Abstract
Small GTPases play important roles in various aspects of cell division as well as membrane trafficking. We and others previously showed that ADP-ribosylation factor 6 (Arf6) is locally activated around the ingressing cleavage furrow and recruited to the Flemming body in late cytokinesis phases, and involved in faithful completion of cytokinesis. However, knockout of the Arf6 gene or Arf6 depletion by siRNAs did not drastically influence cytokinesis. We here show that, in addition to Arf6, Class I Arfs (Arf1 and Arf3) are localized to the Flemming body, and that double knockdown of Arf1 and Arf3 moderately increases the proportion of multinucleate cells and simultaneous knockdown of Arf1, Arf3 and Arf6 leads to severe cytokinesis defects. These observations indicate that Arf1 and Arf3 as well as Arf6 play important roles in cytokinesis. We further show that EFA6 (exchange factor for Arf6) activates not only Arf6 but also Arf1 in the cell. Taken together with our previous data, these Arf GTPases are likely to be locally activated by EFA6 and in turn targeted to the Flemming body to complete cytokinesis.
Keywords: Arf1, Arf6, cytokinesis, EFA6, Flemming body
The ADP-ribosylation factor (Arf) small GTPases serve as molecular switches in regulation of various aspects of membrane trafficking by cycling between a GDP-bound inactive state and a GTP-bound active state. Exchange of bound GDP for GTP on Arfs is catalyzed by guanine nucleotide exchange factors (GEFs), while hydrolysis of bound GTP to GDP and inorganic phosphate is stimulated by GTPase-activating proteins (1, 2). There are six Arf isoforms in mammals. These Arfs are grouped into three classes based on similarity in the primary structure (3, 4): Class I, Arf1–Arf3 (Arf2 is absent in humans); Class II, Arf4 and Arf5; and Class III, Arf6. Among them, Arf1 and Arf6 have been well characterized so far. Arf1 triggers budding of coated carrier vesicles from the Golgi complex and endosomal compartments. Arf6 is divergent from Arf1–Arf5 and localizes to the plasma membrane and endosomal compartments, where it regulates endosomal recycling and remodelling of actin cytoskeleton and membranes (5–7). In addition to these roles in interphase cells, we and others showed that Arf6 is required for faithful completion of cytokinesis (8–12).
We have recently shown that Arf6 is transiently colocalized with and activated by exchange factor for Arf6 (EFA6; (13)) around the ingressing cleavage furrow, and subsequently recruited to the Flemming body in late cytokinetic phases (9, 14, 15). The Flemming body, also known as midbody matrix or midbody dark zone, is a dense structure in the middle of antiparallel microtubule bundles of the central spindle, and serves as a platform for assembly of a variety of structural and regulatory proteins required for completion of cytokinesis, including proteins involved in membrane trafficking (16–18); hereafter, we refer to the middle dense structure of the central spindle as the Flemming body, since in several published studies, the term ‘midbody’ has been confusingly used to represent the entire central spindle or midbody microtubules. Furthermore, we and others have shown that a GTP-bound active, but not a GDP-bound inactive, form of Arf6 is recruited to the Flemming body through interacting with mitotic kinesin-like protein 1 (MKLP1) (8, 9); MKLP1, which forms the central spindlin complex along with MgcRacGAP (also known as CYK-4), localizes to the Flemming body and plays a crucial role in completion of cytokinesis (19, 20).
Although previous studies of our and other research groups suggested a role of Arf6 in completion of cytokinesis, an increase in the proportion of multinucleate cells induced by depletion of Arf6 by siRNAs was not so drastic (9, 11, 12). Furthermore, although some fraction of embryonic fibroblasts derived from an Arf6-knockout mouse exhibit typical multinucleate phenotype, a large fraction of the knockout cells appear to divide without remarkable defects (9). Taken together with the fact that all Arf isoforms can interact with MKLP1 (9), these data make it possible that Arf isoform(s) other than Arf6 also participate in regulation of cytokinesis. To address this possibility, we here examined localization of all Arf isoforms during mitosis and impact of individual and combinatorial depletion of these Arf isoforms by siRNAs on cytokinesis, and found that not only Arf6 but also Class I Arfs (Arf1 and Arf3) are localized to the Flemming body in late cytokinetic phases and contribute to faithful completion of cytokinesis.
