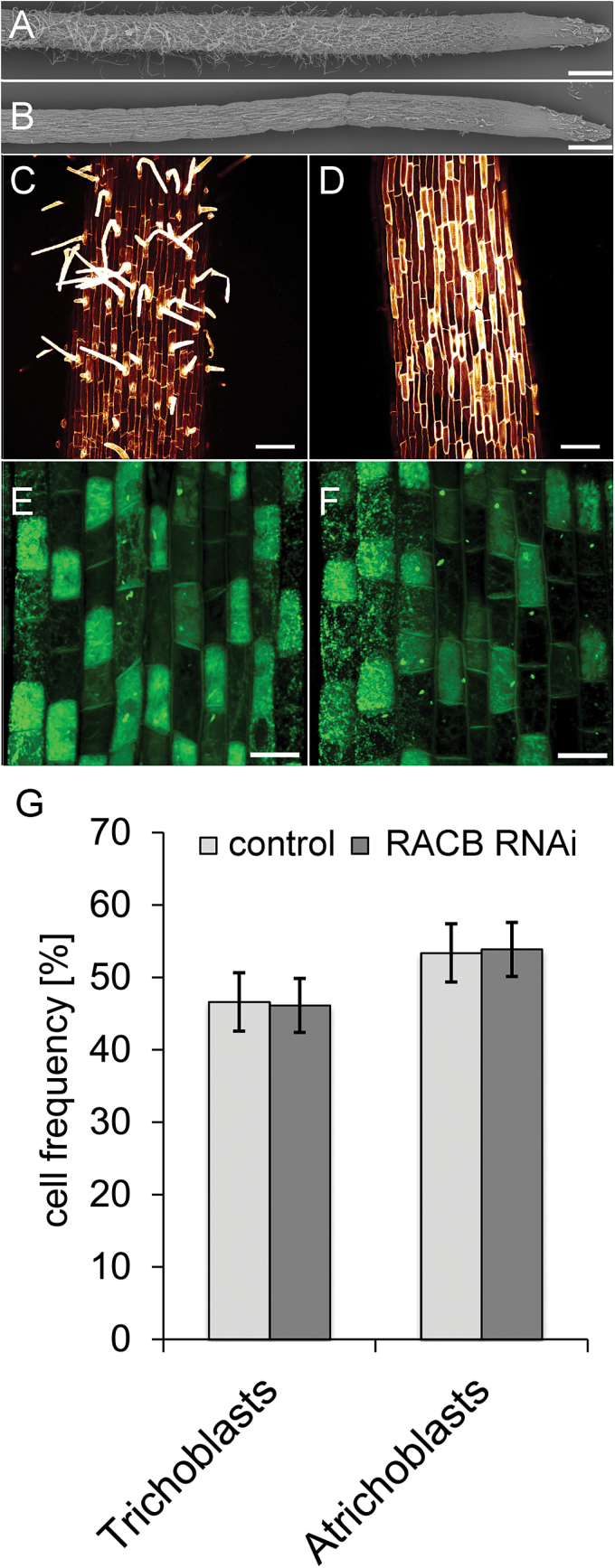
Because RACB did not regulate basal immune responses in a way that would explain its function as a susceptibility factor, we hypothesized that RACB’s role in plant cell development could support pathogenesis. We therefore studied RACB RNAi-mediated developmental failure in more detail. We first confirmed by SEM that knock down of RACB mediates inability to form root hairs (Fig. 4A, B). The roots showed a dramatic reduction of root hair outgrowth and, even if occasionally trichoblast protrusions were formed, they remained short. This has been similarly reported before for two independent transgenic RACB RNAi events (Hoefle et al., 2011). Since barley root hairs can only develop from short epidermal trichoblasts, we compared the epidermal cell size pattern and number of trichoblasts in RACB RNAi plants and non-transgenic sister plants that lost the silencing cassette due to segregation (azygous control). Trichoblasts are shorter than atrichoblasts at late stages of root hair outgrowth (Marzec et al., 2013). PI staining of comparable root sections of root hair initiation showed the occurrence of a typical pattern of shorter and longer cells in both RACB RNAi roots and the control (Fig. 4C, D). However, identification of short cells as trichoblasts was only possible after root hair outgrowth. Thus we used the fact that trichoblasts differ from atrichoblasts in size of vacuoles, density of cytoplasm, and number of cell organelles (Marzec et al., 2013), and stained barley roots with the mitochondrial dye R123. Trichoblasts showed a more intense staining by R123 due to their higher number of mitochondria and denser cytoplasm compared with atrichoblasts. This allowed for the identification and quantification of trichoblasts before any obvious differences in cell expansion occurred. Qualitative and quantitative evaluation revealed that RACB RNAi plants were able to form trichoblasts in the same amount and pattern as azygous controls (Fig. 4E–G). Hence, RACB RNAi plants are able to specify root epidermal cells as trichoblasts but fail at bulging or tip growth at subsequent stages of root hair development (Schiefelbein, 2000).
