Abstract
Hybrid complexes are composed of organisms with multiple combinations of parental genomes (genomotypes) that interconnect through nets of crosses. Although several such complexes are well established without speciation or extinction, mechanisms shaping their dynamics remain poorly understood. In this study, we quantified the reproductive success of the allopolyploid Iberian fish Squalius alburnoides in experimental free-access and directional crosses involving the most common genomotypes. Specifically, we analysed the paternity of the offspring produced when females had free access to male genomotypes and quantified variations in egg allocation, fertilization rate, and offspring survival among crosses involving each male genomotype. The composition of the offspring produced from free-access crosses varied significantly from that expected from random mating, suggesting that offspring production and viability are not independent of parental male genomotype. Moreover, directional crosses producing the genomotype most commonly found in wild populations appeared to be the most successful, with females laying more eggs, and fertilization rate and offspring survival being the highest. These results suggest that reproductive dynamics plays a relevant role in structuring the genomotype composition of populations and opens a path to future research on the ecology and evolutionary biology of allopolyploids and their multiplicity of possible evolutionary pathways.
Keywords: free-access crosses, directional crosses, egg allocation, fertilization rate, offspring survival, paternity analysis
1. Introduction
Successful homoploid hybrids and allopolyploid complexes have been reported in various taxonomic groups, showing stable population dynamics or even evolving into new species through hybrid speciation [1–4]. Multiple mechanisms, such as mate choice, egg and sperm allocation, and offspring survival at early ontogenetic stages, may shape the dynamics of such hybrid populations to variable extents [4–11]. Clarifying those mechanisms is crucial to advance our knowledge regarding hybrids' ecology and evolution, namely, in allopolyploid vertebrates.
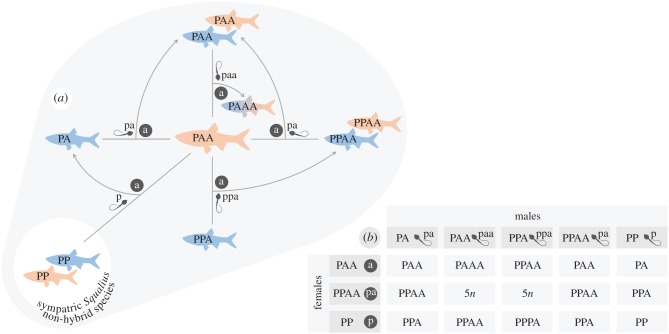
Increasingly recognized as one of the most well-established hybrid vertebrates known to date [4,12], the Squalius alburnoides (Steind. 1866) fish complex is an ideal model to study mechanisms shaping the dynamics of allopolyploids. This Iberian complex arose from intergeneric hybridization, involving S. pyrenaicus (Günther 1868) females (PP genome) and males from an already extinct species of the Anaecypris hispanica (Steind. 1866) lineage (AA genome; reviewed in [4]). It includes hybrid males and females with several ploidies (2n = 50, 3n = 75, and 4n = 100) and various combinations of the parental genomes (i.e. genomotypes) [4]. All these genomotypes are fertile and able to cross to produce offspring and to breed with sympatric bisexual Squalius species. The vast majority of populations are dominated by allotriploids, namely, by the PAA genomotype in central and southern rivers. This highly female-biased genomotype is entirely maintained by crosses with other genomotypes (figure 1), because neither spontaneous parthenogenesis nor gynogenesis occurs in this complex and, thus, PAA females cannot restock their own genomotype without male genome incorporation (reviewed in [4]). This strict genomotype interdependency suggests that the structure of S. alburnoides populations may depend on an intricate reproductive dynamics, promoting the prevalence of the PAA genomotype, although empirical evidence to corroborate this is still mostly lacking. Previous studies have suggested that S. alburnoides females may display differential mate preferences among male genomotypes [13], favouring the ones with which they produce PAA offspring [14]. However, it is unclear whether mate preferences actually also influence reproductive success, offspring production, and genomotype composition.
Figure 1.