Materials and Methods
Antibodies and reagents
Sources of antibodies and reagents are as follows: polyclonal rabbit anti-Arf6 (21), a kind gift from Yasunori Kanaho (University of Tsukuba, Japan); monoclonal mouse anti-Arf1 (3F1), Thermo Scientific; monoclonal mouse anti-Arf3 (41/ARF3), BD Transduction Laboratories; polyclonal rabbit-Arf4, ProteinTech Group; monoclonal mouse anti-Arf5 (1B4), Abnova; monoclonal rat anti-α-tubulin (YL1/2), Abcam; monoclonal mouse anti-β-tubulin (KMX-1), Millipore; monoclonal mouse anti-GFP (JL-8), BD Transduction Laboratory; monoclonal rat anti-HA (3F10), Roche Applied Science; AlexaFluor-conjugated secondary antibodies and DAPI, Molecular Probes and Cy5-conjugated and horseradish peroxidase-conjugated secondary antibodies, Jackson ImmunoResearch Laboratories.
Plasmids
Expression vectors for C-terminally HA- or enhanced green fluorescent protein (EGFP)-tagged human Arfs (except for Arf4) were constructed as described previously (9); the Arf4 cDNA was cloned into a pCAG-based vector (22). Construction of expression vectors for N-terminally EGFP-tagged EFA6 was described previously (15).
Cell culture, immunofluorescence analysis and RNA interference
Culture of HeLa cells, transfection of expression plasmids and immunofluorescence analysis were performed as described previously (23, 24), except for detection of endogenous Arf proteins. For detection of endogenous Arf proteins at the Flemming body, cells were fixed with 10% trichloroacetic acid on ice for 15 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min and processed for immunofluorescence analysis as described previously (9, 14).
For RNA interference experiments, pools of siRNAs for Arfs were generated using a BLOCK-iT RNAi TOPO transcription kit and a BLOCK-iT Dicer RNAi kit (Invitrogen; note that these kits had been discontinued by the manufacturer), and transfected into cells as described previously (9, 22, 25, 26); a set of primers used for preparation of each Arf cDNA fragment for synthesis of double-stranded RNA is shown in Supplementary Table S1. Briefly, HeLa cells were transfected with a pool of siRNAs for each Arf isoform or with a combination of Arf siRNA pools using Lipofectamine 2000 (Invitrogen) and incubated overnight. The transfected cells were then transferred to a new culture dish containing coverslips, and further incubated for up to 120 h. The cells were then processed for immunofluorescence and immunoblot analyses.
Live cell imaging
Live cell imaging was performed as described previously (9, 15, 27). Cells were seeded on a collagen-coated 35-mm glass bottom dish (Mat Tek) and grown to 30% confluence. Cells were then transfected with an expression plasmid for Arf-EGFP and, after 24 h of transfection, placed on the microscope stage that had been pre-warmed at 37°C in 5% CO2 atmosphere. The cells were observed with an A1R-MP confocal microscope (Nikon). Images were acquired sequentially every 5 min and analysed using MetaMorph imaging software (Molecular Devices).
Pull-down assay for active Arfs
GEF activity of EFA6 towards Arfs in cells was determined as previously described for detection of Arf-GEF activity of BIG2 (15, 23, 28). Briefly, lysates were prepared from HeLa cells transfected with an Arf-HA expression vector together with an expression vector for either wild-type or mutant EGFP-EFA6A, or a control EGFP vector, and subjected to pull-down using a glutathione S-transferase (GST) fusion of the GGA1-GAT domain pre-bound to glutathione-Sepharose 4B beads (GE Healthcare Bioscience). Specifically bound proteins were then processed for immunoblot analysis using an anti-HA antibody.