Fig. 4.
Root hair phenotype of RACB RNAi plants. (A) SEM of barley roots. Root hairs develop on azygous controls (non-transgenic segregants from RNAi plants). (B) RACB RNAi plants do not show root hair outgrowth. (C) Detailed view of the azygous control barley root stained with propidium iodide. Propidium iodide intensely stains root hairs. (D) Detailed view on a root segment of RACB RNAi plants, which corresponds to that of the control in Fig. 3C. (E) Rhodamin-123 staining of a root segment close to the tip of an azygous control root in which plasma-rich trichoblasts differentiate from more vacuolized atrichoblasts. (F) Rhodamin-123 staining of a root segment of RACB RNAi plants, which corresponds to that of the control in Fig. 3E. (G) Counting of trichoblasts reveals no differences in relative frequencies of cell types in azygous versus RACB RNAi barley roots. Columns show the mean of 30 root samples with a total of 1844 (control) or 1846 cells (RACB RNAi) counted. Error bars show the SD of the mean. Scale bars: A, B, 1mm; C, D, 100 µm; E, F, 25 µm. (This figure is available in colour at JXB online.
RACB is involved in leaf stomatal subsidiary cell formation
Type I ROPs are required for the asymmetric cell division of the subsidiary mother cell (SMC) resulting in the formation of a stomatal subsidiary cell and a pavement cell in Zea mays (Humphries et al., 2011). We hence visualized patterns of epidermal cells in barley leaves using SEM. The subsidiary cells of RACB RNAi barley showed deformations of different degrees of severity or were often completely lacking. The SMC-derived pavement cells also exhibited serious defects in shape (Fig. 5B, D; Supplementary Fig. S1 at JXB online). The guard cells and other cell types of the leaf epidermis, however, were normally developed. We detected defective subsidiary cell formation on the leaf blade as well as on the leaf sheath, which develop from different meristems. Quantification of cell shape defects revealed that RACB RNAi barley failed to form normal stomatal subsidiary cells in ~20% of stomatal complexes whereas in azygous sister plants, this failure was observed in only 3.5% of stomata and was restricted to moderately distorted subsidiary and pavement cells in most cases (Fig. 5).
Fig. 5.
Stomatal subsidiary cell phenotypes of RACB RNAi plants. (A) Overview of barley second leaf epidermis by SEM. Stomata with subsidiary cells (green) develop properly on azygous controls. (B) RACB RNAi plants show frequent defects in formation of stomata subsidiary cells (red). (C) Detailed view of an azygous control barley leaf stoma. (D) Detailed view of a RACB RNAi barley leaf stoma. (E) Counting of stomatal defects reveals significant differences of frequencies in azygous control versus RACB RNAi barley roots. Columns show the mean of four leaf samples with 10 171 stomata counted on the controls and 9689 stomata counted on RACB RNAi leaves. Error bars show the SD of the mean (**; Student’s t-test P<0.01). Scale bars: A, B, 200 µm; C, D, 20 µm. A similar phenotype was observed in an independent transgenic RACB RNAi event 15/1-16 (Supplementary Fig. S1).