(a) Simplified reproductive framework of the S. alburnoides allopolyploid complex in the Ocreza River (Tagus drainage, Central Portugal), showing the core of its reproductive dynamics and its relationship with the sympatric bisexual S. pyrenaicus. Males and females are represented in blue and pink, respectively. (b) Table of all crosses that could theoretically occur in the referred population. Reproductive modes include meiotic hybridogenesis in PAA females, clonal spermatogenesis in PA, PAA, and PPA males, and regular meiosis in PPAA and PP individuals; 5n offspring is unviable. Dark grey circles represent eggs. Capital letters refer to fish genomes, and small letters to gamete genomes: A, a from the Anaecypris-like paternal ancestor of the complex; P, p from the S. pyrenaicus maternal ancestor of the complex. (Online version in colour.)
In an attempt to clarify the mechanisms shaping the structure of S. alburnoides populations, we analysed the reproductive success of PAA females in free access and directional crosses involving S. alburnoides males with distinct genomotypes (PA, PAA, PPA, and PPAA) and S. pyrenaicus males (PP). Specifically, we conducted two sets of experiments in order to (i) assess the paternity of the offspring produced when PAA females have free access to all male genomotypes and (ii) quantify egg allocation, fertilization, and offspring production by individual mating pairs involving PAA females and males of each genomotype. Results obtained in both experiments were assessed to explore the interplay between genomotype composition and reproductive dynamics, and the way natural selection acting on early ontogenetic stages may shape population structure in S. alburnoides.
2. Material and methods
(a). Fish sampling and genomotype assessment
Mature individuals used in free-access and directional crosses were sampled in the Ocreza River (Portugal), where diverse genomotypes of S. alburnoides co-occur with S. pyrenaicus (figure 1). Sampling was conducted early in the reproductive season (April), using short pulses and moderate voltage electrofishing (300 V, 2–4 A). During this period, individuals could be easily sexed by applying a mild pressure on the abdomen and observing gamete discharge. Fish showing no physiological stress or injuries were transported to the laboratory in separate aerated vats, and the remaining returned to the river.
Because S. alburnoides genomotypes are morphologically similar, the ploidy and genome combination of each individual were unknown until assessment. In the laboratory, individuals were anaesthetized (0.1 g l−1 MS-222, 0.2 g l−1 NaHCO3), measured for standard length (SL, mm) and photographed on their left and right sides for further scale pattern interpretation and individual recognition [15]. Small clips of the caudal fin were collected for genomotype assessment through flow cytometry [16] and Sanger sequencing of the β-actin gene (PCR conditions: 35 cycles of 94°C, 30 s; 55°C, 40 s; 72°C, 90 s) [17]. DNA extraction followed an adapted phenol–chloroform protocol [18].
(b). Assessment of the offspring produced in free-access crosses
An experimental population of 33 S. alburnoides (5.6 cm mean SL, 4.0–7.2 cm) and 19 S. pyrenaicus (7.6 cm mean SL, 5.3–10.6 cm) was established in an artificial pond, under natural light and temperature conditions, in January 2011. The genomotype of S. alburnoides individuals was assessed as described above. The experimental population included 23 PAA females, representing the dominant genomotype in central and southern rivers [4,14], and a high diversity of male genomotypes, that approached the proportions found in the wild, namely, six PA, one PAA, two PPA, one PPAA, and also 10 PP individuals. In addition, nine PP females were translocated to the pond to assess whether the eventual absence of offspring from PP males was due to their lack of interest towards PAA relative to PP females or due to a general failure in reproduction. The experimental population is illustrated in the electronic supplementary material, figure S1.
The artificial pond had a volume of 4 200 l (300 cm length × 200 cm width × 50 cm mean depth (25–90 cm)). The bottom of the pond was covered with a layer of cobbles (2–15 cm), to provide adequate substrate for fish spawning [19], and around 25% of the surface was occupied with the macrophytes Ranunculus sp. and Juncus sp., to provide cover and shelter for the fish [20]. Two pumps and a UV lamp were used to prevent water stagnation and deterioration. Overall, habitat conditions in the pond were close to those found in Iberian rivers during seasonal drought, when fish concentrate in isolated pools [21]. Fish were fed twice a day with commercial flakes for one month to prevent eventual lows in prey availability and facilitate adaptation to the pond conditions. The pond was monitored weekly for water pH (7–10) and inspected for dead fish (never detected) and larvae (first spotted on April). In October, parental fish and offspring were captured using electrofishing and transported to the laboratory in aerated vats. The pond was then emptied to assure complete fish collection.