Results
Arf1 and Arf3 as well as Arf6 are localized at the Flemming body during cytokinesis
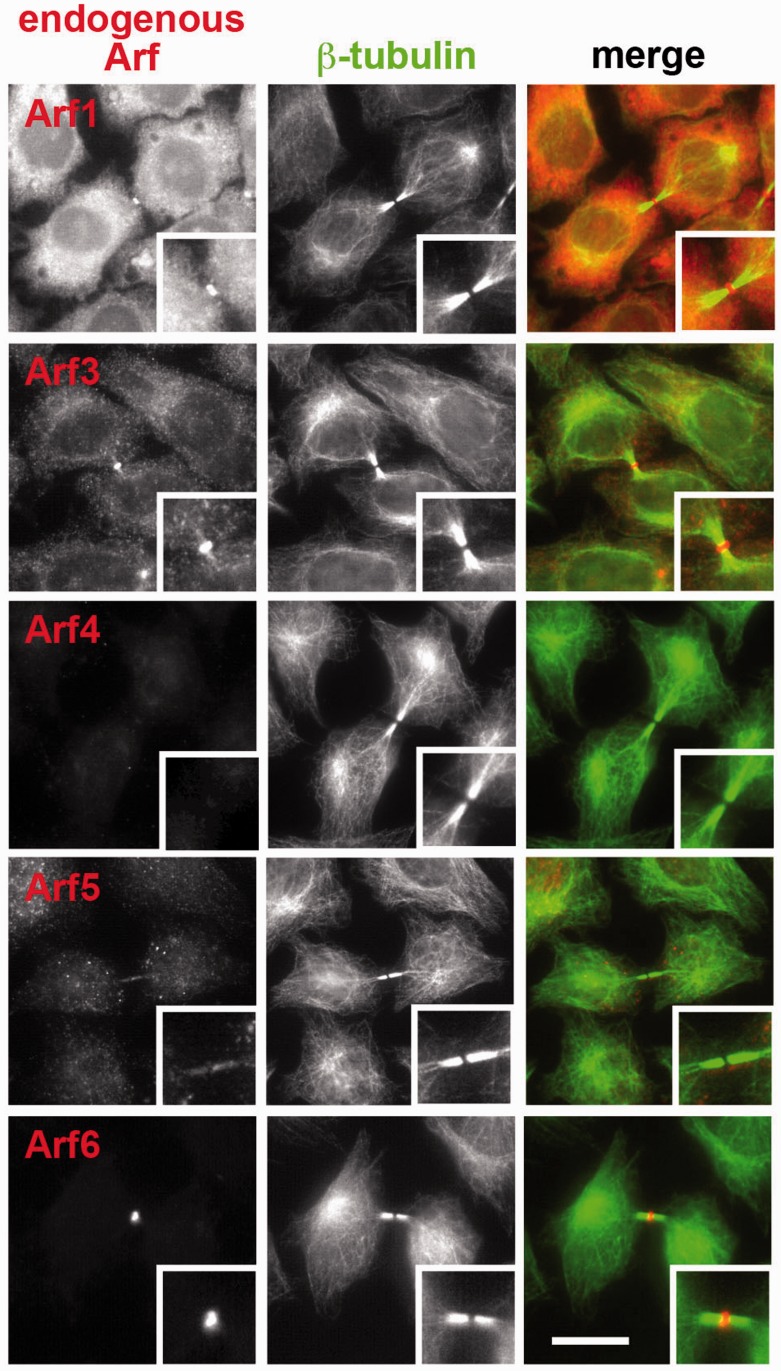
Previous studies of us and others suggested that Arf6 is localized at the Flemming body through interacting with MKLP1 and contributes to the final phase of cytokinesis (8, 9, 11, 12). However, knockout of the Arf6 gene or siRNA-mediated Arf6 depletion did not completely abrogate cytokinesis; namely, a significant proportion of cells depleted or knockout of Arf6 appeared to undergo normal cytokinesis (9) (also see Fig. 3B). Furthermore, in Drosophila melanogaster, although an arf6 null mutation results in male sterility due to a cytokinesis defect during spermatocyte meiosis, somatic cell mitosis appears to be normal (10). We therefore explored the possibility that other Arf isoform(s) also participate in cytokinesis and/or compensate for defects in the absence of Arf6. However, in our initial study using EGFP- or mCherry-tagged constructs, we failed to show significant localization of exogenously expressed Arf1, Arf3, Arf4 and Arf5 at the Flemming body (9). On the other hand, we previously established a procedure for immunologically detecting endogenous Arf6 at the Flemming body (see Materials and Methods) (14). Therefore, we then applied this procedure to determine localization of endogenous proteins of other Arf isoforms during cytokinesis. Among commercially available antibodies examined, we found that those described in Materials and Methods specifically recognize corresponding Arf isoforms: the specificity of each antibody was confirmed by immunoblot and immunofluorescence analyses of cells treated with siRNAs for each Arf isoform (see below). As shown in Fig. 1, using these antibodies, we found that endogenous Arf1 and Arf3 as well as Arf6 are localized at the Flemming body. On the other hand, we failed to observe significant staining for Arf4 and Arf5 at the Flemming body (Fig. 1), although these antibodies can stain intracellular structures in interphase cells (Supplementary Fig. S1). These observations are in keeping with our previous data showing that Arf1 and Arf3 can also interact with MKLP1, and lead to the possibility that Arf1 and Arf3, as well as Arf6, contribute to completion of cytokinesis.