RACB is involved in positioning of the nucleus in cells attacked by Bgh
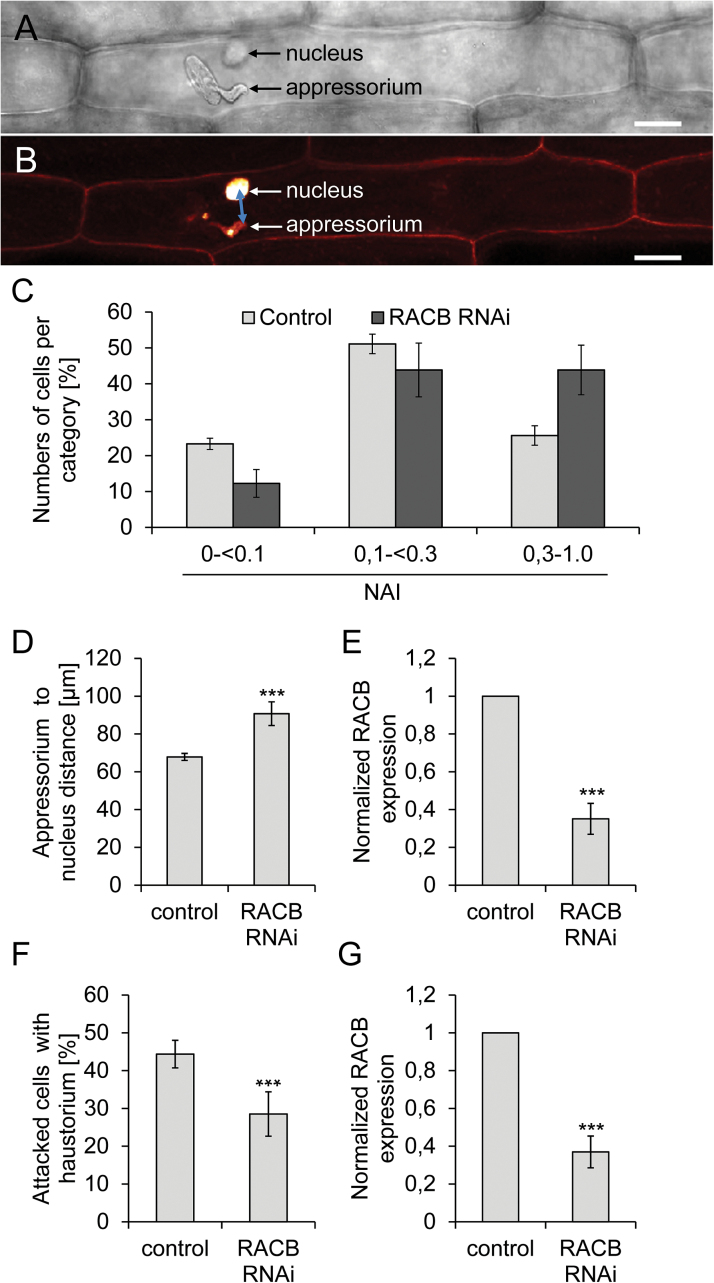
Positioning of the polarized nucleus is a common element of root hair outgrowth, subsidiary cell formation, and powdery mildew infection (Kita et al., 1981; Opalski et al., 2005; Humphries et al., 2011; Griffis et al., 2014). To analyse the influence of RACB on the position of nuclei in spatial association with fungal attack, we measured distances between nuclei and fungal appressoria in controls and RACB RNAi barley (Fig. 6A, B). To avoid mistakes due to cell shape effects, we focused on epidermal B-cell files between stomata and stomata-associated A-cell files (Koga et al., 1990). Additionally, we normalized distances to the cell sizes to obtain an index (NAI, see material and methods) for each attacked cell that displays the distances in a cell size-independent manner. We chose 8 HAI as pre-penetration stage when appressoria are fully developed but haustoria are not yet established. At this point in time, nuclei were more distant from appressoria in RACB RNAi plants when compared with azygous control sister plants. This is evident by grouping the attacked cells by their individual NAIs (Fig. 6C). However, even without normalization to the cell sizes, the reduced attraction of the nucleus to the site of attempted penetration is apparent (Fig. 6D). To confirm that RACB RNAi plants are indeed less susceptible already at early stages of cellular interaction with Bgh, we scored frequencies of immature haustoria at 16 HAI, when haustoria reach a size that can be readily detected after staining with fluorescent wheat germ agglutinin (WGA). Similar to that observed previously for 48 HAI (Hoefle et al., 2011), RACB RNAi plants allowed 35% less frequent establishment of haustoria when compared with azygous control sister plants (28.5% instead of 44.3% in the control) (Fig. 6F). We also confirmed that the RACB transcript amount was indeed reduced in the RACB RNAi plants when compared with azygous sister plants at 8 and 16 HAI. Therefore, we had cut the leaves at the mid rib and fixed one half of the leaf for determining the nucleus to appressorium distance or the haustorium frequency, respectively. The other halves of the leaves were used to measure RACB transcript abundance by RT–qPCR. RACB transcript abundance in RACB RNAi plants corresponded to 28% of the control level at 8 HAI and to 31% of the control level at 16 HAI (Fig. 6E, G). Together, the susceptibility factor RACB is involved in positioning of the nucleus in cells, which Bgh attempts to penetrate. In plants in which RACB expression is reduced by RNAi, the nucleus is more distant from the appressorium before the fungus penetrates and the fungus is subsequently less successful in penetration.
Fig. 6.
