Highlight
Arabidopsis UBC22 functions in the atypical Lys11-linked ubiquitination, which differs from the more common Lys48-linked ubiquitination, and its knockout results in degeneration of the functional megaspore and female gametes.
Key words: Arabidopsis, female gametophyte, functional megaspore, gametogenesis, protein ubiquitination, ubiquitin-conjugating enzyme, UBC22.
Abstract
Protein ubiquitination is critical for numerous processes in eukaryotes. The ubiquitin-conjugating enzyme (E2) is required for ubiquitination. The Arabidopsis genome has approximately 37 E2 genes, but in vivo functions for most of them remain unknown. In this study we observed that knockout mutants of Arabidopsis UBC22 had much-reduced silique length and seed number, with nearly 90% of ovules aborted. Analyses revealed that the majority of mutant embryo sacs displayed severe defects and often contained no gamete nuclei. There was no difference between mutant and wild-type Arabidopsis at the megaspore mother cell stage; however, the functional megaspore was either not present or appeared abnormal in a large portion of mutant ovules, suggesting that the defect started with functional megaspore degeneration in the mutants. Degeneration continued during megagametogenesis, such that the percentage of mature embryo sacs without any gamete nuclei was much greater than the percentage of developing ovules without a functional megaspore and, in addition, various abnormalities in megagametogenesis were observed. Additionally, heterozygous plants had only 13.1% of ovules aborted, indicating that the heterozygous sporophytic tissues could affect the development of the mutant female gametophyte. UBC22 is the sole member of an Arabidopsis E2 subfamily, and is more closely related to one type of E2s in animals that catalyzes Lys11-specific ubiquitination. Indeed, our results showed that Arabidopsis UBC22 could catalyze ubiquitin dimer formation in vitro in a Lys11-dependent manner, suggesting that it likely catalyzes Lys11-linked ubiquitination in plants. This study has thus identified one biochemical property of UBC22 and revealed a novel function in female gametophyte development.
Introduction
Protein ubiquitination, the process through which the small protein ubiquitin (Ub) or a Ub chain is attached to a substrate protein, is involved in diverse cellular processes in eukaryotes, including cell division, cell growth, signaling, apoptosis, and DNA repair (Pickart, 2001). Ubiquitination typically consists of three steps in forming an isopeptide bond, requiring the activities of three enzymes, named E1, E2, and E3 (Hershko and Ciechanover, 1998).
Substrate proteins can be ubiquitinated in different ways. In monoubiquitination, only one Ub molecule is attached to a target protein. Monoubiquitination has functions in histone regulation, endocytosis, and virus budding (Hicke, 2001). In polyubiquitination, substrate proteins are modified by various polyubiquitin chains (Haglund and Dikic, 2005; Pickart, 2001). Since Ub contains seven lysine (Lys, K) residues, K6, K11, K27, K29, K33, K48, and K63, a polyubiquitin chain can be linked through the C-terminal Gly76 of one Ub and one of the seven K residues of the next Ub. In Arabidopsis, diverse forms of polyubiquitin chains have been found (except for the K27-linked chain), with the K48-linked Ub chain being the most abundant, followed by K63- and K11-linked chains (Kim et al., 2013; Maor et al., 2007). It is well known that K48-linked polyubiquitination targets proteins for degradation by the 26S proteasome in both animals and plants (Vierstra, 2009). Some initial understanding has been gained regarding the E2s responsible for K63-linked ubiquitination and the role of K63-linked ubiquitination in plants (see below). However, little is known about the other forms of polyubiquitination in plants.
The Arabidopsis genome is predicted to encode approximately 37 E2s, which are grouped into 14 subfamilies (Bachmair et al., 2001; Kraft et al., 2005). This large number of E2s may reflect functional diversity in terms of different ubiquitination reactions (transferring a single Ub or forming polyubiquitin chains of various types) and interactions with different E3s and other factors required for their activities. On the other hand, the fact that some subfamilies have multiple members also suggests functional redundancy among the members (Callis, 2014). This suggestion is supported by available, albeit limited, experimental evidence.
It is important to understand the specific functions of each of the E2s. Although in vitro studies have shown that most of them possess the classical E2 activity in forming the E2-Ub complex through a thioester linkage, or could catalyze the formation of a Ub chain (reviewed by Callis, 2014), very limited information is available on the specific ubiquitination reactions in which the E2s are involved.
Further, only a few of the E2s have been characterized in terms of their biological functions. Arabidopsis UBC1 and UBC2, belonging to subfamily III, are homologs of the yeast RAD6, which is known to play a critical role in DNA repair and histone monoubiquitination for transcriptional activation (Hoege et al., 2002; Kao et al., 2004). UBC1 and UBC2, together with two closely related RING-type E3s called HUB1 (HISTONE MONOUBIQUITINATION1) and HUB2, have been shown to be involved in histone 2B monoubiquitination and regulation of flowering time (Cao et al., 2008; Xu et al., 2009). However, UBC3, the other member of subfamily III, does not show redundancy with UBC1 and UBC2, since only the ubc1 ubc2 double mutant, and not double mutants with ubc3, showed the early flowering phenotype (Cao et al., 2008).
UBC21 was initially identified as PEX4 through screening for mutants defective in peroxisomal processes (Zolman et al., 2005). It is tethered to the peroxisome membrane through a peroxisomal protein, PEX22, and is suggested to function in the ubiquitination of the peroxisome matrix protein receptor PEX5. In yeast, Pex5 can be monoubiquitinated for translocation and polyubiquitinated, mainly through the K48-linked Ub chain, for degradation by a complex consisting of three E3 ligases, Pex2, Pex10, and Pex12 (Platta et al., 2004). Studies of the Arabidopsis homologs of the E3 ligases indicate the existence of a similar mechanism in plants (Hu et al., 2012). UBC24, one of the four members of subfamily IX, functions in inorganic phosphate (Pi) signaling and was originally identified as PHO2. pho2/ubc24 mutant plants overaccumulate Pi in leaves and when grown in Pi-rich soil display signs of Pi toxicity such as chlorosis and necrosis (Aung et al., 2006; Bari et al., 2006). More recent studies have shown that UBC24/PHO2 works with an E3 named NLA (Nitrogen Limitation Adaptation) in polyubiquitination, with a Pi transporter, PT2, as a downstream target, leading to its degradation via the 26S proteasome (Park et al., 2014). UBC32, a member of E2 subfamily XIV, was found to be a component of the endoplasmic reticulum-associated protein degradation (ERAD) complex, and to be induced by salt stress and involved in brassinosteroid-mediated growth promotion (Cui et al., 2012). UBC32 has been shown to be involved in the polyubiquitination and degradation of a known ERAD substrate, MLO-12 (Cui et al., 2012).