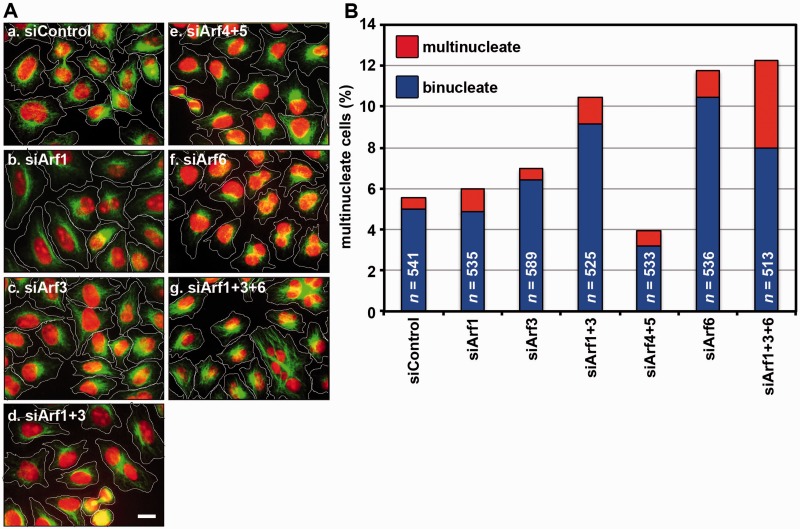
Fig. 3.
Multinucleate phenotype of cells knocked down of Arf isoforms. (A) HeLa cells were treated with a pool(s) of control siRNAs (for LacZ) (a), or siRNAs for Arf1 (b), Arf3 (c), Arf1+Arf3 (d), Arf4+Arf5 (e), Arf6 (f) and Arf1+Arf3+Arf6 (g), and stained for microtubules with anti-β-tubulin antibody and for nucleus with DAPI. Bar = 10 µm. (B) HeLa cells treated with Arf siRNAs in (A) were classified as mono-nucleate, bi-nucleate, and multi-nucleate (≥3 nuclei), and counted. Percentages of bi-nucleate and multi-nucleate cells are expressed as bar graphs.
Fig. 1.
Localization of Arf isoforms during cytokinesis. HeLa cells were fixed and permeabilized as described in Materials and Methods doubly stained with anti-β-tubulin antibody (middle panels) and antibody against Arf1, Arf3, Arf4, Arf5 or Arf6 (left panels). Merged images are shown in right panels. Bar = 10 µm.
Simultaneous knockdown of Arf1, Arf3 and Arf6 causes cytokinesis defects
Next, we determined which Arf isoforms are required for cytokinesis by assessing multinucleate phenotype of HeLa cells treated with siRNAs for one of the Arf isoforms or their combinations. Immunoblot analyses revealed that Arf1, Arf3, Arf4, Arf5 and Arf6 were specifically and almost completely depleted after treatment of cells with corresponding siRNAs for 120 h (Fig. 2A). In addition, treatment of cells with siRNAs for Arf1, Arf3 and Arf6 specifically abolished corresponding immunoreactive signals at the Flemming body (Fig. 2B–D); note that Arf1 and Arf3 retained their Flemming body localization in cells knocked down of Arf6 alone (Fig. 2C and D). When the siRNA-treated cells were stained for microtubules and nucleus and classified as mono-nucleate, bi-nucleate and multi-nucleate (≥3 nuclei), knockdown of either of Class I Arfs (Arf1 and Arf3) alone did not increase the proportion of bi- or multi-nucleate cells, compared with control cells treated with siRNAs for LacZ (Fig. 3A and B). However, simultaneous knockdown of Arf1 and Arf3 resulted in a substantial increase in the proportion of bi-nucleate cells, comparable to that of Arf6 knockdown. In contrast to the Arf1+Arf3 knockdown, simultaneous knockdown of Class II Arfs (Arf4 and Arf5) did not cause a substantial increase in the proportion of bi-nucleate or multi-nucleate cells. When Arf6 was knocked down in combination with Arf1 and Arf3, the proportion of bi- plus multi-nucleate cells was only slightly increased in the Arf1+3+6 triple knockdown cells, compared with cells subjected to single knockdown of Arf6 or double knockdown of Arf1 and Arf3 (Fig. 3B). However, compared with the Arf6 single knockdown and the Arf1+Arf3 double knockdown, the Arf1+3+6 triple knockdown led to a substantial increase in the proportion of multi-nucleate (≥3 nuclei) cells, indicating more severe cytokinetic defects (see Discussion). In contrast to the Arf1+Arf3 knockdown, simultaneous knockdown of Class II Arfs (Arf4 and Arf5) did not significantly increase the proportion of bi- or multi-nucleate cells. We also attempted to examine the effect of simultaneous knockdown of all Arf isoforms (Arf1 + Arf3 + Arf4 + Arf5 + Arf6), but the attempts have so far been unsuccessful, because the viability of cells subjected to the quintuple Arf-knockdown was extremely decreased.