RACB influences positioning of the nucleus relative to the site of fungal attack. (A) Barley epidermal cell attacked by Bgh with the fungal appressorium and the plant nucleus (out of focus) visible in the transmission channel of the confocal laser scanning microscope. Size bar=25 µm. (B) The same barley epidermal cell as in (A) imaged by confocal laser scanning fluorescence microscopy. The fixed leaf was stained with propidium iodide to stain the fungus, the plant cell wall, and nucleus. The double-headed arrow indicates the distance from the site of fungal attack to the nucleus. Size bar=25 µm. (C) Considering that the nucleus is attracted by fungal attack, a nucleus attraction index (NAI) was calculated for 8h after inoculation (10 spores mm−2). First, the distance of the nucleus from the appressorium to the centre of the nucleus was calculated based on the horizontal distance (x/y-position, line in B) and the position of the nucleus in z. Subsequently, the NAI was calculated after normalizing to cell sizes (see the Materials and methods). The NAI was categorized into three groups representing the nucleus in close proximity of the fungus (0 to <0.1), in proximity to the fungus (0.1 to <0.3), and distant from the fungus (0.3 to <1). The NAI was measured on each of five or more azygous controls and RACB RNAi leaves at a minimum of 85 attacked cells per leaf. The χ2 test P-value for genotype-dependent differential distribution into the three NAI categories is P<0.001 for 8 HAI. A similar phenotype was observed in an independent transgenic RACB RNAi event 15/1-16 (Supplementary Fig. S1). (D) Absolute appressorium to nucleus distances in azygous controls and RACB RNAi plants at 8 HAI. (E) Expression level of RACB in leaf segments sampled in parallel to that used for the experiment in (D). (F) Frequencies of successful haustorium formation in azygous controls and RACB RNAi plants at 16 HAI. (G) Expression level of RACB in leaf segments sampled in parallel to that used for the experiment in (F). ***, Student′s t-test, P-value<0.001. Error bars show the SD of the mean of five individual leaves. (This figure is available in colour at JXB online.)
Discussion
Susceptibility factors are plant components that serve the demands of a pathogen during disease development. However, the mode of action of susceptibility factors is often not well understood. They might be classified as negative regulators of host immunity or as host factors that support metabolic or developmental processes required for successful pathogenesis (Hückelhoven, 2005; Hückelhoven et al., 2013; Lapin and Van den Ackerveken, 2013; van Schie and Takken, 2014). The latter, however, is challenging to provide evidence for, since it is difficult to distinguish whether failure of a pathogen to infect is because the host mutant does not properly support pathogenesis or, is due to an enhanced basal defence of that mutant. Our data support that RACB acts as a susceptibility factor through its function in cell polarity rather than by suppressing early MAMP-triggered immune responses or defence gene expression. Indeed, CA RACB enhanced PR gene expression in non-infected plants, and silencing of RACB lowered the level of PR gene expression. This is, however, not reflected in the resistance status of RACB-transgenic barley (Schultheiss et al., 2005; Hoefle et al., 2011) (Fig. 6) and therefore counterintuitive. Interestingly, expression of a dominant negative form of the Arabidopsis type I RAC/ROP protein DN ROP6 also causes enhanced defence gene expression. DN ROP6 expression further led to reduced penetration success and reduced reproductive success of the powdery mildew fungus Golovinomyces orontii in Arabidopsis. Genetic experiments with salicylic acid biosynthesis and signalling mutants suggested that defence gene expression can be uncoupled from powdery mildew resistance in this case (Poraty-Gavra et al., 2013). Arabidopsis ROP6 might be involved in susceptibility to adapted powdery mildew independent of its function in salicylic acid signalling and defence gene expression. Together, perturbation of RAC/ROP signalling appears to alter plant defence gene expression but this cannot explain enhanced or reduced susceptibility of RAC/ROP mutants to powdery mildew.
Developmental host reprogramming is observed for mutualistic symbiosis such as root nodule development. However, there are few examples of pathogenic interaction with plant developmental programmes (Evangelisti et al., 2014). The barley susceptibility factor RACB is involved in plant development and cytoskeleton organization (Opalski et al., 2005; Pathuri et al., 2008; Hoefle et al., 2011). However, other RAC/ROP proteins are involved in regulating typical immune responses (Kawano et al., 2014). We therefore studied typical early MAMP responses in RACB-misexpressing barley. This showed that barley reacts to MAMPs like other plants by early ROS production and MAPK activation. However, neither expression of CA RACB nor suppression of RACB expression greatly influences the ability of barley to respond quickly to fungal MAMP chitin or to the bacterial MAMP flg22. This suggests that RACB and the co-silenced RACD do not regulate canonical MAMP-triggered immunity in barley.