In the laboratory, YOYs (youngs-of-the-year) were identified using length frequency distributions and maintained in a 500 l tank. All individuals were assessed for ploidy level, and a sample of 100 YOYs was randomly selected for sex and paternity assessment. Individuals were sacrificed with an overdose of MS-222 anaesthetic and sexed as described in [22]. Paternity was assessed through microsatellite genotyping, using nine microsatellites with high variability among cyprinids [23–25]. Moreover, we used an extra microsatellite found by MM Coelho team (2013, unpublished data), after sequencing a genomic fragment containing the intron region of the aminomethyltransferase gene (AMT), from which the primers were designed. Excepting LCO1, LCO3, and LCO4, all microsatellites were genotyped using primers with a M13 tail, following [26]. Complete information on the 10 microsatellites is in the electronic supplementary material, table S1.
Paternity was primarily assessed by comparing microsatellite alleles between offspring and parental individuals. When microsatellites were unable to distinguish between pairs of female progenitors, a mitochondrial fragment including the d-loop/control region, the tRNA–Phe gene, and the beginning of the 12S gene (PCR conditions: 35 cycles of 94°C, 30 s; 50°C, 30 s; 72°C, 90 s) [27] was amplified and sequenced for ambiguous individuals. Overall, male progenitors were identified for all YOYs and female progenitors for 91 YOYs.
(c). Assessment of the offspring produced in directional crosses
Directional crosses were conducted during 2012 and 2013, using additional fish samples collected in the Ocreza River. In total, 29 mating pairs involving PAA females and 12 PA males, five PAA males, four PPAA males, two PPA males, and six PP males were used in directional crosses conducted in three experimental outdoor tanks, under natural conditions of light and temperature. To control for size effects on fecundity, females were selected to show the least variation in length as possible, ranging between 5.7 and 7.2 cm. No similar size selection was possible for males, which generally differ in length among genomotypes [4], with PP males being the largest.
Because spontaneous reproduction is hard to accomplish in captivity for isolated pairs of both S. alburnoides and S. pyrenaicus [19], outdoor tanks were compartmentalized for holding six mating pairs each. Tanks were 130.0 cm × 70.0 cm × 50.0 cm (length × width × height) and compartments (43.3 cm × 25.0 cm) were divided by transparent perforated acrylic plates (8.0 mm diameter holes). This allowed all sorts of stimuli in the water to be shared among fishes, but breeding to occur only between mates in each compartment. Because S. alburnoides is a multiple bottom spawner [13,28], whose eggs stick to the substrate after fertilization, the holes were located on the top half of the acrylic plates, near the water surface, to prevent cross fertilization, and the bottom of each compartment was delimited by an acrylic net (1 cm2 square holes) to avoid egg predation. Moreover, thin malleable acrylic sheets (0.5 mm) were put below the bottom nets, so that the stuck eggs of each batch could be removed through independent lateral compartments (10 cm width) without disturbing the mates.
Each year, experiments lasted three months (May–July). Water temperature in the tanks was similar between years for average (20.5°C versus 19.9°C, t182 = 1.44, p = 0.152) and maximum (25.4°C versus 24.9°C, t182 = 0.76, p = 0.449) values, and the pH ranged between 7.0 and 8.0. Water was filtered using pumps connected to UV lamps. In general, tanks held mating pairs with the same combination of genomotypes. In the only case in which two distinct male genomotypes were held in the same tank, the perforated acrylic plates were replaced by watertight opaque glass and separate filters and UV lamps were used to avoid water and stimuli mixture. In all cases, fish were fed twice a day with frozen bloodworms and brine shrimp.
Eggs produced by each mating pair were collected the day after spawning, rinsed with water, counted, and the bottom of each compartment was vacuumed to guarantee complete egg collection. Eggs laid in consecutive days were considered as a batch and were transferred to Petri dishes in groups of 100. Daily, groups of eggs were inspected under a stereo-microscope, and unfertilized and dead eggs and embryos were counted and removed. Larvae feeding exogenously and swimming properly were transferred to aerated containers (3 l) and fed daily with ground commercial flakes. A month after spawning, larvae were counted and the experiments were concluded.