Fig. 2.
Knockdown of Arf isoforms. HeLa cells were treated with control siRNAs (for LacZ) or a pool(s) of siRNAs for Arf isoform(s) indicated as described in Materials and Methods and processed for immunoblot analyses using antibodies indicated (A), or double immunostaining analyses using antibodies against α-tubulin (middle panels) and Arf6 (B), Arf1 (C) or Arf3 (D) (left panels). Merged images are shown in right panels. Bar = 10 µm.
We then observed the division process of HeLa cells treated with Arf siRNAs by time-lapse recording. As shown in Fig. 4A and Supplementary Video S1, control cells (cells treated with siRNAs for LacZ) progressed normally through the cell cycle. In striking contrast, cells subjected to simultaneous knockdown of Arf1, Arf3 and Arf6 exhibited aberrant phenotypes. A substantial fraction of the Arf1 + Arf3 + Arf6-knockdown cells proceeded to divide into two, yet underwent apoptotic cell death before abscission (Fig. 4B and Supplementary Video S2); this observation can explain why the apparent proportion of multinucleate cells was not very high when Arf1, Arf3 and Arf6 were triply knocked down. It is also noteworthy that another fraction of the triple knockdown cells underwent multipolar cell division (Fig. 4C and Supplementary Video S3), suggesting that, in addition to cytokinesis, these Arfs participate in regulation of replication and/or clustering of centrosomes. The pleiotropic phenotypes observed for the Arf1 + Arf3+Arf6-knockdown cells may reflect the variable extent of depletion of each Arf isoform in individual cells, although we did not further pursue this issue.
Fig. 4.
Triple knockdown of Arf1, Arf3 and Arf6 exhibits pleiotropic effects on cell division. HeLa cells treated with control siRNAs (A) or siRNAs for Arf1, Arf3 and Arf6 (B and C) were subjected to time-lapse recording. Image sequences from Supplementary Videos S1 (A), S2 (B) and S3 (C) are shown.
Intracellular activation of Arf isoforms by EFA6
We have recently shown that, among Arf-GEFs examined, Arf6 is transiently colocalized with and activated by EFA6 (exchange factor for Arf6) in the ingressing cleavage furrow region, and only the activated Arf6 is in turn recruited to the Flemming body through interaction with MKLP1 (15). Although initial in vitro studies on EFA6 revealed that it can activate Arf6 robustly and Arf1 very weakly (29, 30), none of the studies addressed whether it could activate other Arf isoforms. Therefore, we then addressed whether EFA6 can activate other Arf isoforms intracellularly. To this end, HeLa cells were cotransfected with expression vectors for EFA6A and any of Arf isoforms, and the cell lysates were subjected to a pulldown assay with a GST fusion of the GGA1-GAT domain, which can interact with GTP-bound, but not GDP-bound, Arfs (23, 28). As described previously, coexpression of EGFP-EFA6A(WT) robustly increased the amount of Arf6-HA pulled down with GST-GGA1-GAT when compared with the control (Fig. 5, compare lanes 13 and 14), indicating that coexpressed EFA6A can activate Arf6 in the cells. Moreover, the increase in the pulldown amount of Arf6-HA was much smaller in cells coexpressing EGFP-EFA6A(R765E/K766E), which has mutations in two conserved basic residues in the PH domain and thereby cannot associate with membranes (15, 31), than that in cells coexpressing EGFP-EFA6A(WT) (Fig. 5, compare lanes 14 and 15), indicating that membrane association of EFA6A is important for its ability to activate Arf6. The pulldown amount of Arf1-HA was also substantially increased by coexpressing EGFP-EFA6A(WT) (Fig. 5, compare lanes 1 and 2), although the increase was less prominent than the pulldown amount of Arf6-HA (lanes 13 and 14). On the other hand, the pulldown amounts of other Arf isoforms were not significantly increased by EGFP-EFA6A(WT) coexpression (Fig. 5, lanes 4–12). These results thus indicate that, in addition to Arf6, at least Arf1 can be activated by EFA6 in the cells.