RACB RNAi effects on fungal success to develop haustoria can be observed in a cell-autonomous manner after transient induced gene silencing or after transient overexpression of CA RACB in single barley epidermal cells (Schultheiss et al., 2002, 2003). This shows that RACB is required for fungal entry in a WT background, and reduced susceptibility of RACB RNAi barley is not a secondary consequence of developmental alterations. However, we considered developmental effects of RACB RNAi as instrumental to better understand the physiological role of RACB from which Bgh might profit. The involvement of RACB-like RAC/ROPs of dicots in development of root hairs and pollen tubes provoked the ‘inverted tip growth’ hypothesis (Schultheiss et al., 2003). According to this, Bgh profits from RACB’s function in polar cell growth for inward growth of the fungal haustorium into an intact epidermal cell that surrounds the haustorium with a host-derived extrahaustorial membrane and matrix. Stable transgenic RNAi-mediated silencing of RACB provided first evidence for this hypothesis because a reduction of the RACB transcript level in RACB RNAi plants caused a dramatic reduction of frequency and size of hairs on the root epidermis and a strong reduction of frequency and size of Bgh haustoria in the leaf epidermis at 48 HAI (Hoefle et al., 2011). In contrast, stable or transiently overexpressed CA RACB supports establishment of haustoria but induces isotropic instead of polar root hair growth (Schultheiss et al., 2003; Pathuri et al., 2008, 2009). In Arabidopsis, root hair development is dependent on ROP signalling. ROP proteins accumulate at the site of root hair initiation, and constitutively activated ROPs abolish root hair polarity whereas dominant negative ROPs restrict root hair development (Molendijk et al., 2001; Jones et al., 2002). The receptor-like kinase Feronia activates ROP2 for root hair development (Duan et al., 2010). The positioning of root hairs is spatially controlled by the ROP-GDP dissociation inhibitor SCN1, and different ROP-GEFs influence the number, localization, and length of root hairs (Carol et al., 2005; Huang et al., 2013). The NADPH oxidase RBOHC is a potential ROP effector protein and required for root hair formation (Foreman et al., 2003; Jones et al., 2007). Also in barley a SCN1 homologue and ROS might function in root hair development (Kwasniewski et al., 2010, 2013). Actin microfilaments, microtubules, Ca2+ gradients, and ROS together appear to orchestrate polar root hair initiation and growth. Interestingly, all these components are influenced by ROP signalling in root hairs (Molendijk et al., 2001; Jones et al., 2002; Carol and Dolan, 2006; Yang et al., 2007; Takeda et al., 2008) and play a role in interactions of plants with powdery mildew fungi (Kobayashi et al., 1997; Kim et al., 2002; Hückelhoven and Kogel, 2003; Felle et al., 2004; Hoefle et al., 2011; Dörmann et al., 2014). In the interaction of barley with Bgh, microfilament and microtubule organization are strongly influenced by RACB or by RACB-associated signalling components (Opalski et al., 2005; Hoefle et al., 2011; Huesmann et al., 2012). In barley, only short plasma-rich epidermal cells, which gained identity as trichoblasts, are capable of initiating root hairs, whereas long expanding and highly vacuolized atrichoblasts remain hairless (Marzec et al., 2013). We analysed cell identity of epidermis cells in the root differentiation zone and root transition zone of RACB RNAi barley (Verbelen et al., 2006). This suggests that the knock down of RACB does not change trichoblast identity (Fig. 4E, G) but limits the ability of trichoblasts to undergo root hair initiation, bulging, and tip growth. Concerning the role of ROPs in root hair development (Molendijk et al., 2001; Jones et al., 2002; Singh et al., 2008), this is probably caused by the inability to establish and maintain cell polarity in the trichoblasts, which is required for root hair development. Together, this supports that RACB acts in cell polarization during root hair initiation and tip growth. Bgh might profit from a similar function for RACB during initiation of and progressive ingrowth of the fungal haustorium into the leaf epidermis. Root hair formation always goes along with specific nucleus positioning in the trichoblasts throughout all phases of root hair development (Ketelaar et al., 2002; Čiamporová et al., 2003) (Fig. 7). Similar to this, the precise positioning of the nucleus of the SMC next to the guard mother cell (GMC) is the first visible indication for SMC polarization during stoma development in barley and maize. This SMC polarization is required for asymmetric cell division, resulting in a small-volume subsidiary cell and a large-volume epidermal pavement cell (Fig. 7) (Facette and Smith, 2012). The site-directed nucleus migration and polarization of the SMC is highly ROP regulated. It is thought that maize ROP2 and ROP9 stimulate the formation of a polar actin patch after their own accumulation at the anticlinal interface of the GMC. Maize rop2/rop2;rop9/+ mutants show a similar subsidiary cell formation defect to that which we examined on our barley RACB RNAi plants (Fig. 5) (Humphries et al., 2011; Facette and Smith, 2012). Considering that ROP2 and ROP9 are extremely similar homologues of barley RACB (98% and 99% amino acid sequence identity, respectively), we suggest that RACB is involved in similar processes of SMC polarization and nuclear direction during subsidiary cell formation. The observed defects in subsidiary cell formation of RACB RNAi lines are best explained by a failure of asymmetric cell division. As a result of this, in most cases one cell, which originates from the undivided SMC (Fig. 5D), instead of two developed. In some other cases, the cell wall between the subsidiary cell and the SMC did not show the usual longitudinal orientation but was twisted, such that the cell wall of the subsidiary cell did not have a typically convex shape.
Fig. 7.