UBC35 and UBC36, two members of subfamily XV, are closely related to Ubc13 proteins of non-plant species, and they were able to complement the yeast ubc13 mutant for spontaneous mutagenesis and sensitivity to DNA-damaging agents (Wen et al., 2006); accordingly, these two E2s are also referred to as UBC13A and UBC13B, respectively. Ubc13 and its homologs are the only known E2s responsible for K63-linked ubiquitination in yeast and humans (Andersen et al., 2008; Chen and Sun, 2009). Arabidopsis UBC13A (UBC35) has been shown to catalyze K63-linked ubiquitination in vitro (Wen et al., 2006). The double mutant of Arabidopsis UBC13A/B displays strong phenotypes including shortened primary roots, a reduced number of lateral roots, and few and short root hairs (Li and Schmidt, 2010; Wen et al., 2014). A tomato UBC13 homolog, Fni3, has been found to affect plant immunity positively (Mural et al., 2013). Further, in yeast and humans, Ubc13 proteins require partnering with the ubiquitin-conjugating enzyme variant (UEV), which is a UBC domain-containing protein but lacks the catalytic cysteine. Four Arabidopsis UEV genes have been identified, with UEV1D implicated in the DNA damage response (Wen et al., 2008). Results from these studies indicate that UBC13A/B (UBC35/36) have a function in different processes and likely catalyze K63-linked ubiquitination in plants, although in vivo evidence for involvement of Arabidopsis UBC13 protein in K63-linked ubiquitination remains to be obtained.
Apart from the E2s described above, little is known about the other E2s in plants, and there is a great need to understand them and their functions. To this end, in this study we characterized Arabidopsis UBC22, identified its important role in female gametophyte development, and obtained experimental evidence of its ability to catalyze a K11-specific ubiquitination reaction.
Materials and methods
Plant growth
Arabidopsis thaliana ecotype Columbia and its mutant lines were grown in a growth room or chamber (20 °C constant temperature, 16h day/8h night photoperiod with a fluence rate of 90±10 µmoles m–2 min–1). Arabidopsis UBC22 T-DNA insertion lines SALK_011800 and GK_642C08 were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu) and Nottingham Arabidopsis Stock Centre (http://arabidopsis.info), respectively.
Isolation and analysis of plant genomic DNA and RNA
Genomic DNA was isolated from Arabidopsis leaf tissues as described by Edwards et al. (1991) and 1.0 µl of the DNA sample was used in a standard 20 µl PCR, with the following program: 94 °C for 3min followed by 30 cycles of 94 °C for 45s, 55 °C for 45s, and 72 °C for 2min. The primers used are listed in Supplementary Table S2 at JXB online.
Total RNA was isolated using TRIzol reagent (Invitrogen) and RNA concentration was determined with a NanoVue Plus Spectrophotometer (GE Healthcare) following the manufacturer’s instructions. For reverse transcription (RT)-PCR, the first-strand cDNA was synthesized using the ThermoScript RT-PCR system (Invitrogen). The final input amount of cDNA used in the RT-PCR was normalized based on the level of the reference gene At4g33380 (Czechowski et al., 2005).
Complementation of ubc22 mutants
The full-length coding sequence of UBC22 was amplified from wild-type (WT) Arabidopsis cDNA by RT-PCR with the primers HW1089 and HW1090. The fragment was cloned into a plant expression vector modified from pCambia1300 (http://www.cambia.org/daisy/cambia/585.html) and carrying an HA (influenza hemagglutinin) protein fusion tag, resulting in the construct Pro35S::HA-UBC22. A 3.2kb promoter region (–3219 to –18 positions upstream of ATG) of UBC22 was amplified from WT Arabidopsis genomic DNA by PCR and cloned as a HindIII-BamHI fragment to replace the 35S promoter in Pro35S::HA-UBC22, resulting in ProUBC22::HA-UBC22. The construct was then introduced into ubc22-1 mutant plants, and progeny (T2) plants were used in the phenotype analyses.
Light microscopy
For observation of female gametophytes, the pistil was removed and cuts made on both sides of the pistil replum using a scalpel to expose the ovules. The samples were fixed in FAA solution (1:2:10:7 formaldehyde:acetic acid:ethanol:water) for 1h, washed in 10–20% ethanol three times, each for 5–10min, and transferred into water. The ovules were detached, mounted in clearing solution (8:2:1 chloral hydrate:water:glycerol) and observed under a Leica DM2500 microscope equipped with differential interference contrast (DIC) optics. Photographs were taken using a Leica DFC450C digital microscope camera.
Histochemical analysis
For preparing a GUS (β-glucuronidase) fusion construct, first, a HindIII-EcoRI fragment containing a promoter::GUS reporter was cut from a pBI121-based construct and cloned into pCambia1300. The 3.2kb UBC22 promoter region was cloned into a HindIII-BamHI fragment to replace the existing promoter in the modified pCambia1300, resulting ProUBC22::GUS. This construct was introduced into WT plants. Transformants were selected on half-strength Murashige and Skoog plates containing 1% sucrose, 0.7% phytoagar, and 40 μg/ml hygromycin. The plates were placed in a tissue culture chamber. A number of transformants were transferred to, and grown in, soil. Histochemical GUS staining was performed as described by Jefferson (1987) with minor modifications. Seedlings and flowers were vacuum-infiltrated for 1min in GUS staining solution [50mM PO4 3− (pH 7.0), 2mM K3(Fe(CN)6), 2mM K4(Fe(CN)6), 10mM Na2EDTA (pH 8.0), 0.08% Triton X-100, 0.5mg/ml X-gluc] and incubated overnight at 37 °C. The pistils were first fixed in 3.7% formaldehyde in GUS staining solution for 15min and then washed with GUS staining solution for 5–10min. They were then incubated at 37 °C for 12–16h (ovules at early stages of development for approximately 12h and ovules at late stages of development, e.g. FG7, for 16h). After fixation in FAA solution for 10min and washing with 10–25% ethanol three times, each for 5–10min, ovules were mounted in the clearing solution (described above) and examined under the microscope equipped with DIC optics.
Protein expression and purification
The coding sequence of UBC22 was cloned into pET28c (Novagene). The resulting construct pET28-UBC22 was used to transform Escherichia coli strain BL21 (DE3) (Stratagene). For protein expression, 2.5ml of overnight culture was added to 100ml fresh Luria-Bertani liquid medium containing 50 μg/ml kanamycin. The culture was grown at 37 °C with vigorous shaking until the optical density at 600nm reached approximately 0.6; then, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5mM, and the culture was grown for another 4h. Cells were then spun down and resuspended in lysis buffer [50mM PO4 3− (pH 8.0), 300mM NaCl, 10mM imidazole]. Lysozyme (Fisher, BP535-1) was added to a final concentration of 1mg ml–1 and nuclease (Piece Universal Nuclease through Fisher Scientific) was added to a concentration of 3 units ml–1 culture, and the suspension was incubated on ice for 15–30min. The lysate was centrifuged at 12000 × g for 20min at 4 °C and the recombinant protein was purified from the supernatant using Ni-NTK spin columns (Qiagen) following the manufacturer’s instructions.
Ubiquitin conjugation reaction
In vitro Ub conjugation reactions were performed using the purified His-tagged UBC22 as described above. The Ub thioester/conjugation initiation reagents were purchased from Boston Biochem (K-995). Unless noted otherwise, the reaction mixture contained 5 μM purified His-UBC22 and 62.5 μM Ub in the supplied reaction buffer; concentrations of other components were used following the manufacturer’s instructions. K11R, K48R, and K63R mutant Ub proteins were purchased from Boston Biochem (UM-K11R, UM-K48R, and UM-K63R). The conjugation reactions were performed at 30 °C for 4h. Samples were added with 4X non-reducing loading buffer [200mM Tris-HCl (pH 6.8), 8% SDS, 0.2% bromophenol blue, 40% glycerol] and heated at 95 °C for 5min, then subjected to SDS-PAGE (15% gel). Ub and Ub dimers were detected by western blotting using a monoclonal mouse anti-Ub antibody (P4D1, Cell Signaling Technology) and a horseradish peroxidase-conjugated goat anti-mouse antibody (Bio-Rad). The signal was visualized with ECL Prime reagent (GE Health) according to the manufacturer’s instructions.