Fig. 5.
Intracellular activation of Arf isoforms by EFA6. Lysates prepared from HeLa cells co-expressing C-terminally HA-tagged Arf isoform indicated and either EGFP-EFA6A(WT) or EFA6A(R765E/K766E) were subjected to pulldown using GST-GGA1(GAT), which interacts with GTP-bound Arfs, and processed for immunoblot analysis using anti-HA antibody.
Discussion
We have recently shown that Arf6 is recruited to the Flemming body in its GTP-bound state via interaction with MKLP1 in the late phase of cytokinesis and plays a role in successful completion of cytokinesis (9), and that EFA6, an Arf guanine-nucleotide change factor, is transiently colocalized with Arf6 in the ingressing cleavage furrow region and involved in its local activation in late cytokinetic phases (15). Here, we extended these results and showed that Class I Arfs (Arf1 and Arf3) are also recruited to the Flemming body and participate in regulation of cytokinesis.
In our previous study (9), we reported that Arf6 is localized at the Flemming body in late cytokinetic phases and cells derived from Arf6-knockout mice and HeLa cells knockdown of Arf6 by siRNAs exhibit moderate cytokinesis defects; namely, although knockout or knockdown of Arf6 approximately doubled the proportion of bi-nucleate and multi-nucleate (≥3 nuclei) cells compared with control cells (also see Fig. 2), a large proportion of cells appeared to be normal (mono-nucleate). However, the data showing that Arf1 and Arf3 are also localized at the Flemming body and that simultaneous knockdown of Arf1, Arf3 and Arf6 caused more severe defects lead to the possibility that, in the absence of Arf6, Arf1 and/or Arf3 can compensate for its function in cytokinesis to some extent. It is also possible that Arf1 and Arf3 participate in cytokinesis in a different aspect from that of Arf6, although they also play overlapping roles with that of Arf6, since double knockdown of Arf1 and Arf3 also caused a modest increase in the proportion of bi-nucleate cells.
The proportion of bi-nucleate and multi-nucleate (≥3 nuclei) cells increased by the Arf knockdown might not reflect the actual proportion of cells suffering cytokinesis defects, because cytokinesis failure potentially reduces cell viability. Indeed, our live cell imaging analyses revealed that a substantial fraction of cells triply knocked down of Arf1, Arf3 and Arf6 underwent apoptotic cell death immediately before cytokinetic abscission, although these cells initially proceeded to divide into two daughter cells (Fig. 4B and Supplementary Video S2). This observation suggests that cells that have not accomplished the final step of cytokinesis might be destined for cell death. Furthermore, a substantial fraction of the triple Arf-knockdown cells underwent multipolar cell division (Fig. 4C and Supplementary Video S3). Since we treated HeLa cells with Arf siRNAs for 120 h, cycles of cell division preceding the observed multipolar division were likely to occur normally. This makes it possible that Arf1, Arf3 and/or Arf6 also participate in certain aspects of cell division other than cytokinesis, such as replication of centrioles and segregation of centrosomes, although we could not precisely determine the proportion of cells exhibiting aberrant phenotypes for technical reasons; because the populations of cells undergoing apoptosis and abnormal division tended to be clustered probably due to uneven distribution of cells with similar knockdown extents of individual Arf isoforms, wide-field live-cell imaging must be repeated dozens of times to estimate the proportion of abnormally dividing cells. In the context of non-cytokinetic roles for Arfs, it is noteworthy that knockdown of components of the endosomal sorting complex required for transport (ESCRT) machinery, which are also localized around the Flemming body in late cytokinetic phases and responsible for the final abscission step of cytokinesis, was reported to not only inhibit abscission but also alter centrosome and spindle pole numbers and cause defects in chromosome segregation (32). Thus, at least some of the phenotypes observed in triple Arf-knockdown cells closely resemble those observed in ESCRT-knockdown cells, suggesting that Arfs and ESCRT may regulate some common steps of cell division other than completion of cytokinesis.
