Abstract
Phytochrome A (phyA) is a light labile phytochrome that mediates plant development under red/far-red light condition. Degradation of phyA is initiated by red light-induced phyA-ubiquitin conjugation through the 26S proteasome pathway. The N-terminal of phyA is known to be important in phyA degradation. To determine the specific lysine residues in the N-terminal domain of phyA involved in light-induced ubiquitination and protein degradation, we aligned the amino acid sequence of the N-terminal domain of Arabidopsis phyA with those of phyA from other plant species. Based on the alignment results, phytochrome over-expressing Arabidopsis plants were generated. In particular, wild-type and mutant (substitutions of conserved lysines by arginines) phytochromes fused with GFP were expressed in phyA-211 Arabidopsis plants. Degradation kinetics of over-expressed phyA proteins revealed that degradation of the K206R phyA mutant protein was delayed. Delayed phyA degradation of the K206R phyA mutant protein resulted in reduction of red-light-induced phyA-ubiquitin conjugation. Furthermore, seedlings expressing the K206R phyA mutant protein showed an enhanced phyA response under far-red light, resulting in inhibition of hypocotyl elongation as well as cotyledon opening. Together, these results suggest that lysine 206 is the main lysine for rapid ubiquitination and protein degradation of Arabidopsis phytochrome A.
Keywords: light-induced phyA degradation, phytochrome A, proteasome, ubiquitination
Light is one of the most important environment factors that regulate plant growth and development. Light provides energy for photosynthesis as well as modulates many developmental processes in plants (1, 2). Plants are very sensitive to changes in light conditions because they have several classes of photoreceptors such as red and far-red light-absorbing phytochromes (3), blue/ultraviolet A (UVA) light-absorbing cryptochromes, phototropins, members of the Zeitlupe family (4, 5) and ultraviolet B (UVB) light-absorbing UV resistance eight proteins (6).
Red and far-red light-absorbing phytochromes play many roles in the plant life cycle from seed germination to reproductive development (7, 8). Native phytochromes are synthesized in the dark in an inactive red light-absorbing form (Pr); after red light absorption, their conformation changes into the active far-red absorbing form (Pfr). Active Pfr form can be converted back to the inactive Pr form by absorption of specific far-red light (9, 10). The model plant, Arabidopsis thaliana, contains five genes (PHYA to PHYE) that encode phytochrome apoproteins (11, 12). Phytochromes are classified as light labile phytochrome A (phyA) and light stable phytochromes B–E (phyB-phyE) (13). Among the light stable phytochromes, phyB mediates the red high irradiance response (red HIR) and the red/far-red reversible low fluence response (red/far-red LFR) (14–16). Compared with light stable phytochromes, light labile phyA modulates specific physiological processes through two action modes: the very low fluence response (VLFR) and the far-red HIR (17–19). In addition, phyA also shows high red light irradiance responses (20). To mediate photomorphogenesis, the Pr form of phyA in the cytoplasm is converted to the Pfr form by light absorption, upon which Pfr is translocated into the nucleus with the help of Far-red Hypocotyl 1 (FHY1) and FHY1-Like proteins (21, 22). Phytochrome A is the only phytochrome that translocates to the nucleus under far-red light conditions (23).
Phytochrome A protein is known to be degraded rapidly in response to light absorption after conformational change into the Pfr form. PhyA is expressed at high levels in the cytoplasm in the inactive Pr form in the dark. When a light signal is provided, the light-induced active Pfr form of phyA translocates to the nucleus, but degradation occurs in both the cytoplasm and nucleus (22, 24, 25). This light-induced reduction in phyA protein levels involves light-repressed PHYA gene transcription (26) as well as light-induced protein degradation (27–29). The rate of phyA degradation is decreased in ubiquitin mutant plants, indicating that ubiquitination by the proteasome pathway is involved in phyA degradation (30). In addition, proteasome inhibitor (MG132) has been shown to slow down phyA degradation under red light conditions, confirming that phytochrome degradation occurs mainly via the proteasome pathway (22, 24).
Previous phyA degradation studies (31, 32) found that the N-terminal domain of phyA plays an important role in phyA degradation, with some contribution from the C-terminal domain. The N-terminal domain of potato phyA is required for phyA degradation through the selective recognition and ubiquitination of Pfr function (31). In addition, a truncated Arabidopsis phyA N-terminal mutant (PHYA686-YEP-DD) was imported into the nucleus and degraded under red light, even though its degradation was a little slower than that of wild-type PHYA-YEP; however, PHYA686-YEP-DD did not initiate far-red HIR signalling responses (25). Moreover, studies with chimeric phytochromes between Arabidopsis PHYA and PHYB confirmed that the 415 N-terminal amino acid (N-PAS and GAF domain) of Arabidopsis phy A is critical for rapid Pfr phyA degradation (32).
Our aim in this study was to identify ubiquitination sites in the N-terminal domain of phytochrome A. For this purpose, conserved lysines on the N-terminal domain of Arabidopsis phyA were replaced by arginines and these mutant constructs were expressed in phyA-211 mutant plants, followed by analysis of degradation kinetics. Degradation kinetic studies of wild-type and mutant phyA proteins revealed delayed degradation of the K206R phyA mutant protein. K206R phyA showed less ubiquitination than wild-type phyA in co-immunoprecipitation analysis as well. Furthermore, K206R phyA-expressing plants showed increased biological phyA responses.