Results
Characterization of Arabidopsis UBC22 and ubc22 mutants
The 37 Arabidopsis E2s are grouped into 14 subfamilies, many of which have more than one member (Kraft et al., 2005). UBC22 (At5g05080) is the sole member of subfamily X. Interestingly, sequence analysis showed that Arabidopsis UBC22 is more closely related to a human E2, Ube2S, than to other Arabidopsis E2s (Supplementary Fig. S1), and it shares 65% sequence identity with the human and animal Ube2S homologs (Supplementary Fig. S2).
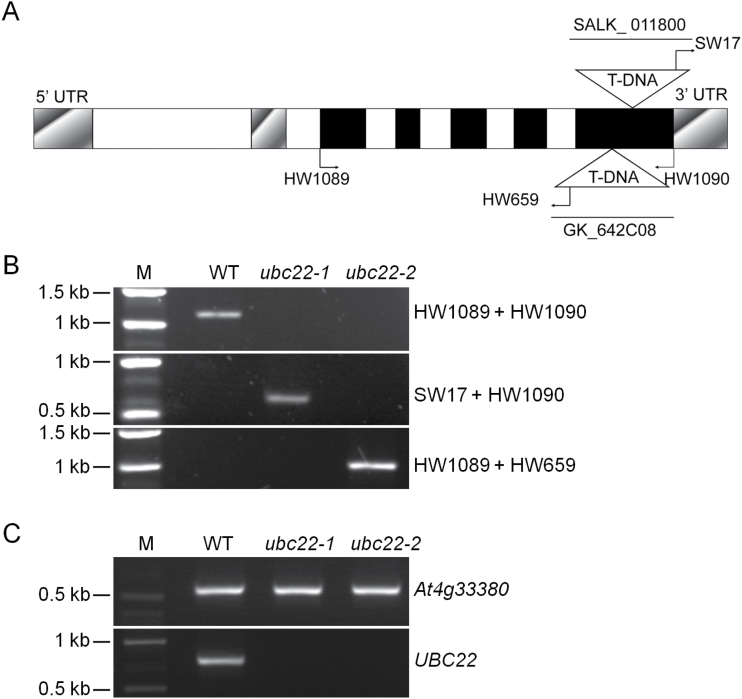
To investigate the function of Arabidopsis UBC22, a T-DNA insertion line (SALK_011800) was obtained from the Arabidopsis Biological Resource Center and genotyped to obtain homozygous lines. Since phenotypes were observed with this line (see below), another independent T-DNA line (GK_642C08) was obtained from the Nottingham Arabidopsis Stock Centre. Sequence analysis of these two T-DNA insertion lines revealed that the T-DNA was in the fifth exon of UBC22 in both lines (Fig. 1A). Genomic DNA PCR and RT-PCR using UBC22 gene-specific primers did not amplify the full-length genomic or cDNA of UBC22 (Fig. 1B, C). Thus, the two alleles are considered null mutation alleles, and are designated ubc22-1 (SALK_011800) and ubc22-2 (GK_642C08) in this study.
Fig. 1.
Confirmation of ubc22 T-DNA insertion mutants. (A) Schematic representation of the genomic structure of UBC22 with the locations of primers and insertion sites of T-DNA in the ubc22-1 and ubc22-2 mutants shown. Closed boxes, exons; open boxes, introns; shadowed boxes, untranslated regions; HW1089 and HW1090, gene-specific primers for UBC22; SW17, T-DNA left border primer for SALK_011800 line; HW659, T-DNA left border primer for GK_642C08 line. (B) Characterization of ubc22 T-DNA lines by genomic PCR. Plant lines used are indicated above the panels, and the primers to the right of the panels. M, DNA size marker. (C) Characterization of ubc22 T-DNA lines by RT-PCR; 30 PCR cycles were used. PCR amplification of a reference At4g33380 cDNA is shown in the upper panel and amplification of UBC22 cDNA is shown in the lower panel.
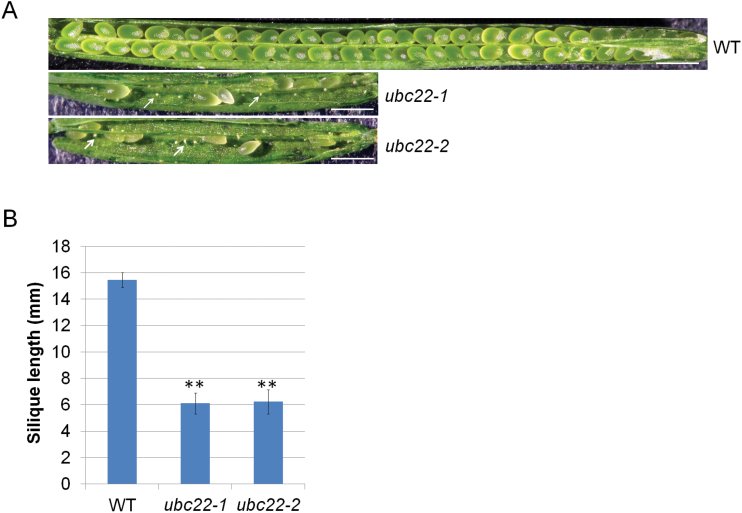
To characterize the phenotypes of ubc22-1 and ubc22-2, plants from these two T-DNA lines were grown side by side with WT plants. While the mutant plants were similar to the WT plants in most aspects, they had considerably shorter siliques than the WT plants (less than 50% of WT length) (Fig. 2A, B). Further examination showed that approximately 88.3% of ovules were aborted in the mutant plants (Fig. 2A, Table 1), whereas the total ovule number was not affected (Table 1). Reciprocal crosses between the WT and mutant revealed that the short silique phenotype was retained when the mutant, but not the WT, was used as the female parent (Table 2).
Fig. 2.
Silique phenotype of ubc22 mutants. (A) Representative siliques of WT, ubc22-1, and ubc22-2 mutant plants. The white arrows indicate aborted ovules. Scale bar, 1mm. (B) Silique length of WT, ubc22-1, and ubc22-2 mutant plants. Eight fully elongated siliques of the main inflorescent stem from each plant and four different plants in each line were measured. Student’s t-test was performed to determine the differences between the mutants and WT. ** Significant difference at P<0.01.
Table 1.
Analysis of ovule and seed development in WT and ubc22-1 mutant plants
| Line | Seeds/ovules counted | Normal seeds | Aborted ovules | Aborted seeds | Number of ovules per silique | Number of seeds per silique |
|---|---|---|---|---|---|---|
| WT | 1536 (100%) |
1531 (99.67%) |
4 (0.26%) |
1 (0.07%) |
61.4±2.6 | 61.2±2.7 |
| ubc22-1 | 1515 (100%) |
156 (10.30%) |
1337 (88.25%) |
22 (1.45%) |
60.6±3.1 | 6.2±2.1 |
Five WT and mutant plants were used, and for each plant the first five siliques were analyzed. For the number of ovules and number of seeds per silique, the data shown are mean±SD. The difference between the WT and the mutant in number of seeds per silique was significant (P<0.01).