In our previous study (15), we showed that EFA6 transiently associates with the ingressing cleavage furrow membrane and locally activates Arf6 in late cytokinesis phase. In this study, we showed that Arf1 can be also activated by EFA6 in the cells, suggesting that EFA6 is required for local activation and subsequent Flemming body targeting of not only Arf6 but also Arf1, although there was no direct evidence for the local activation. It is currently unknown why EFA6 activated Arf1 as well as Arf6, but failed to activate Arf3, although Arf1 shares 96% of amino acids with Arf3 while both Arf1 and Arf3 share 67% with Arf6. If EFA6 cannot activate Arf3, which Arf-GEF(s) participate in the local Arf3 activation? Since most of Arf-GEF studies to date have focused on Arf1 and Arf6 (6), we do not have enough information about Arf-GEFs involved in Arf3 activation. Candidates for such Arf3-activating GEFs are BIG1 and/or BIG2 (brefeldin A-inhibitable GEFs 1 and 2, respectively) for the following reasons: (i) both BIG1 and BIG2 can activate Class I Arfs (Arf1 and Arf3) (23); (ii) although BIG1 and BIG2 associate mainly with the trans-Golgi network, both also associate with recycling endosomes (33); (iii) Arf1 and Arf3 are also associated with recycling endosomes and regulate trafficking through these compartments (25) and (iv) in late cytokinesis phase, recycling endosomal vesicles are transported along the central spindle towards the Flemming body and involved in completion of cytokinesis (14, 34, 35). Anyway, future studies on the substrate specificities of Arf-GEFs and their spatiotemporal changes in the localization during cell division will help our understanding of the roles of Arfs in cytokinesis.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgements
The authors would like to thank Yasunori Kanaho and Hiroyuki Sakagami for kindly providing materials, and Yohei Katoh and Senye Takahashi for technical advice.
Glossary
Abbreviations
- Arf
ADP-ribosylation factor
- BIG
brefeldin A-inhibitable guanine nucleotide exchange factor
- EFA6
exchange factor for Arf6
- EGFP
enhanced green fluorescent protein
- ESCRT
endosomal sorting complex required for transport
- GEF
guanine nucleotide exchange factor
- GST
glutathione S-transferase
- MKLP1
mitotic kinesin-like protein 1
Funding
This work was supported in part by Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20058015 and 25113514 to K.N.) and Grant-in-Aid for Scientific Research (B) from the Japan Society for Promotion of Science (22390013 to K.N.) and the Ono Medical Research Foundation and the Uehara memorial Foundation.
Conflict of Interest
None declared.
References
- 1.Shin H.-W., Nakayama K. (2004) Guanine nucleotide exchange factors for Arf GTPases: Their diverse functions in membrane traffic. J. Biochem. 136, 761–767 [DOI] [PubMed] [Google Scholar]
- 2.Donaldson J.G., Jackson C.L. (2011) ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn R.A., Cherfils J., Elias M., Lovering R.C., Munro S., Schurmann A. (2006) Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillingham A.K., Munro S. (2007) The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 5.D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 6.Casanova J.E. (2007) Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 7.Myers K.R., Casanova J.E. (2008) Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 18, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph N., Hutterer A., Poser I., Mishima M. (2012) ARF6 GTPase protects the post-mitotic midbody from 14-3-3-mediated disintegration. EMBO J. 31, 2604–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makyio H., Ohgi M., Takei T., Takahashi S., Takatsu H., Katoh Y., Hanai A., Ueda T., Kanaho Y., Xie Y., Shin H.-W., Kamikubo H., Kataoka M., Kawasaki M., Kato R., Wakatsuki S., Nakayama K. (2012) Structural basis for Arf6–MKLP1 complex formation on the Flemming body responsible for cytokinesis. EMBO J. 31, 2590–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyer N., Rebollo E., Dominguez P., Elkhatib N., Chavrier P., Daviet L., González C., González-Gaitán M. (2007) Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development 134, 4437–4447 [DOI] [PubMed] [Google Scholar]
- 11.Schweitzer J.K., D’Souza-Schorey C. (2002) Localization and activation of the ARF6 GTPase during cleavage furrow ingression and cytokinesis. J. Biol. Chem. 277, 27210–27216 [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer J.K., D’Souza-Schorey C. (2005) A requirement for ARF6 during the completion of cytokinesis. Exp. Cell Res. 311, 74–83 [DOI] [PubMed] [Google Scholar]