Materials and Methods
Plasmid construction, site-directed mutagenesis and plant transformation
cDNA encoding full-length wild-type AtPHYA was amplified by overlapping polymerase chain reaction to generate a AtPHYA::GFP fusion gene, which was inserted into the XhoI and SacI restriction sites of plant binary vector, pJJ461. Each lysine to arginine (K_R) mutant was generated in a truncated DNA fragment using mutated primers and inserted back into the XhoI and AvrII restriction sites of the truncated DNA fragment of the wild-type AtPHYA::GFP fusion gene. All primer sequences are listed in Supplementary Table S1. All mutated DNA constructs were verified by DNA sequencing. For protein expression, DNA constructs cloned in binary vector pJJ461 were introduced into the phyA-null allele of the Col-0 ecotype (phyA-211) using the Agrobacterium tumefaciens-mediated floral dip method (33).
Plant growth conditions
Arabidopsis thaliana plants were grown in a culture room with a 16 h light/8 h dark cycle at 23°C for general growth, transformation and seed harvesting. For seedling growth, seeds were surface sterilized in 70% ethanol for 5 min, followed by incubation in 20% bleach (sodium hypochlorite: 12% Cl) plus 0.05% Triton X-100 for 5 min and washed five times with sterile water. Seeds were placed on one-half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) without sucrose plus 0.8% (W/V) phyto agar (Duchefa Biochemie) in petri dishes. Plates were stored at 4°C in the dark for 2–4 days for stratification. Germination was induced by exposing seeds to white light at 23°C for 4 h followed by incubation in the dark for 18–24 h. Plates were exposed to different light conditions or continuous dark.
Immunoblot analysis and co-immunoprecipitation assay
Experiments were performed under dim green light. Three-day-old dark-grown seedlings, which had been treated with red light for different periods of time, were homogenized in liquid nitrogen in extraction buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.1 mM dithiothreitol, 0.2% Triton X-100 and 1X complete protease inhibitor cocktail (Roche)). Crude extract was centrifuged at 14,000 rpm for 15 min at 4°C, and the protein concentration was measured. Same protein concentration was mixed with the same volume of Z buffer (125 mM Tris-HCl, 12% (w/v) sodium dodecyl sulfate (SDS), 10% (v/v) glycerol, 22% (v/v) β-mercaptoethanol and 0.001% (w/v) bromophenol blue) and boiled for 5 min. Proteins were separated using 8% SDS polyacrylamide gel electrophoresis (PAGE). Proteins were then transferred to a nitrocellulose membrane (Whatman, Florham Park, NJ) and probed with mouse monoclonal anti-GFP (Santa Cruz) for phyA determination.
For co-immunoprecipitation, samples were homogenized in extraction buffer including 1 mM phenylmethylsulfonyl fluoride. Crude total protein extract was centrifuged and filtered through a 0.45 μm polyvinylidene difluoride membrane filter. Clear total protein extract (500 μg) was mixed with 1 μg of anti-GFP antibody (ab6556) and protein A/G agarose (Santa Cruz) and incubated for 4 h at 4°C. Antibody-bound protein complexes were precipitated with protein A/G agarose. Beads were washed five times with extraction buffer. Proteins were eluted and analysed by western blot using anti-Ubi (ab7254) and anti-phyA antibody.
Proteasome inhibitor treatment
Three-day-old dark-grown seedlings were incubated in one-half-strength MS liquid media including dimethylsulfoxide (DMSO) (Duchefa Biochemie) as a control or 50 µM MG132 (Calbiochem) dissolved in DMSO for 3 h. Seedlings were exposed to red light and protein extraction was performed using the method described earlier.
Phenotypic assay
About 30 seeds per line were sterilized and plated on one-half-strength MS without sucrose and incubated at 4°C in the dark for about 2–4 days. Seeds were then treated with white light at 23°C for 4 h and then incubated in darkness for 18–24 h. Seeds were grown under different fluence continuous far-red light for 4 days or were exposed to a 3 min far-red light pulse every hour for 3 days. Hypocotyl length and cotyledon opening were analysed and standardized.
Results
Lysine 206 of Arabidopsis phyA is the main ubiquitination site
The N-terminal domain of phyA has been shown to be ubiquitinated and degraded as a result of the biochemical response to light (31, 32). Identification of ubiquitination site can be accomplished by either direct analysis of ubiquitinated proteins by mass spectrometry (MS) or protein degradation analysis using lysine mutated proteins (34, 35). Here, we identified ubiquitination sites of phyA by using the lysine mutated phyA proteins because prerequisite purification of ubiquitinated phyA for MS analysis is really difficult process. To identify the critical residues involved in ubiquitination and degradation under red light, the 415 N-terminal amino acids of Arabidopsis phyA were aligned with those of other plant phytochromes. Amino acid alignment showed that 15 of 21 Arabidopsis phyA N-terminal lysines were highly conserved among the 10 plant species analysed (Supplementary Fig. S1). To analyse these 15 conserved lysines, we generated 12 Arabidopsis mutant phyA constructs with lysine replaced by arginine; nine mutants with single lysine to arginine substitutions, and three mutants where two lysines in close proximity were replaced with arginines (202/206, 284/286 and 361/363). The 12 phyA mutant constructs (Supplementary Table S2) and wild-type phyA were fused with GFP (K_R phyA mutants and AtPHYA::GFP) and introduced into a phytochrome A null mutant of Arabidopsis (phyA-211).