Positioning of the nucleus in polar epidermis cell development, which involves RACB in barley. (A) Subsidiary cell formation in stomata of grasses (Facette and Smith, 2012). Subsidiary cells develop from subsidiary mother cells (light blue) in that nuclei (dark-red) position to a guard mother cell. This allows for subsequent asymmetric cell division, in which a new cell wall is built between the subsidiary daughter cell and the adjacent epidermal daughter pavement cell (light blue). Subsequently guard cells develop from the guard mother cell by cell division. (B) Root hairs develop from short epidermal trichoblasts (blue). The nucleus moves to a position close to the future bulging site. The trichoblast bulges and growths out. Subsequently, the nucleus migrates into the tip-growing root hair and keeps a certain distance from the tip (Griffis et al., 2014). Polar deposition of cell wall and membrane material is required for rapid growth of the hair. (C) The nucleus moves to the site of contact with a fungal appressorium and future ingrowth of the fungal haustorium. Polar deposition of membrane and cell wall (extrahaustorial membrane and matrix) is required for rapid accommodation of the fungal haustorium.
There is increasing awareness of an effector-triggered influence of microbes on plant development processes (Evangelisti et al., 2014). Positioning of the nucleus is dynamic in both parasitic and mutualistic plant–microbe interactions. The formation of the pre-penetration apparatus in the response of legumes to hyphopodia formation by arbuscular mycorrhiza fungi involves attraction of the nucleus and its movement in front of the penetration hyphae (Genre and Bonfante, 2007). Therefore, we examined RACB RNAi plants for their capability for single cell polarization after fungal attack. We used positioning of the nucleus as a marker because it is common to root hair and subsidiary cell formation and to cell polarization in plant–microbe interactions (Fig. 7). The data support that nuclei closely associate with fungal appressoria in non-transgenic plants, as observed earlier (Kita et al., 1981; Schmelzer, 2002; Opalski et al., 2005). However, when we observed nuclei at 8 HAI before Bgh actually penetrated, nuclei appeared less attracted by fungal appressoria in RACB RNAi plants because NAIs were shifted to higher values and the absolute nucleus to appressorium distance was higher when compared with azygous controls (Fig. 6). This may indicate reduced single cell polarization of the attacked RACB RNAi cells at an early stage of plant–pathogen interaction and that RACB may be involved in positioning the nucleus in response to a fungal penetration attempt. The nucleus is confined and connected to the site of attack by both microtubules and microfilaments (Opalski et al., 2005; Hoefle et al., 2011). Since RAC/ROP proteins are key regulators of the plant cytoskeleton, and the cytoskeleton is an important target of plant pathogens (Cheong et al., 2014; Porter and Day, 2015), it is logical that RACB mutants show nucleus positioning phenotypes in combination with an altered susceptibility. The role, however, of nucleus positioning during plant–pathogen interaction is hardly understood. Movement of the nucleus correlates with cytoplasmic aggregation at the sites of fungal attack and with subsequent secretion events for formation of cell wall appositions. In interaction with filamentous pathogens, cell polarization, and nuclear attraction is often less frequent and more transient in compatible interactions when compared with resistance (Schmelzer, 2002). Invasive hyphae of the Cowpea rust fungus are connected via host actin microfilaments and microtubules to the host plant nucleus in both compatible and incompatible interactions. However, nuclei are more often close to fungal hyphae in compatible interactions. The actin inhibitor cytochalasin E inhibits positioning of the nucleus close to fungal hyphae and hypersensitive cell death in resistant cultivars (Skalamera and Heath, 1998). Cytochalasin E also inhibits penetration resistance of barley to Bgh and Erysiphe pisi (Kobayashi et al., 1997; Miklis et al., 2007). Barley actin microfilaments and microtubules are strongly re-organized in cells that defend against fungal penetration (Kobayashi et al., 1997; Opalski et al., 2005; Hoefle et al., 2011). Quantification of actin cytoskeleton patterns at 14–36 HAI suggested an association of cell polarity with penetration resistance to Bgh (Opalski et al., 2005). In interaction with Bgh, stability of microtubules and polarization of both microfilaments and microtubules is influenced by RACB or by the RACB-interacting proteins MAGAP1 (MICROTUBULE-ASSOCIATED ROP GTPASE ACTIVATING PROTEIN 1) and RBK1 (ROP BINDING KINASE 1) (Opalski et al., 2005; Hoefle et al., 2011; Huesmann et al., 2012). Together, this strongly suggests a function of polarity in penetration resistance. On the other hand, polar secretory events are also required for fungal accommodation in intact cells. Similar components of membrane transport act in penetration resistance and in formation of perimicrobial compartments in compatible plant–microbe interactions (Dörmann et al., 2014). Additionally, the nucleus is a target of virulence effectors of diverse plant pathogens including powdery mildew (Wessling et al., 2014). Cell polarization and movement of the nucleus may thus be important for basal penetration resistance. However, Bgh might also co-opt this during host cell re-programming for fungal accommodation. Additionally, Bgh might profit from a host cell developmental programme for polar growth including local cell wall remodelling and supply with sufficient building blocks for formation of the haustorial complex. As an obligate biotroph that has lost some essential gene functions during co-evolution with its host (Spanu et al., 2010), Bgh might partially depend on support from its host. Our data support that RACB is a susceptibility factor that supports accommodation of fungal infection structures by its function in polar cell development.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Polar cell development and the nucleus positioning phenotype of RACB RNAi event 15/1-16.