Table 2.
Analysis of seed development in siliques from reciprocal crosses and self-fertilized siliques of heterozygous plants
| Female × male | Seeds/ovules counted | Normal seeds | Aborted ovules | Aborted ovules per silique | Aborted seeds | Silique length (mm) |
|---|---|---|---|---|---|---|
| WT × WT | 1265 (100%) |
1264 (99.92%) |
1 (0.08%) |
0.1±0.2 | 0 | 16.2±0.2 |
| ubc22-1 × WT | 1165 (100%) |
89 (7.64%) |
1057 (90.73%) |
52.9±2.9** | 19 (1.63%) |
6.4±1.0 |
| WT × ubc22-1 | 1228 (100%) |
1154 (93.97%) |
71 (5.78%) |
3.6±2.5** | 3 (0.24%) |
15.7±0.6 |
|
ubc22-1/+ × ubc22-1/+
(self-fertilized) |
1196 (100%) |
1038 (86.79%) |
157 (13.13%) |
7.9±2.9** | 1 (0.08%) |
14.9±0.6 |
Reciprocal crosses were made between WT and homozygous ubc22-1 mutant plants. Heterozygous plants (ubc22-1/+) were also included in the analysis as a comparison. Twenty fully elongated siliques from each type were used for determining the number of normal seeds, aborted ovules, and aborted seeds, as well as silique length. Student’s t-test was performed on the number of aborted ovules per silique from reciprocal crosses and heterozygous plants compared with WT plants. ** Significant difference at P<0.01.
We analyzed the seed setting. The reciprocal cross using the ubc22-1 mutant as the female parent had only approximately 7.6% of ovules developing into normal seeds, while the reciprocal cross using WT as the female parent showed approximately 94% of ovules developing into normal seeds, in comparison to almost 100% in the WT plants. These results confirmed that the major defect was due to the female parent. Further, in the self-pollinated siliques of heterozygous plants, approximately 13.1% of ovules were aborted (Table 2); this proportion was much less than half of that in the self-pollinated siliques of the mutant (88.3%; Table 1), suggesting that majority of female gametophytes carrying the mutant allele could develop normally in the heterozygous plants. In addition, when the WT was used as the female parent and the mutant as the male parent, the siliques contained only 5.8% aborted ovules (Table 2). Thus, we analyzed the mutant pollen and observed that the homozygous mutant plants had a slightly higher frequency of pollen with only one or no sperm nuclei (2.8% versus 1.6% in the WT) (Supplementary Fig. S3 and Table S1). These results indicate a minor defect in male gametogenesis of the mutant.
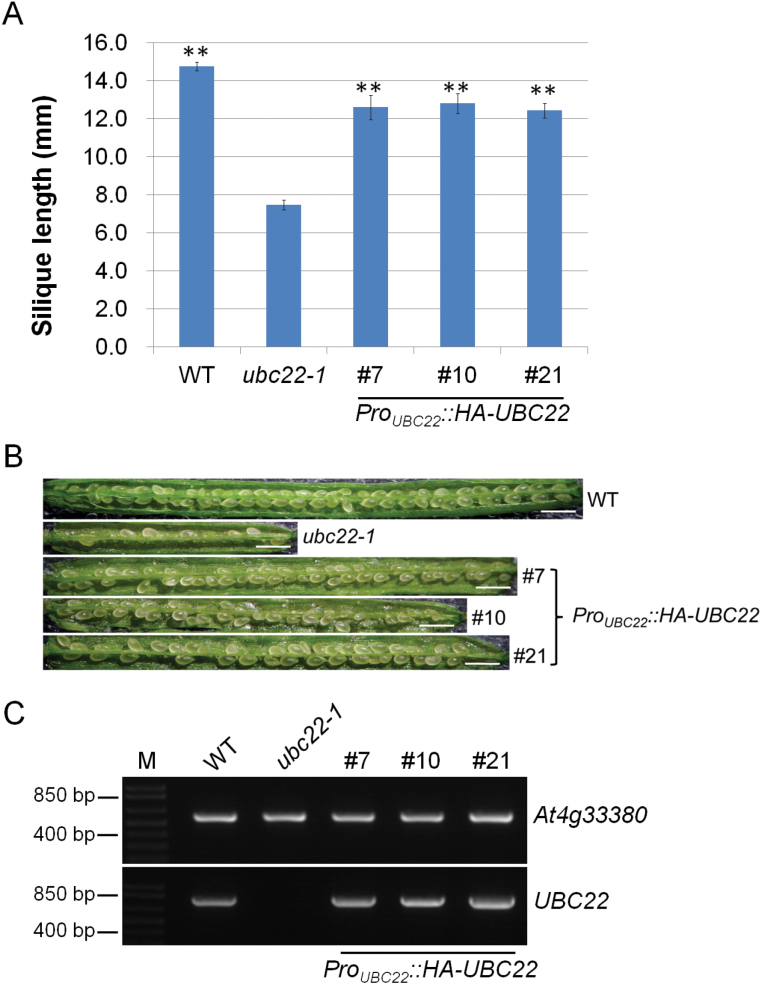
The observation that two independent T-DNA insertion lines showed nearly identical phenotypes strongly suggests that the phenotypes were due to the inactivation of UBC22. To confirm that the phenotype was due to inactivation of UBC22, we expressed UBC22 driven by its own promoter in the ubc22-1 mutant to see whether it could complement the mutant phenotypes. Among the 21 T1 transformants obtained, three independent transformants showed nearly full complementation. Quantitative measurements using T2 plants of these lines showed that the silique length in those lines was approximately 85% of that in the WT plants (Fig. 3A). In addition, the majority of the seeds in these HA-UBC22 lines developed normally (Fig. 3B). The transcript level of HA-UBC22 in these lines was found to be similar to the level of endogenous UBC22 in the WT plants (Fig. 3C).
Fig. 3.
Complementation of the ubc22-1 mutant by Pro UBC22 ::HA-UBC22. For complementation, Pro UBC22 ::HA-UBC22 was introduced into the ubc22-1 mutant. T2 plants of independent lines were used for further analysis. (A) Silique length of WT, ubc22-1, and three individual complementation lines (#7, #10 and #21). Approximately six siliques from each plant were measured, from at least four different plants in each line. Student’s t-test was performed to determine whether there was a significant difference in silique length between the complementation lines and the mutant. ** Significant difference at P<0.01. (B) Representative siliques from WT, ubc22-1, and three complementation lines. Scale bar, 1mm. (C) RT-PCR analysis of WT, ubc22-1, and three independent complementation lines. PCR amplification of a reference At4g33380 cDNA is shown in the upper panel and amplification of UBC22 cDNA is shown in the lower panel. Plant lines used are indicated above the panels. M, DNA size marker.