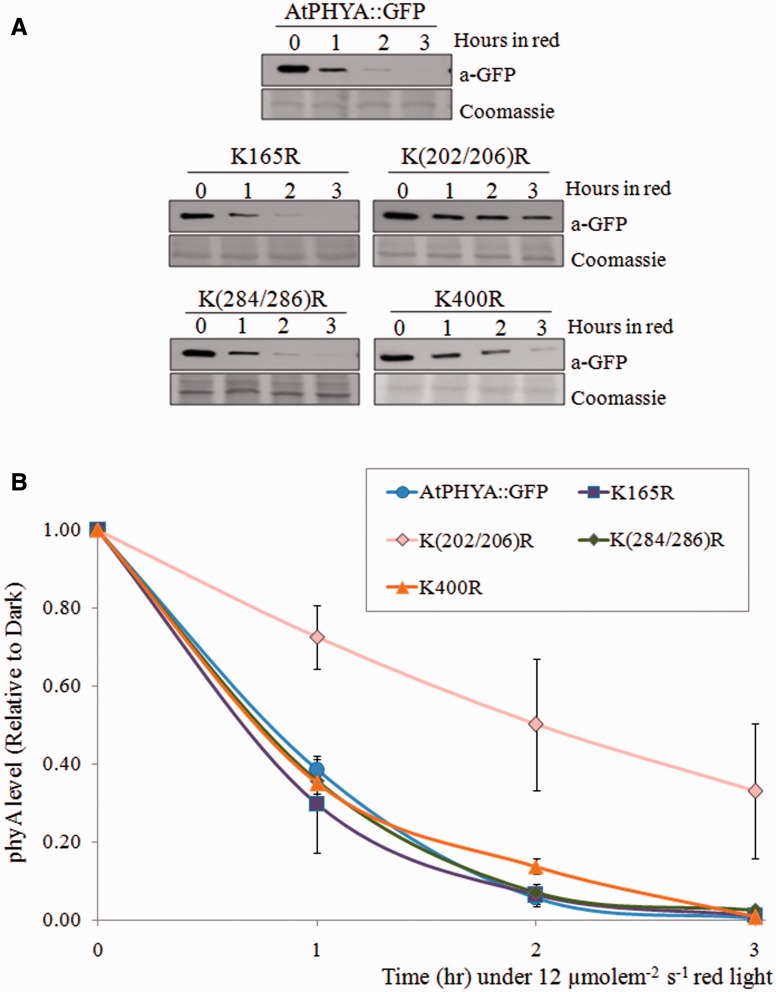
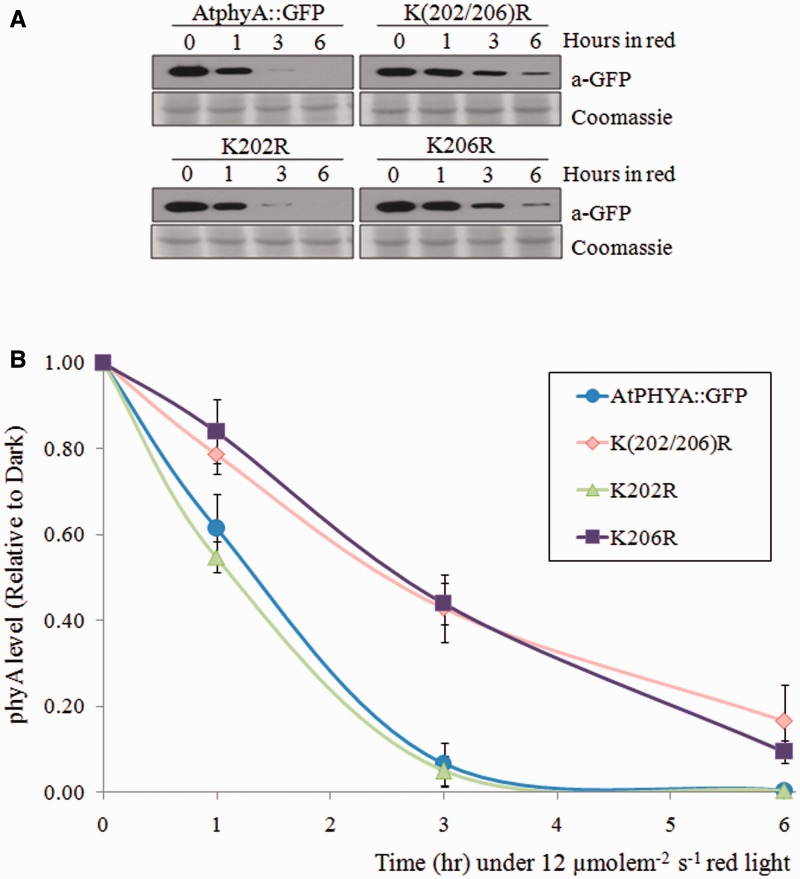
The degradation kinetics of the mutant phyA proteins were compared with those of wild-type phyA. Levels of AtPHYA::GFP control protein decreased rapidly by ∼50–60% after 1 h of red light treatment, and complete degradation was observed after 3 h (Fig. 1). Most K_R phyA mutants had similar degradation kinetics to AtPHYA::GFP; the exception was the K(202/206)R phyA mutant. This phyA double mutant degraded more slowly than wild-type phyA protein under red light conditions (Fig. 1 and Supplementary Fig. S2). To further investigate the effects of the other conserved N-terminal lysines of phyA on degradation, triple or quadruple K_R phyA mutants were generated in combination with K(202/206)R (Supplementary Table S3). No additive degradation effects were observed for these triple or quadruple mutants relative to K(202/206)R phyA (Supplementary Fig. S3). Therefore, single K202R and K206R phyA mutants were generated. Degradation kinetics of K202R phyA were similar to those of AtPHYA::GFP, while the degradation kinetics of K206R phyA were similar to those of dual K(202/206)R phyA (Fig. 2).