Acknowledgements
We are grateful to undergraduate student Christin Gebhardt for contributing to the ROS assays, and to Caroline Hoefle, Christopher McCollum, and Mathias Nottensteiner for technical advice. This study is supported by a research grant to RH in the framework of the German Research Foundation Collaborative Research Centre SFB924 (TP B08).
References
- Carol RJ, Dolan L. 2006. The role of reactive oxygen species in cell growth: lessons from root hairs. Journal of Experimental Botany 57, 1829–1834. [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L. 2005. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438, 1013–1016. [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. 2010. Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant and Cell Physiology 51, 585–595. [DOI] [PubMed] [Google Scholar]
- Cheong MS, Kirik A, Kim J-G, Frame K, Kirik V, Mudgett MB. 2014. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathogens 10, e1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čiamporová M, Dekánková K, Hanáčková Z, Peters P, Ovečka M, Baluška F. 2003. Structural aspects of bulge formation during root hair initiation. Plants and Soil 255, 1–7. [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Kim H, Ott T, Schulze-Lefert P, Trujillo M, Wewer V, Hückelhoven R. 2014. Cell-autonomous defense, re-organization and trafficking of membranes in plant–microbe interactions. New Phytologist 204, 815–822. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu H-M. 2010. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences, USA 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Rey T, Schornack S. 2014. Cross-interference of plant development and plant–microbe interactions. Current Opinion in Plant Biology 20, 118–126. [DOI] [PubMed] [Google Scholar]
- Facette MR, Smith LG. 2012. Division polarity in developing stomata. Current Opinion in Plant Biology 15, 585–592. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Felle HH, Herrmann A, Hanstein S, Hückelhoven R, Kogel KH. 2004. Apoplastic pH signaling in barley leaves attacked by the powdery mildew fungus Blumeria graminis f. sp hordei . Molecular Plant-Microbe Interactions 17, 118–123. [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, et al. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Genre A, Bonfante P. 2007. Check-in procedures for plant cell entry by biotrophic microbes. Molecular Plant-Microbe Interactions 20, 1023–1030. [DOI] [PubMed] [Google Scholar]
- Griffis AH, Groves NR, Zhou X, Meier I. 2014. Nuclei in motion: movement and positioning of plant nuclei in development, signaling, symbiosis, and disease. Frontiers in Plant Science 5, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Boernke F, Hensel G, Kumlehn J, Hückelhoven R. 2011. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. The Plant Cell 23, 2422–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G-Q, Li E, Ge F-R, Li S, Wang Q, Zhang C-Q, Zhang Y. 2013. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytologist 200, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. 2005. Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiology Letters 245, 9–17. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Eichmann R, Weis C, Hoefle C, Proels RK. 2013. Genetic loss of susceptibility: a costly route to disease resistance? Plant Pathology 62, 56–62. [Google Scholar]
- Hückelhoven R, Kogel K-H. 2003. Reactive oxygen intermediates in plant–microbe interactions: who is who in powdery mildew resistance? Planta 216, 891–902. [DOI] [PubMed] [Google Scholar]
- Huesmann C, Reiner T, Hoefle C, Preuss J, Jurca ME, Domoki M, Feher A, Hückelhoven R. 2012. Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiology 159, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG. 2011. ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. The Plant Cell 23, 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Yang Z, Smirnoff N. 2007. NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. Journal of Experimental Botany 58, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang ZB, Grierson CS. 2002. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. The Plant Cell 14, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, et al. 2010. Activation of a Rac GTPase by the NLR family disease resistance protein pit plays a critical role in rice innate immunity. Cell Host and Microbe 7, 362–375. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kaneko-Kawano T, Shimamoto K. 2014. Rho family GTPase-dependent immunity in plants and animals. Frontiers in Plant Science 5, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NC, Grierson CS, Dogterom M, Emons AM. 2002. Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. The Plant Cell 14, 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. 2002. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. [DOI] [PubMed] [Google Scholar]
- Kita N, Toyoda H, Shishiyama J. 1981. Chronological analysis of cytological responses in powdery-mildewed barley leaves. Canadian Journal of Botany 59, 1761–1768. [Google Scholar]
- Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H. 1997. Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. The Plant Journal 11, 525–537. [Google Scholar]
- Koga H, Bushnell WR, Zeyen RJ. 1990. Specificity of cell type and timing of events associated with papilla formation and the hypersensitive reaction in leaves of Hordeum vulgare attacked by Erysiphe graminis f. sp. hordei . Canadian Journal of Botany 68, 2344–2352. [Google Scholar]
- Kogel KH, Ortel B, Jarosch B, Atzorn R, Schiffer R, Wasternack C. 1995. Resistance in barley against the powdery mildew fungus (Erysiphe graminis f. sp. hordei) is not associated with enhanced levels of endogenous jasmonates. European Journal of Plant Pathology 101, 319–332. [Google Scholar]