Female gametophyte development in the ubc22 mutant
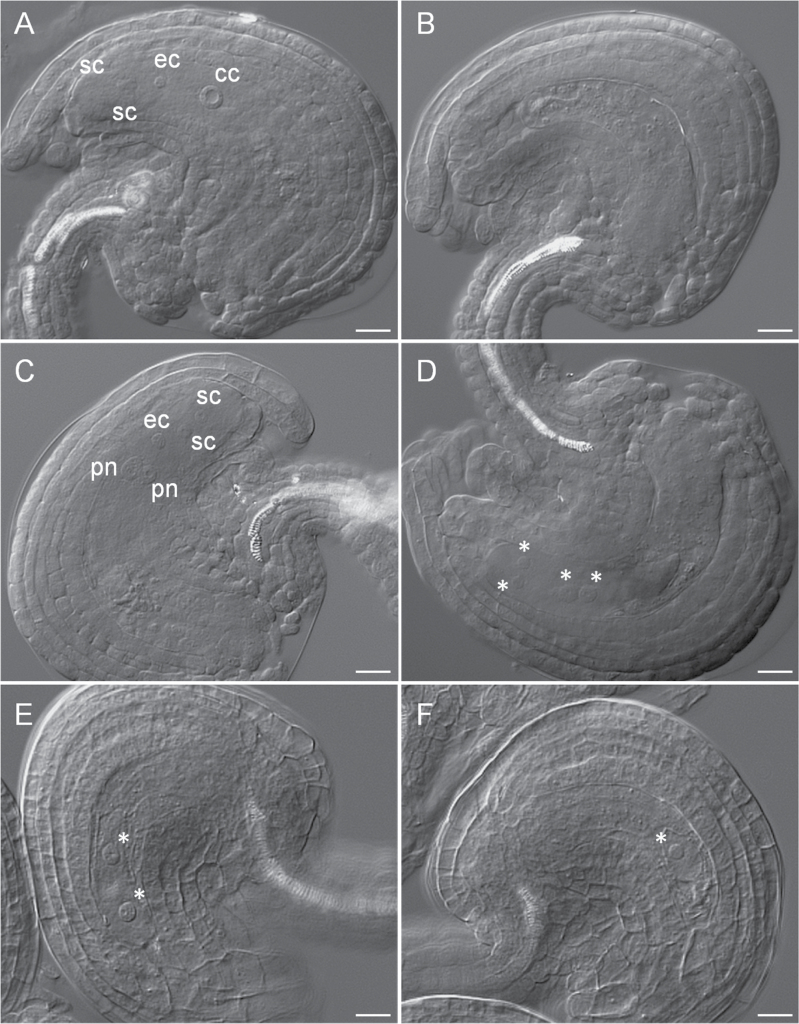
To further characterize the ovule abortion phenotype of the mutant, the development of the female gametophyte in ubc22-1 mutant plants was examined. First, embryo sacs just before flowering were investigated. In the WT Arabidopsis plants, the typical embryo sac at FG7, with egg, central, and synergid cells (Christensen et al., 1997), could be easily distinguished under a microscope with DIC (Fig. 4A). By contrast, in the majority of mutant embryo sacs (63.8%; Table 3), no egg, central, and synergid nuclei could be seen. Generally, these embryo sacs were smaller and narrower than the WT embryo sacs. In addition, the contents of the mutant embryo sacs appeared grainier under the microscope (Fig. 4B). These cellular features suggest that either there was no gametogenesis or the gamete nuclei had degenerated in these embryo sacs. Most of the mutant embryo sacs with observable gamete nuclei were also abnormal. In terms of developmental stage, only a small proportion of them (5.7%) were at the typical FG7 stage, with one central, one egg, and two synergid nuclei, similar to the WT embryo sacs (Fig. 4A). A slightly larger proportion (7.0%) were at the FG5 stage, with two polar, one egg, and two synergid nuclei (Fig. 4C). Some embryo sacs were still at the FG4 stage, with four nuclei and one large vacuole (Fig. 4D).
Fig. 4.
Female gametophyte development in WT and ubc22-1. Ovules from flowers just before opening were prepared and observed under a microscope with DIC. (A) A typical WT embryo sac (FG7 stage) showing one central cell (cc), one egg cell (ec), and two synergid cells (sc). (B–F) Abnormal embryo sacs in ubc22-1 plants. (B) Mutant embryo sac without any nucleus. The embryo sac was smaller and narrower than the WT embryo sac. (C) Mutant embryo sac at the FG5 stage showing two polar nuclei (pn), two synergid cells, and one egg cell. (D) Mutant embryo sac at the FG4 stage, with four nuclei (indicated by asterisks). (E) Mutant embryo sac at the FG3 stage, with two nuclei and a larger vacuole between them. (F) Mutant embryo sac with only one nucleus. Scale bars, 10 μm.
Table 3.
Female gametophyte development in WT and ubc22-1 mutant plants
| Line | Total number | Female gametophyte stage | No nucleus |
One nucleus |
Two nuclei |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | |||||
| WT (%) |
270 (100) |
0 | 0 | 0 | 0 | 1 (0.4) |
0 | 268 (99.3) |
1 (0.4) |
0 | 0 |
|
ubc22-1
(%) |
331 (100) |
2 (0.6) |
2 (0.6) |
4 (1.2) |
8 (2.4) |
23 (7.0) |
3 (0.9) |
19 (5.7) |
211 (63.8) |
47 (14.2) |
12 (3.6) |
Embryo sacs from flowers just before opening were dissected, prepared, and then observed under a microscope equipped with DIC optics. The stages of embryo sacs were determined based on the number of cells and their configurations according to the description of Christensen et al. (1997).