Fig. 1.
Degradation kinetics of AtPHYA::GFP and K_R phyA mutants under red light. (A) Western blot of AtPHYA::GFP and K_R phyA mutant proteins extracted from 3-day-old dark-grown seedling treated with continuous red light (12 µmole m−2 s−1). Twenty microgram aliquots of crude extract was separated on 8% SDS-PAGE gel and the western blot was probed with anti-GFP antibody (top). The same blots were subjected to Coomassie blue staining (bottom) as a control. (B) Quantification of phyA levels in western blot bands. PhyA protein level was compared with the dark levels of each genotype as a control. Data are mean of biological triplicates ± SD.
Fig. 2.
Degradation kinetics of K202R and K206R phyA mutants. (A) Western blot of AtPHYA::GFP, K(202/206)R, K202R and K206R phyA proteins extracted from 3-day-old dark-grown seedling treated with continuous red light (12 µmole m−2 s−1). Twenty microgram aliquots of crude extract was separated on 8% SDS-PAGE gel and the western blot was probed with anti-GFP antibody (top). The same blots were subjected to Coomassie blue staining (bottom) as a control. (B) Quantification of phyA levels in western blot bands. PhyA protein level was compared with the dark levels of each genotype as a control. Data are means of biological quintuplicates ± SD.
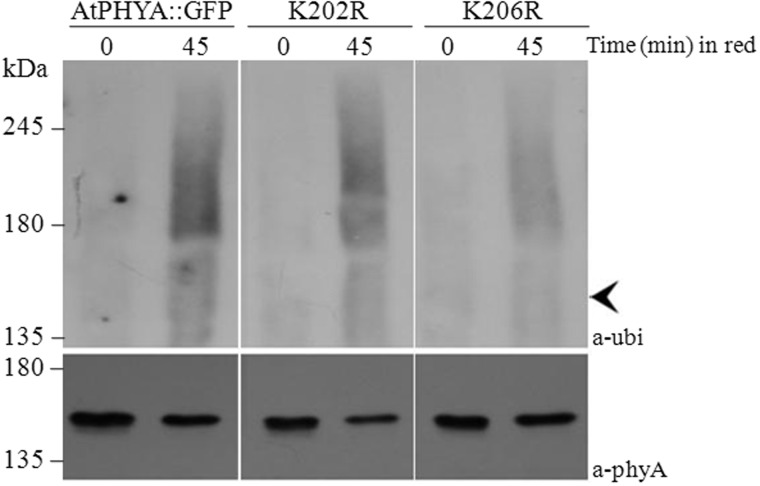
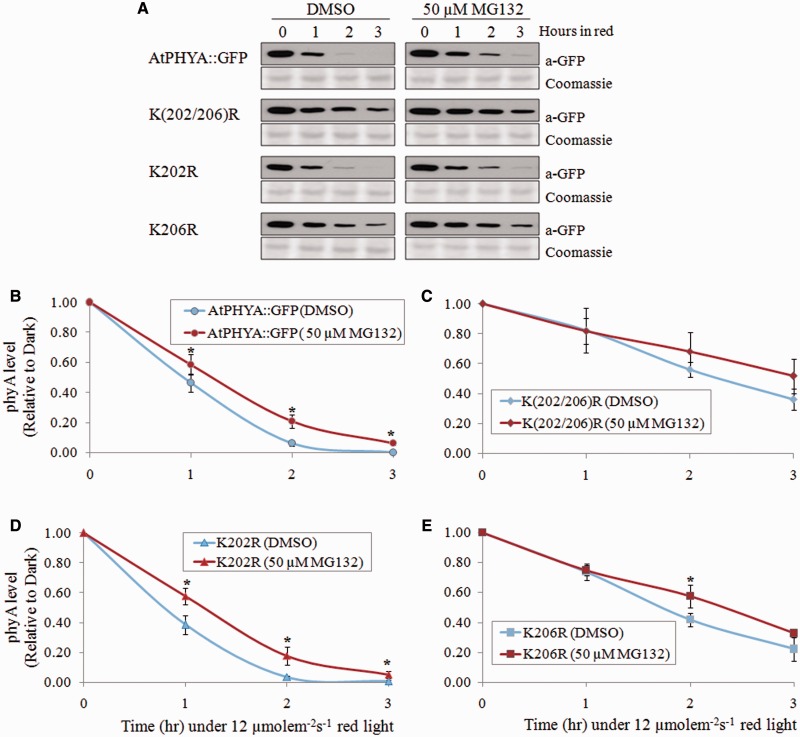
Ubiquitination is a prerequisite process for phyA degradation (28, 29). To confirm that the lysine at 206 was the main target for ubiquitination, ubiquitination levels of wild type, K202R and K206R phyA proteins were analysed. Seedlings expressing AtPHYA::GFP, K202R and K206R phyA were exposed to red light for different periods of time, and phyA proteins were purified by co-immunoprecipitation. Ubiquitinated phyA protein conjugate was detected by western blot analysis with anti-ubiquitin antibody. Ubiquitinated phyA protein was detected after red light treatment in all phyA samples. However, the level of ubiquitinated K206R phyA was much lower than levels of AtPHYA::GFP and K202R phyA (Fig. 3). These results indicate that K206 plays a greater role than the other conserved N-terminal lysines in rapid ubiquitination and degradation of phyA. Proteasome inhibitor MG132 has been reported to partially inhibit the phyA degradation in response to red light (22, 24, 29). To investigate the effect of MG132 on the degradation of K(202/206)R, K202R and K206R phyA mutant proteins, 3-day-old dark-grown seedlings were treated with MG132 (50 µM), and phyA protein degradation was analysed after red light treatment (Fig. 4A). The K202R phyA mutant protein showed delayed degradation similar to that of wild-type phyA protein in response to MG132 treatment (Figs. 4B and D), consistent result with a previous study that MG132 slows down phyA degradation (22, 24). However, K(202/206)R and K206R phyA mutant proteins had a similar degradation pattern with and without MG132 treatment (Fig. 4C and E), suggesting that K206 is required for the degradation of phyA through the proteasome pathway.