- Lapin D, Van den Ackerveken G. 2013. Susceptibility to plant disease: more than a failure of host immunity. Trends in Plant Science 18, 546–554. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Marzec M, Melzer M, Szarejko I. 2013. Asymmetric growth of root epidermal cells is related to the differentiation of root hair cells in Hordeum vulgare (L.). Journal of Experimental Botany 64, 5145–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. 2007. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiology 144, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CSV, Friml J, Braun M, Gilroy S, Palme K. 2001. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO Journal 20, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, Pons KHJ, Samans B, Kogel KH, Waller F. 2011. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Molecular Plant-Microbe Interactions 24, 1427–1439. [DOI] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Hückelhoven R. 2005. The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp hordei . The Plant Journal 41, 291–303. [DOI] [PubMed] [Google Scholar]
- Ovesna J, Kucera L, Vaculova K, Strymplova K, Svobodova I, Milella L. 2012. Validation of the beta-amy1 transcription profiling assay and selection of reference genes suited for a RT-qPCR assay in developing barley caryopsis. PLoS One 7, e41886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathuri IP, Imani J, Babaeizad V, Kogel K-H, Eichmann R, Hückelhoven R. 2009. Ectopic expression of barley constitutively activated ROPs supports susceptibility to powdery mildew and bacterial wildfire in tobacco. European Journal of Plant Pathology 125, 317–327. [Google Scholar]
- Pathuri IP, Zellerhoff N, Schaffrath U, Hensel G, Kumlehn J, Kogel K-H, Eichmann R, Hückelhoven R. 2008. Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Reports 27, 1877–1887. [DOI] [PubMed] [Google Scholar]
- Pavan S, Jacobsen E, Visser RF, Bai Y. 2010. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Molecular Breeding 25, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhänsel C, Freialdenhoven A, Kurth J, Kolsch R, Schulze-Lefert P. 1997. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. The Plant Cell 9, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poraty-Gavra L, Zimmermann P, Haigis S, Bednarek P, Hazak O, Stelmakh OR, Sadot E, Schulze-Lefert P, Gruissem W, Yalovsky S. 2013. The Arabidopsis Rho of Plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiology 161, 1172–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter K, Day B. 2015. From filaments to function: the role of the plant actin cytoskeleton in pathogen perception, signaling, and immunity. Journal of Integrative Plant Biology (in press). [DOI] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. 2011. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. The Plant Journal 68, 100–113. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P. 2009. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO Journal 28, 3439–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Friml J. 2010. Immunolocalization of proteins in plants. Methods in Molecular Biology 655, 253–263. [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW. 2000. Constructing a plant cell. The genetic control of root hair development. Plant Physiology 124, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E. 2002. Cell polarization, a crucial process in fungal defence. Trends in Plant Science 7, 411–415. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. 2002. A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiology 128, 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. 2003. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. The Plant Journal 36, 589–601. [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Hensel G, Imani J, Broeders S, Sonnewald U, Kogel KH, Kumlehn J, Hückelhoven R. 2005. Ectopic expression of constitutively activated RACB in barley enhances susceptibility to powdery mildew and abiotic stress. Plant Physiology 139, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Fischer U, Singh M, Grebe M, Marchant A. 2008. Insight into the early steps of root hair formation revealed by the procuste1 cellulose synthase mutant of Arabidopsis thaliana. BMC Plant Biology 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalamera D, Heath MC. 1998. Changes in the cytoskeleton accompanying infection-induced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. The Plant Journal 16, 191–200. [DOI] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, et al. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330, 1543–1546. [DOI] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. 2008. Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Research 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Takken FL. 2014. Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Verbelen JP, De Cnodder T, Le J, Vissenberg K, Baluska F. 2006. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signaling and Behavior 1, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling R, Epple P, Altmann S, et al. 2014. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FS. 1987. Localization of mitochondria in plant cells by vital staining with rhodamine 123. Planta 171, 346–357. [DOI] [PubMed] [Google Scholar]
- Yalovsky S. 2015. Protein lipid modifications and the regulation of ROP GTPase function. Journal of Experimental Botany 66, 1617–1624. [DOI] [PubMed] [Google Scholar]
- Yang G, Gao P, Zhang H, Huang S, Zheng Z-L. 2007. A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS One 2, e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2008. Cell polarity signaling in Arabidopsis. Annual Review of Cell and Developmental Biology 24, 551–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.