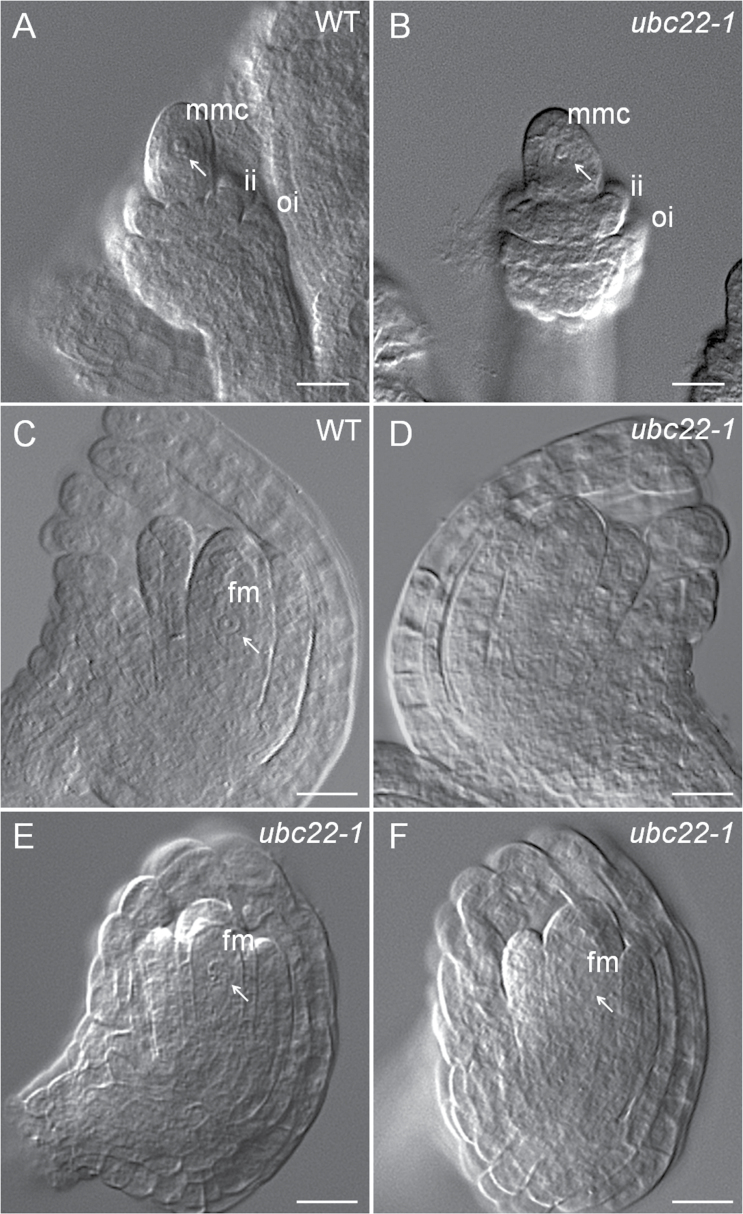
The ovules at earlier stages of development were examined to determine the stage at which ovule development of the mutants departed from that of WT. During megasporogenesis, a megaspore mother cell (MMC) is produced through differentiation of one subepidermal archesporial cell. Careful examination of MMC development in ovules at stage 2-II (Schneitz et al., 1995), with the inner integument initiating, did not reveal any obvious difference between WT and mutant ovules (Fig. 5A, B, Table 4). Therefore, the development of the functional megaspore (FM) was examined. In the WT ovule, following meiosis, the megaspore closest to the chalazal end enlarges and gives rise to the FM, whereas the other three megaspores degenerate. We examined ovules at about stage 3-I according to Schneitz et al. (1995), with the inner integument near and just below the top of the nucellus. Most of the WT ovules examined contained a typical FM, with the nucleus having a clear edge under DIC, a round shape, and often an easily recognizable nucleolus (Fig. 5C). In contrast, in approximately 37% of the mutant ovules examined, no FM nucleus was observed (Table 5, Fig. 5D). In 35% of the mutant ovules, FMs could be identified but appeared abnormal; their nuclei did not have an easily recognized boundary, appeared thin in nuclear content, and/or were not spherical in shape (Fig. 5E). In some cases, the nuclei of FMs appeared thin and ‘flat’ under DIC, with the nuclear–cytoplasm boundary barely visible (Fig. 5F), suggesting that they might be at late stages of degeneration. These data indicate that the FM in some mutant ovules was undergoing degeneration or had degenerated, resulting in no nucleus being observed. Thus, these results indicate that the abnormality during female gametophyte development in the ubc22-1 mutant likely started with the degeneration of the FM in some ovules.
Fig. 5.
Megaspore mother cell (MMC) and functional megaspore (FM) development in WT and ubc22-1. Ovules from the flower buds were prepared and observed under a microscope with DIC. (A, B) WT (A) and ubc22-1 mutant (B) ovules with a typical MMC. ii, inner integument; oi, outer integument. (C–F) Ovules at the FM stage. (C) WT ovule with a typical FM. (D) A ubc22-1 ovule without an observable FM. (E, F) ubc22-1 ovules with an abnormal FM. The FM in (E) is visible, but the boundary is not clear and the shape is abnormal. The FM in (F) is barely visible under the microscope. White arrows indicate the MMC in (A, B) or the FM in (C, E, F). Scale bars, 10 μm.
Table 4.
Megaspore mother cell (MMC) development in WT and ubc22-1 mutant ovules
| Line | Total number | No nucleus | One nucleus | Two nuclei |
|---|---|---|---|---|
| WT | 267 (100%) |
40 (15.0%) |
211 (79.0%) |
16 (6.0%) |
| ubc22-1 | 138 (100%) |
20 (14.5%) |
112 (81.2%) |
6 (4.3%) |
Flowering buds were first checked for ovule developmental stage, and the sixth bud from the newly opened flower was used. The buds were dissected, prepared, and then observed under a microscope equipped with DIC optics. The number of ovules in different categories, in terms of the presence and appearance of MMC, was determined.
Table 5.
Functional megaspore (FM) development in WT and ubc22-1 mutants
| Line | Total number | Typical FM (Type I) | Visible FM with abnormalities (Type II) | FM not observable (Type III) |
|---|---|---|---|---|
| WT (%) |
114 (100) |
99 (86.8) |
10 (8.8) |
5 (4.4) |
|
ubc22-1
(%) |
123 (100) |
34 (27.6) |
43 (35.0) |
46 (37.5) |
Flowering buds (the fifth bud from the newly opened flower) were dissected, prepared, and then observed under a microscope equipped with DIC optics. Ovules at the functional megaspore (FM) stage (with the inner integument near, but below, the top of the nucellus) were included in the analysis. Three types of ovules were observed. Type I (normal): A typical FM had one prominent nucleus with a round shape and clear nuclear–cytoplasm boundary. Type II: The FM was visible. However, the nuclear–cytoplasm boundary was not clear, and/or the nucleoplasm appeared to be disorganized. In the ubc22-1 mutant, the FM was often barely visible (see Fig. 5E, F). Type III: No FM could be observed.
Expression of UBC22 in different tissues and developmental stages
To understand the expression of UBC22, we first examined the microarray data available from Genevestigator (Zimmermann et al., 2004). As shown in Supplementary Fig. S4, UBC22 is expressed in different tissues, with a higher level of expression in seeds. To determine the locations of UBC22 expression more specifically, we fused its promoter to the GUS reporter and the patterns of expression were analyzed by histochemical GUS staining. In 10-day-old seedlings, a strong GUS signal was detected in vascular tissues, root tips, and lateral root primordia (Supplementary Fig. S5A–C). Similar results were obtained from 20-day-old seedlings. In the reproductive organs, a GUS signal was observed in stigmas, filaments, and veins of sepals, but very little was detected in petals and pollen (Supplementary Fig. S5D). Additionally, a GUS signal was detected in ovules of different developmental stages (Supplementary Fig. S5E–G). These data indicate that UBC22 is expressed in a variety of tissues.
UBC22 catalyzing K11-linked ubiquitin dimer formation in vitro
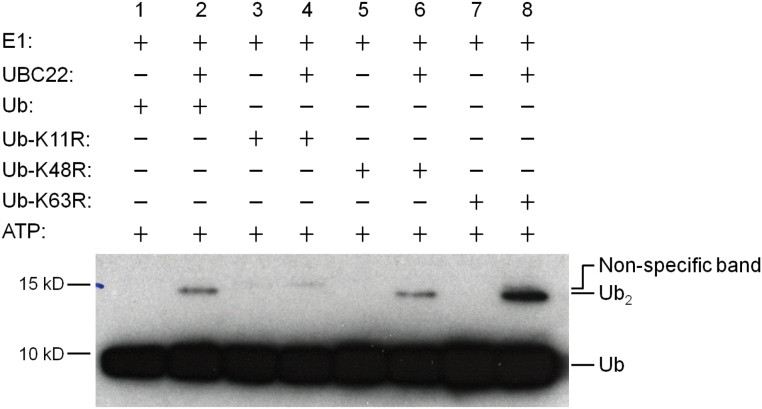
The observation that Arabidopsis UBC22 is more closely related to human Ube2S (65% identity) than to other Arabidopsis E2s (Supplementary Fig. S1) suggests functional conservation between the Arabidopsis and human proteins. In addition, the N-terminal region of approximately 160 amino acids shares high levels of similarity with the human Ube2S and putative Ube2S homologs from other species (Supplementary Fig. S2). It was recently shown that human Ube2S is capable of generating K11-linked Ub dimers and chains (Wickliffe et al., 2011a). We thus determined whether UBC22 could catalyze Ub dimer formation in vitro using recombinant UBC22. As shown in Fig. 6 (lane 2), Ub dimers were formed when His-UBC22 was present in the reaction. Further, when the Ub-K11R mutant, which lacks the K11 residue, was used (lanes 3 and 4 in Fig. 6), little dimer was produced; in contrast, dimer formation was not affected when the Ub-K63R mutant, which lacks the K63 residue, was used (lanes 5 and 6 in Fig. 6). These results indicate that UBC22 was able to catalyze dimer formation in vitro specifically through the K11 residue of Ub.