- 13.Sakagami H. (2008) The EFA6 family: Guanine nucleotide exchange factors for ADP ribosylation factor 6 at neuronal synapses. Tohoku J. Exp. Med. 214, 191–198 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi S., Takei T., Koga H., Takatsu H., Shin H.-W., Nakayama K. (2011) Distinct roles of Rab11 and Arf6 in the regulation of Rab11-FIP3/Arfophilin-1 localization in mitotic cells. Genes Cells 16, 938–950 [DOI] [PubMed] [Google Scholar]
- 15.Ueda T., Hanai A., Takei T., Kubo K., Ohgi M., Sakagami H., Takahashi S., Shin H.-W., Nakayama K. (2013) EFA6 activates Arf6 and participates in its targeting to the Flemming body during cytokinesis. FEBS Lett. 587, 1617–1623 [DOI] [PubMed] [Google Scholar]
- 16.Douglas M.E., Mishima M. (2010) Still entangled: Assembly of the central spindle by multiple microtubule modulators. Sem. Cell Dev. Biol. 21, 899–908 [DOI] [PubMed] [Google Scholar]
- 17.Otegui M.S., Verbrugghe K.J., Skop A.R. (2005) Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 15, 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu C.-K., Coughlin M., Mitchison T.J. (2012) Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 23, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotzer M. (2005) The molecular requirements for cytokinesis. Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 20.Mishima M., Kaitna S., Glotzer M. (2002) Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41–54 [DOI] [PubMed] [Google Scholar]
- 21.Akiyama M., Zhou M., Sugimoto R., Hongu T., Furuya M., Funakoshi Y., Kato M., Hasegawa H., Kanaho Y. (2010) Tissue- and development-dependent expression of the small GTPase Arf6 in mice. Dev. Dyn. 239, 3416–3435 [DOI] [PubMed] [Google Scholar]
- 22.Nakai W., Kondo Y., Saitoh A., Naito T., Nakayama K., Shin H.-W. (2013) ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol. Biol. Cell 24, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin H.-W., Morinaga N., Noda M., Nakayama K. (2004) BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: Its localization to recycling endosomes and implication in the endosome integrity. Mol. Biol. Cell 15, 5283–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin H.-W., Shinotsuka C., Torii S., Murakami K., Nakayama K. (1997) Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J. Biochem. (Tokyo) 122, 525–530 [DOI] [PubMed] [Google Scholar]
- 25.Kondo Y., Hanai A., Nakai W., Katoh Y., Nakayama K., Shin H.-W. (2012) ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct. Funct. 37, 141–154 [DOI] [PubMed] [Google Scholar]
- 26.Takashima K., Saitoh A., Hirose S., Nakai W., Kondo Y., Takasu Y., Kakeya H., Shin H.-W., Nakayama K. (2011) GBF1–Arf–COPI–ArfGAP-mediated Golgi-to-ER transport involved in regulation of lipid homeostasis. Cell Struct. Funct. 36, 223–235 [DOI] [PubMed] [Google Scholar]
- 27.Takatsu H., Katoh Y., Ueda T., Waguri S., Murayama T., Takahashi S., Shin H.-W., Nakayama K. (2013) Mitosis-coupled, microtubule-dependent clustering of endosomal vesicles around centrosomes. Cell Struct. Funct. 38, 31–41 [DOI] [PubMed] [Google Scholar]
- 28.Shin H.-W., Shinotsuka C., Nakayama K. (2005) Expression of BIG2 and analysis of its function in mammalian cells. Methods Enzymol. 404, 206–215 [DOI] [PubMed] [Google Scholar]
- 29.Franco M., Peters P.J., Boretto J., van Dondelaar E., Neri A., D'Souza-Schorey C., Chavrier P. (1999) EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18, 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia E., Chabre M., Franco M. (2001) Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J. Biol. Chem. 276, 24925–24930 [DOI] [PubMed] [Google Scholar]
- 31.Macia E., Partisani M., Favard C., Mortier E., Zimmermann P., Carlier M.-F., Goumon P., Luton F., Franco M. (2008) The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with PI(4,5)P2 and F-actin. J. Biol. Chem. 283, 19836–19844 [DOI] [PubMed] [Google Scholar]
- 32.Morita E., Colf L.A., Karren M.A., Sandrin V., Rodesch C.K., Sundquist W.I. (2010) Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl Acad. Sci. USA 107, 12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishizaki R., Shin H.-W., Mitsuhashi H., Nakayama K. (2008) Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell 19, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montagnac G., Echard A., Chavrier P. (2008) Endocytic traffic in animal cell cytokinesis. Curr. Opin. Cell Biol. 20, 454–461 [DOI] [PubMed] [Google Scholar]
- 35.Schiel J.A., Prekeris R. (2013) Membrane dynamics during cytokinesis. Curr. Opin. Cell Biol. 25, 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.