Fig. 3.
Western blot analysis of phytochrome-ubiquitin conjugates of wild type, K202R and K206R phyA proteins. Proteins were extracted from 3-day-old etiolated seedlings exposed to continuous red light (12 µmole m−2 s−1) and immunoprecipitated using anti-GFP polyclonal antibody. Immunoprecipitated samples were separated on 6% SDS-PAGE gels and probed with anti-Ubi or anti-phyA antibodies. Arrowhead indicates unubiquitinated AtPhyA::GFP (∼150 kDa).
Fig. 4.
The effects of proteasome inhibitor (MG132) on phyA degradation. (A) Degradation kinetics of AtPhyA::GFP and K_R phyA mutant proteins. Three-day-old dark-grown seedling were incubated with DMSO or 50 µM MG132 for 3 h and then exposed to red light (12 µmole m−2 s−1). Twenty microgram aliquots of crude extract were separated on 8% SDS-PAGE gel, and the western blot was probed with anti-GFP antibody (top). The same blots were subjected to Coomassie blue staining (bottom) as a control. (B–E) Quantification of phyA levels in western blot bands. PhyA protein level was compared with the dark levels of each genotype as a control. Data are means ± SDs of three replicates. Statistical significance of the difference in expression according to DMSO or MG132 was determined using the t-test as implemented in IBM SPSS Statistics 21 with P < 0.05 considered statistically significant.
Seedlings expressing K206R phyA showed inhibition of hypocotyl elongation and cotyledon opening
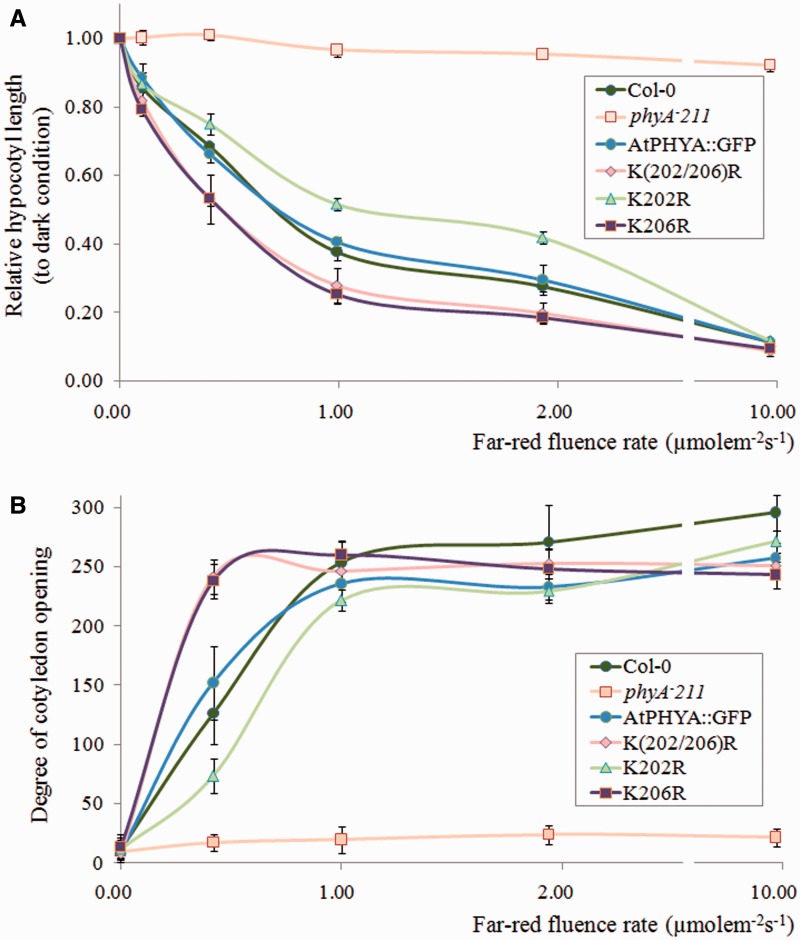
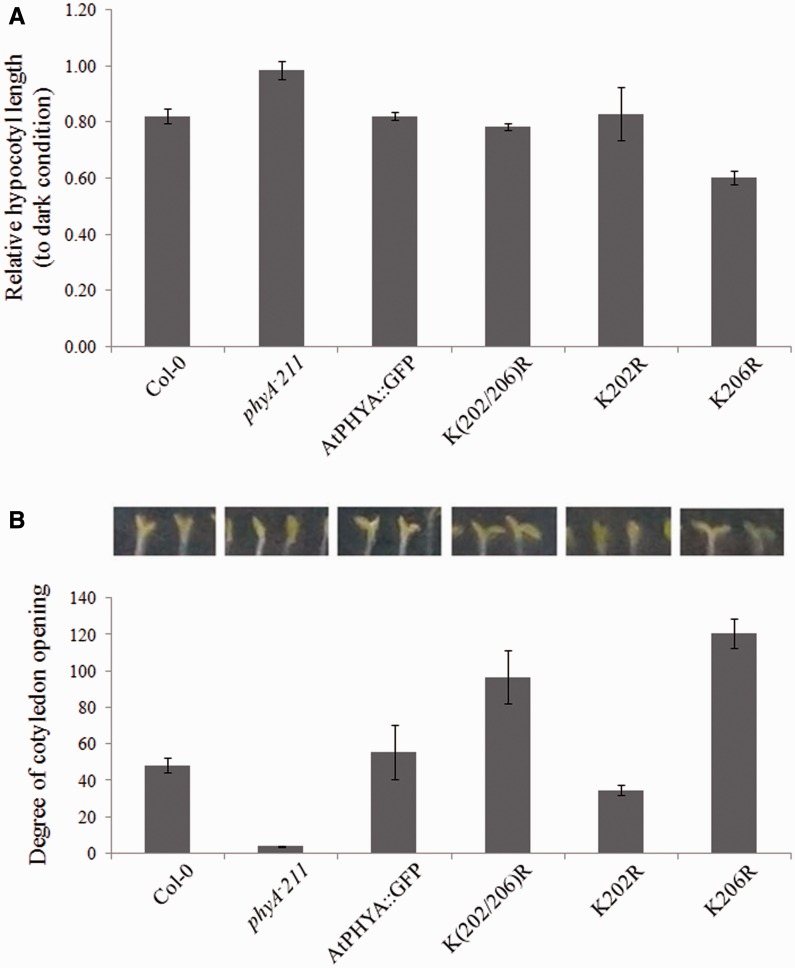
Unlike light stable phytochromes (phyB-E), phytochrome A mediates VLFR and far-red HIR responses such as germination, hypocotyl elongation and cotyledon opening under very low fluence red light conditions or far-red light conditions (17, 18, 36, 37). To investigate the biological effects of the K206R mutation of phyA, VLFR and far-red HIR photomorphogenesis, such as inhibition of hypocotyl elongation under continuous far-red light, were analysed. In contrast to a null phyA mutant (phyA-211) seedling, the hypocotyl elongation of all phyA expressing seedlings including Col-0, AtPHYA::GFP, K(202/206)R, K202R and K206R was inhibited by an increase in far-red light intensity (Fig. 5A). However, hypocotyl elongation was inhibited to a greater extent in K(202/206)R and K206R phyA mutant seedlings than Col-0 and AtPHYA::GFP seedlings (Fig. 5A). Because cotyledon opening is a well-known phyA phenotype under far-red light conditions (36–38), the degree of cotyledon opening under different fluence far-red light conditions was investigated. The phyA null plant phyA-211 did not display cotyledon opening under dark or far-red light conditions, whereas all phyA-expressing plants showed increased cotyledon opening as the fluence of far-red light increased (Fig. 5B). Under low fluence (below 1 µmolem−2s−1) far-red light, cotyledon opening of K(202/206)R and K206R phyA mutant plants occurred to a greater extent than that of Col-0 and AtPHYA::GFP plants, consistent with increased phyA activity due to delayed phyA degradation (Fig. 5B). However, K202R phyA mutant plants showed longer hypocotyl length and inhibited cotyledon opening compared with AtPHYA::GFP plants, indicating that the K202R phyA mutant protein showed decreased phyA function (Fig. 5A and B). These phenotypic results were duplicated even under very low fluence far-red light pulses (Fig. 6A and B). It is unclear why K202R shows decreased photomorphogenesis. Nevertheless, these results suggested that delayed phyA degradation resulted in an increase in phyA effects on early photomorphogenic responses such as inhibition of hypocotyl elongation and cotyledon opening under far-red light conditions.
Fig. 5.
Phenotypic analysis of phyA-expressing seedlings grown under continuous far-red conditions. (A) Hypocotyl length and (B) angle of cotyledon opening of seedlings grown under continuous far-red light for 4 days. Hypocotyl lengths relative to those of dark grown seedlings are reported. Data are means ± SDs (n = 30). Col-0, Columbia.
Fig. 6.
Phenotypic analysis of phyA-expressing seedlings under hourly far-red pulse conditions. (A) Hypocotyl length and (B) angle of cotyledon opening of seedlings grown under 3-min far-red light pulses hourly (5 µmole m−2 s−1) for 3 days. Hypocotyl lengths relative to those of dark grown seedlings are reported. Data are means ± SDs (n = 30). Col-0, Columbia.