Fig. 6.
Ub dimer formation assay by His-UBC22. An in vitro Ub dimer formation assay was performed with (lanes 2, 4, 6, and 8) or without (lanes 1, 3, 5, and 7) the addition of His-UBC22. The components added in the different reactions are indicated at the top of the figure. The conjugation reactions were performed at 30 °C for 4 hours. Assay samples were subjected to SDS-PAGE. Ub and Ub dimer were detected by western blotting using an anti-Ub antibody. Free Ub and Ub dimer (Ub2) are indicated at the right side of the figure. For the K11R mutant, no new Ub dimer formation was seen when His-UBC22 protein was added. Due to slight impurity in the recombinant K11R protein, one weak band was present which is slightly higher than the Ub dimer synthesized when His-UBC22 was added. Three experiments were performed and produced similar results.
Discussion
UBC22 plays an important role in female gametophyte development
During megasporogenesis, the MMC develops from one subepidermal archesporial cell. In the great majority of angiosperms, following meiosis, the megaspore closest to the chalazal end enlarges and gives rise to the FM, while the other three megaspores degenerate (Yadegari and Drews, 2004). The developmental feature that only one megaspore survives ensures that only one egg cell and one central cell will be produced in each ovule. In the ubc22 mutants in this study, the MMC developed normally, based on the observations of developing ovules at stage 2_II (Schneitz et al., 1995); however, approximately 37% of ovules at stage 3-I (with the outer integuments near the top of nucellus) did not have an FM. In a small proportion of WT ovules (4.4%), the FM could not be observed; this was likely due to developmental variation among ovules or simply a limitation of the microscope optics. Since an FM was present in most of the WT ovules at this developmental stage, but absent in a large proportion of the mutant ovules, we infer that a key point for UBC22 function is during female megasporogenesis, specifically from MMC to the development of the FM.
There are two possible causes for the absence of FMs in the ubc22 mutant: (i) arrest or inhibition of meiosis and (ii) degeneration of the FM together with the other three megaspores. Inhibition of meiosis usually leads to abnormal meiosis and meiotic products, which were not observed in the ubc22 mutant. It has been shown that the non-functional megaspores degenerate, likely through programmed cell death (Citterio et al., 2005). In the ubc22 mutant, many FMs appeared to be at different stages of degeneration, from having an indistinct nuclear–cytoplasm boundary to almost completely disappearing (Fig. 5E, F). This observation, together with the absence of an FM in a large percentage of ovules, suggests that the FM degenerates along with the three non-functional megaspores in the mutant.
In addition to the absence of an FM, defects also occurred during female gametogenesis in the ubc22 mutant. Over 60% of mutant embryo sacs at the flowering stage did not contain any gamete nuclei, likely due to degeneration during either megasporogenesis or gametogenesis. Further, in approximately 30% of embryo sacs in which gamete nuclei were observed, the embryo sacs were at various stages of gametogenesis, with a relatively high proportion in stage FG5 (Table 3), in contrast to the WT embryo sacs, which were almost all at stage FG7; this result indicates that megagametogenesis in those mutant ovules was delayed to various extents. The defects in female megasporogenesis and gametogenesis in the ubc22 mutant indicate that UBC22 plays a critical role in female gametophyte development. It could be postulated that the inactivation of UBC22 inhibits female gametophyte development, likely due to impaired degradation of certain substrate proteins. Since the extent of inhibition varies among ovules, it is possible that strong inhibition in some ovules leads to degeneration at the FM stage, while weaker inhibition allows FM development but affects female gametogenesis.
Although major defects were observed in female gametophyte development in the ubc22 mutant plants, the results of reciprocal crossing experiments and the analysis of mutant pollen also revealed a relatively minor effect of UBC22 inactivation on male gametogenesis. In addition, the results from promoter-GUS expression (Fig. 6) and microarray data (Supplementary Fig. S4) showed that UBC22 is expressed in various tissues, suggesting that UBC22 may have a function in other tissues. The widespread expression of UBC22 also provides an explanation for the influence of heterozygous sporophytic tissues on the development of female gametophytes carrying the mutant gene.
UBC22 likely catalyzes K11-linked protein ubiquitination
UBC22 is the sole member of subfamily X among the 37 Arabidopsis E2s (Kraft et al., 2005), implying that it has a unique function in plants. Several studies have suggested that UBC22 may be involved in polyubiquitination in Arabidopsis and could catalyze E3-independent ubiquitination in vitro (Kraft et al., 2005; Takahashi et al., 2009; Zhao et al., 2013). Additionally, an E2 Ub conjugation assay showed thioester linkage formation between Ub and UBC22 (Zhao et al., 2013). The present results further show that UBC22 can catalyze Ub dimer formation in vitro independent of the presence of an E3.
The close sequence similarity of Arabidopsis UBC22 with the human and animal Ube2S homologs (Supplementary Fig. S1) suggests functional conservation. Results from different studies have shown that human Ube2S plays a unique function in K11-linked polyubiquitination together with the anaphase-promoting complex/cyclosome (APC/C), a large complex consisting of approximately 13 subunits, which targets cell cycle proteins for degradation by the 26S proteasome (Garnett et al., 2009; Wickliffe et al., 2011a; Williamson et al., 2009). So far, Ube2S is the only E2 that has been shown to be able to catalyze K11-linked ubiquitination. A major role for K11-linked ubiquitination is in the regulation of mitosis (Wickliffe et al., 2011b). In this study, we showed that the formation of Ub dimer in vitro is K11-specific. This finding, together with the sequence conservation across UBC22 and the mammalian Ube2S homologs, strongly supports the notion that UBC22 catalyzes K11-linked ubiquitination in plants.
APC/C acts as a platform to recruit substrates to E2s for their polyubiquitination. In mammalian cells, Ube2S conjugates the C-terminus of a donor Ub to the K11 of an acceptor Ub (Wickliffe et al., 2011a). APC/C provides acceptor-Ub binding sites for Ube2S. The APC/C is highly conserved in plants, and different APC/C subunits from Arabidopsis are able to complement the corresponding yeast mutants (Capron et al., 2003; Eloy et al., 2011; Wang et al., 2012). Results from studies in Arabidopsis indicate that APC/C functions in regulating the cell cycle by targeting cell cycle proteins containing the specific D- or KEN/GxEN-box destruction signals (Heyman and De Veylder, 2012). In Arabidopsis, the APC/C consists of at least 11 components, and some Arabidopsis APC/C mutants display defects in female gametogenesis. The ovules of apc2 and apc6 mutants are arrested at the two-nucleus stage of megagametogenesis (Capron et al., 2003; Kwee and Sundaresan, 2003), while defects at different stages of megagametogenesis are found in apc1 and apc4 mutants (Wang et al., 2012, 2013). Thus, it would be interesting to determine whether Arabidopsis UBC22 interacts with the APC/C. However, there are important differences between the phenotypes of ubc22 mutants and apc mutants. ubc22 mutants show clear defects in both megasporogenesis and megagametogenesis, while the reported defects in apc mutants are only in megagametogenesis. Thus, it is unlikely that the ubc22 mutant phenotypes are entirely due to impaired ubiquitination through the APC/C.