Discussion
Light-induced phyA degradation by ubiquitination via the 26S proteasome pathway is an important biochemical process that controls phyA signalling (27–29). E3 ligase, a key protein in the proteasome pathway, recognizes the target protein for ubiquitination and consequently transfers ubiquitin to specific target lysine residues. COP1 have been identified as E3 ligases in the proteasome pathway, which with CUL1 is involved in phyA degradation under different growth media conditions (29, 30). To define specific ubiquinated lysines in phyA, C-terminal conserved lysines within the region required for Pfr signal transduction of oat phyA were replaced with arginines but these mutant phyA proteins showed the same degradation kinetics as wild-type phyA (31). Recent phyA functional studies showed that the N-terminal domain of phyA is important for ubiquitination and degradation (25, 31, 32). We identified lysines in the N-terminal of phyA involved in ubiquitination and degradation. We found that dual mutation of lysines 202 and 206 delayed phyA degradation under red light (Supplementary Fig. S2). The single mutation of lysine 206 was sufficient to delay phyA degradation. To determine if additional lysines were important for phyA degradation, we generated a construct in which all 15 conserved lysines were replaced with arginines (15K_Rs phyA mutant). The degradation kinetics of the 15K_Rs phyA mutant was similar to those of the K206R phyA mutant (Supplementary Fig. S4). These results suggested that only lysine 206 in the N-terminal of phyA plays an important role in phyA degradation.
Ubiquitination is a prerequisite process for all proteins targeted for degradation through the proteasome pathway; less ubiquitinated proteins are degraded to a lesser extent than more ubiquitinated proteins. PhyA is ubiquitinated before degradation but phyA-ubiquitin conjugation was reduced in the K206R mutant (Fig. 3). This result indicates that lysine 206 of Arabidopsis phyA is an important N-terminal lysine for ubiquitination. Only the K206R phyA mutant showed delayed phyA degradation as well as reduced phyA-ubiquitin conjugation. The K206R mutant phyA protein was still degraded by ubiquitination, although degradation was delayed. We expected phyA to be stable when all 15 conserved N-terminal lysines were replaced with arginines, but the mutant was still degraded. In addition, missense mutations in the N-terminal of phyA resulting in amino acid changes or interruption of autophosphorylation reduced phyA degradation, suggesting that there might be other degradation mechanisms for phyA degradation in addition to the proteasome pathway (39–41). These data suggest that if the main ubiquitination site in the N-terminal domain is excluded, E3 ligase ubiquitinates alternative lysines to degrade phyA and/or other unknown mechanisms are involved in phyA degradation. To evaluate these possibilities, we used a proteasome inhibitor to confirm that phyA is degraded via the proteasome pathway. MG132 is a protease inhibitor, and treatment of plant with MG132 has been shown to delay phyA degradation under red light conditions (22, 24). In this study, MG132 had no effect on the degradation of K206R phyA protein, even though wild-type phyA degradation was delayed (Fig. 4). These data strengthen our hypothesis that lysine 206 is the main N-terminal lysine in phyA for ubiquitination and degradation. Therefore, we should think about there might be other major degradation mechanisms except proteasome pathway for phyA degradation process.
N-terminal domain of phyA is important for the biological activities of phytochrome A (32, 42). Some point mutations in the N-terminal domain of phyA adversely affect the biological functions of phyA (43). However, most point mutations of phyA that involved substitution of conserved lysines by arginines in the N-terminal domain had normal phyA activity, with the exception of K202R. K202R phyA had reduced phyA activity, even though the degradation kinetics of this mutant protein was normal (Fig. 2). The reason for this is unclear. There may have been some structural change in the phytochrome A protein due to this substitution.
PhyA signalling during early development is regulated by protein stability (41, 42). Therefore, if phyA degradation is delayed, phyA function could be enhanced during the early stage of development. We analysed the effect of mutating K206 of phyA to R206 on the inhibition of hypocotyl elongation and cotyledon opening under far-red light. PhyA null mutant seedlings are characterized by loss of inhibition of hypocotyl elongation and cotyledon opening under far-red light conditions, which can be rescued by introduction of full-length phytochrome A (25, 42, 44). Stable phyA mutants exhibit hypersensitive responses to light due to increased phyA activity (41). Our results were consistent with these previous reports; delayed phyA degradation due to mutation of an ubiquitination site (lysine 206) enhanced phyA function in terms of inhibition of hypocotyl elongation and cotyledon opening (Figs 6 and 7). These data indicate that lysine 206 of Arabidopsis phyA is the main ubiquitination site among the N-terminal conserved lysines and that phyA stability is important for the biological functions of phyA.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
This research was supported by a grant from the Next-Generation BioGreen 21 Program (Project No: PJ01107501), Rural Development Administration, Republic of Korea.
Conflict of Interest
None declared.
References
- 1.Deng X.W., Quail P.H. (1999) Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129 [DOI] [PubMed] [Google Scholar]
- 2.Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010) Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66 [DOI] [PubMed] [Google Scholar]
- 3.Neff M.M., Fankhauser C., Chory J. (2000) Light: an indicator of time and place. Genes Dev. 14, 257–271 [PubMed] [Google Scholar]
- 4.Franklin K.A., Larner V.S., Whitelam G.C. (2005) The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49, 653–664 [DOI] [PubMed] [Google Scholar]
- 5.Demarsy E., Fankhauser C. (2009) Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12, 69–74 [DOI] [PubMed] [Google Scholar]
- 6.Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 [DOI] [PubMed] [Google Scholar]
- 7.Franklin K.A., Quail P.H. (2010) Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasser B., Sánchez-Lamas M., Yanovsky M.J., Casal J.J., Cerdán P.D. (2010) Arabidopsis thaliana life without phytochromes. Proc. Natl Acad. Sci. USA. 107, 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockwell N.C., Su Y.S., Lagarias J.C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae G., Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311 [DOI] [PubMed] [Google Scholar]
- 11.Sharrock R.A., Quail P.H. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757 [DOI] [PubMed] [Google Scholar]
- 12.Clack T., Mathews S., Sharrock R.A. (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427 [DOI] [PubMed] [Google Scholar]
- 13.Sharrock R.A., Clack T. (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinomura T., Nagatani A., Hanzawa H., Kubota M., Watanabe M., Furuya M. (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 93, 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 [DOI] [PubMed] [Google Scholar]
- 17.Nagatani A., Reed J.W., Chory J. (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neff M.M., Chory J. (1998) Genetic interactions between phytochrome A, phytochrome B, and crytochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin K.A., Whitelam G.C. (2007) Phytochrome A function in red light sensing. Plant Signal. Behav. 2, 383–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debrieux D., Fankhauser C. (2010) Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol. Biol. 73, 687–695 [DOI] [PubMed] [Google Scholar]
- 23.Chen M. (2008) Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Curr. Opin. Plant Biol. 11, 503–508 [DOI] [PubMed] [Google Scholar]
- 24.Toledo-Ortiz G., Kiryu Y., Kobayashi J., Oka Y., Kim Y., Nam H.G., Mochizuki N., Nagatani A. (2010) Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol. 51, 1648–1660 [DOI] [PubMed] [Google Scholar]
- 25.Wolf I., Kircher S., Fejes E., Kozma-Bognár L., Schäfer E., Nagy F., Ádám E. (2011) Light-regulated nuclear import and degradation of Arabidopsis phytochrome-A N-terminal fragments. Plant Cell Physiol. 52, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantón F.R., Quail P.H. (1999) Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis . Plant Physiol. 121, 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanklin J., Jabben M., Vierstra R.D. (1987) Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc. Natl Acad. Sci. USA. 84, 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabben M., Shanklin J., Vierstra R.D. (1989) Red light-induced accumulation of ubiquitin-phytochrome conjugates in both monocot and dicot. Plant Physiol. 90, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18, 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debrieux D., Trevisan M., Fankhauser C. (2013) Conditional involvement of constitutive photomorphogenic1 in the degradation of phytochrome A. Plant Physiol. 161, 2136–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough R.C., Jordan-Beebe E.T., Lohman K.N., Marita J.M., Walker J.M., Gatz C., Vierstra R.D. (1999) Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167 [DOI] [PubMed] [Google Scholar]
- 32.Oka Y., Ono Y., Toledo-Ortiz G., Kokaji K., Matsui M., Mochizuki N., Nagatani A. (2012) Arabidopsis phytochrome A is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome. Plant Cell 24, 2949–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 34.Xu G., Jaffrey S.R. (2013) Proteomic Identification of protein ubiquitination events. Biotechnol. Genet. Eng. Rev. 29, 73–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hau Z., Kao T. (2008) Identification of major lysine residues of S3-RNase of Petunia inflate involved in ubiquitin-26S proteasome-mediated degradation in vitro. Plant J. 54, 1094–1104 [DOI] [PubMed] [Google Scholar]
- 36.Parks B.M., Quail P.H. (1993) hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casal J.J., Davis S.J., Kirchenbauer D., Viczian A., Yanovsky M.J., Clough R.C., Kircher S., Jordan-Beebe E.T., Schäfer E., Nagy F., Vierstra R.D. (2002) The serine-rich N-terminal domain of oat phytochrome A helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol. 129, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerdán P.D., Yanovsky M.J., Reymundo F.C., Nagatani A., Staneloni R.J., Whitelam G.C., Casal J.J. (1999) Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana . Plant J. 18, 499–507 [DOI] [PubMed] [Google Scholar]
- 39.Weller J.L., Batge S.L., Smith J.J., Kerckhoffs L.H.J., Sineshchekov V.A., Murfet I.C., Reid J.B. (2004) A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135, 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dieterle M., Bauer D., Büche C., Krenz M., Schäfer E., Kretsch T. (2005) A new type of mutation in phytochrome A causes enhanced light sensitivity and alters the degradation and subcellular partitioning of the photoreceptor. Plant J. 41, 146–161 [DOI] [PubMed] [Google Scholar]
- 41.Han Y.J., Kim H.S., Kim Y.M., Shin A.Y., Lee S.S., Bhoo S.H., Song P.S., Kim J.I. (2010) Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 51, 596–609 [DOI] [PubMed] [Google Scholar]
- 42.Trupkin S.A., Debrieux D., Hiltbrunner A., Fankhauser C., Casal J.J. (2007) The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol. Biol. 63, 669-678 [DOI] [PubMed] [Google Scholar]
- 43.Mateos J.L., Luppi J.P., Ogorodnikova O.B., Sineshchekov V.A., Yanovsky M.J., Braslavsky S.E., Gärtner W., Casal J.J. (2006) Functional and biochemical analysis of the N-terminal domain of phytochrome A. J. Biol. Chem. 281, 34421–34429 [DOI] [PubMed] [Google Scholar]
- 44.Yanovsky M.J., Luppi J.P., Kirchbauer D., Ogorodnikova O.B., Sineshchekov V.A., Adam E., Kircher S., Staneloni R.J., Schäfer E., Nagy F., Casal J.J. (2002) Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.