In humans, Ube2S is considered to be an ‘elongation’ E2, which extends the Ub chain through K11-linkages following the initial ubiquitination of the substrate by another E2 (Ube2C/UbcH10 or Ube2D/UbcH5) together with APC/C (Primorac and Musacchio, 2013). In Arabidopsis, UBC19 and UBC20 have been identified as the homologs of UbcH10, based on sequence relatedness and their ability to complement a yeast mutant (Criqui et al., 2002). However, currently there is no information regarding the specific ubiquitination processes with which UBC19 and UBC20 are involved in plants. The present results have provided initial experimental evidence for the existence of the atypical K11-linked form of protein ubiquitination in plants, and identified a critical role for UBC22 in female gametophyte development. It will be interesting to determine whether a module similar to the animal Ube2S-APC/C module is conserved in plants, as well as possible differences between the plant and animal E2 homologs involved in K11-specific ubiquitination.
Supplementary data
Figure S1. Phylogenetic analysis of Arabidopsis E2s.
Figure S2. Alignment of putative UBC22 homologs from different species.
Figure S3. Pollen of WT and ubc22 mutants.
Figure S4. Developmental expression profile of UBC22.
Figure S5. GUS staining of transgenic Pro UBC22 ::GUS plants.
Table S1. Analysis of pollen development in WT and ubc22-1 mutant plants.
Table S2. Sequences of primers used for confirming ubc22 mutants and transcript level of UBC22 in the complementation lines.
Acknowledgements
We thank the Arabidopsis Biological Resource Center (ABRC) and Nottingham Arabidopsis Stock Centre for T-DNA mutant lines, and also two anonymous reviewers for their helpful comments. We gratefully acknowledge financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC) (Discovery grant) to HW.
References
- Andersen PL, Xu F, Xiao W. 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Research 18, 162–173. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. 2006. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a MicroRNA399 target gene. Plant Physiology 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. 2001. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends in Plant Science 6, 463–470. [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J. 2014. The ubiquitination machinery of the ubiquitin system. The Arabidopsis Book 12, e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. 2008. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. The Plant Cell 20, 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A, Serralbo O, Fulop K, et al. 2003. The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit. The Plant Cell 15, 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Molecular Cell 33, 275–286. [DOI] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN. 1997. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sexual Plant Reproduction 10, 49–64. [Google Scholar]
- Citterio S, Albertini E, Varotto S, Feltrin E, Soattin M, Marconi G, Sgorbati S, Lucchin M, Barcaccia G. 2005. Alfalfa Mob1-like genes are expressed in reproductive organs during meiosis and gametogenesis. Plant Molecular Biology 58, 789–807. [DOI] [PubMed] [Google Scholar]
- Criqui MC, de Almeida Engler J, Camasses A, Capron A, Parmentier Y, Inze D, Genschik P. 2002. Molecular characterization of plant ubiquitin-conjugating enzymes belonging to the UbcP4/E2-C/UBCx/UbcH10 gene family. Plant Physiology 130, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q. 2012. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. The Plant Cell 24, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GT, Inze D, Ferreira PC. 2011. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. The Plant Journal 68, 351–363. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nature Cell Biology 11, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. 2005. Ubiquitylation and cell signaling. EMBO Journal 24, 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. 1998. The ubiquitin system. Annual Review of Biochemistry 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Heyman J, De Veylder L. 2012. The anaphase-promoting complex/cyclosome in control of plant development. Molecular Plant 5, 1182–1194. [DOI] [PubMed] [Google Scholar]
- Hicke L. 2001. Protein regulation by monoubiquitin. Nature Reviews Molecular Cell Biology 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. 2012. Plant peroxisomes: biogenesis and function. The Plant Cell 24, 2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405. [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes & Development 18, 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD. 2013. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. The Plant Cell 25, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. 2005. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiology 139, 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee HS, Sundaresan V. 2003. The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. The Plant Journal 36, 853–866. [DOI] [PubMed] [Google Scholar]
- Li W, Schmidt W. 2010. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. The Plant Journal 62, 330–343. [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nuhse TS, Studholme DJ, Peck SC, Shirasu K. 2007. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Molecular & Cellular Proteomics 6, 601–610. [DOI] [PubMed] [Google Scholar]
- Mural RV, Liu Y, Rosebrock TR, Brady JJ, Hamera S, Connor RA, Martin GB, Zeng L. 2013. The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. The Plant Cell 25, 3615–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH. 2014. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. The Plant Cell 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. 2001. Ubiquitin enters the new millennium. Molecular Cell 8, 499–504. [DOI] [PubMed] [Google Scholar]
- Platta HW, Girzalsky W, Erdmann R. 2004. Ubiquitination of the peroxisomal import receptor Pex5p. Biochemical Journal 384, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Musacchio A. 2013. Panta rhei: the APC/C at steady state. Journal of Cell Biology 201, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE. 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. The Plant Journal 7, 731–749. [Google Scholar]
- Takahashi H, Nozawa A, Seki M, Shinozaki K, Endo Y, Sawasaki T. 2009. A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biology 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. 2009. The ubiquitin-26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biology 10, 385–397. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hou Y, Gu H, Kang D, Chen Z, Liu J, Qu LJ. 2012. The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. The Plant Journal 69, 227–240. [DOI] [PubMed] [Google Scholar]
- Wang YB, Hou YN, Gu HY, Kang DM, Chen ZL, Liu JJ, Qu LJ. 2013. The Arabidopsis anaphase-promoting complex/cyclosome subunit 1 is critical for both female gametogenesis and embryogenesis. Journal of Integrative Plant Biology 55, 64–74. [DOI] [PubMed] [Google Scholar]
- Wen R, Newton L, Li G, Wang H, Xiao W. 2006. Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Molecular Biology 61, 241–253. [DOI] [PubMed] [Google Scholar]
- Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W. 2008. Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. The Plant Cell 20, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Wang S, Xiang D, Venglat P, Shi X, Zang Y, Datla R, Xiao W, Wang H. 2014. UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. The Plant Journal 80, 424–436. [DOI] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. 2011. a. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M. 2011. b. K11-linked ubiquitin chains as novel regulators of cell division. Trends in Cell Biology 21, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. 2009. Identification of a physiological E2 module for the human anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America 106, 18213–18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Menard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH. 2009. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana . The Plant Journal 57, 279–288. [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN. 2004. Female gametophyte development. The Plant Cell 16 Suppl, S133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q. 2013. A plant-specific in vitro ubiquitination analysis system. The Plant Journal 74, 524–533. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. 2005. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. The Plant Cell 17, 3422–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.