(d). Data analyses
Analyses focused on variation in reproductive success of PAA females in relation to PA, PAA, PPA, PPAA, and PP males (henceforth designated as male genomotypes). Whenever relevant, regression analyses were conducted to account for effects of fish size on the response variables characterizing reproductive success (see below). Likewise, t-tests were used for assessing variation in response variables between two samples (e.g. sexes), after verification of conformity to assumptions (see below). Significance of statistical testing was assessed at p < 0.05, and analyses were performed using StatSoft Statistica software [29]. For clarity, variables are presented in original units in all figures.
To assess whether male genomotypes varied in reproductive success when PAA females had free access to mates, we compared the proportion of YOYs produced by each male against the proportions expected if females paired randomly with them. Likewise, we evaluated the extent of variation in reproductive success among PAA females through comparisons of YOYs produced against those that would be expected if the reproductive success of PAA females was similar. Using this approach, we assumed that offspring viability and survival were similar among crosses, and recognized that variation in reproductive success may reflect the influence of multiple factors, such as intersexual and intrasexual selection, egg allocation, fertilization rate, and larval survival. Nevertheless, measurable deviations from randomness in the proportion of YOYs produced would indicate variation in reproductive success irrespective of the exact mechanisms involved. The analyses were performed with observed versus expected χ2-tests.
Reproductive success in directional crosses was assessed from the variation in egg allocation, fertilization rate, and larval survival among mating pairs involving different male genomotypes. Egg allocation was estimated from the total number of eggs laid by females throughout the reproductive season and from the number and average size of each batch. Fertilization rate was assessed as the proportion of fertilized eggs in batches with, at least, 100 eggs, which generally averaged 93% (±12%, ranging from minimum 50 to maximum 100%) of the total number of eggs laid by each female. Offspring survival was calculated from the proportion of larvae at the end of the experiments for samples with, at least, 50 fertilized eggs, which included, on average, 74% (±39%, 0–100%) of the total fertilized eggs of each female. Prior to analyses, variables were inspected for skewness and transformed to dampen the influence of exceptionally large numbers whenever necessary. Each variable was assessed for normality and homogeneity of variances using the Shapiro–Wilk and Levene tests, respectively, and tested for variation among male genomotypes using one-way ANOVA and Tukey honestly significant difference (HSD) post hoc tests. Tukey HSD multiple comparisons and grouping were performed following the procedure described in [30], i.e. first comparing the largest mean against the smallest, then against the next smallest and so on, until the largest has been compared with the second largest, and, thereafter, performing the same procedure for the second largest and so on.
3. Results
(a). Offspring produced in free-access crosses
No YOYs of S. pyrenaicus were captured in the artificial pond, but there were 261 YOYs of S. alburnoides. All but one of these individuals were triploid (99.6%), averaging 5.9 cm SL (±0.8, 4.2–7.7). The only diploid was an allodiploid PA male which was 5.1 cm, that was produced androgenetically by a PA male. It showed no female nuclear genomic contribution, but the sequence of the mitochondrial gene differed from that of the progenitor male, matching one of the PAA females, likely the mother [31].
Among the YOYs analysed for sex, genomotype, and paternity, there was a strong prevalence of females (6 : 1), which tended to be significantly larger than males (5.6 ± 0.7 cm, 4.3–6.9 versus 4.9 ± 0.5 cm, 4.2–6.3; t97 = 3.88, p < 0.001). With the exception of the allodiploid male (PA), all the remaining individuals were PAA.
The 13 males PAA, PPA, and PP fathered no offspring, with the six PA and the single PPAA males fathering 89.0% and 11.0% of the YOYs analysed, respectively (table 1). These proportions varied significantly from those expected if crosses were independent of male genomotype, i.e. including all five male genomotypes and all 20 male individuals in the analysis (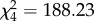 , p < 0.001). Considering PA and PPAA male genomotypes only, these proportions did not vary significantly from those expected from random mating (
, p < 0.001). Considering PA and PPAA male genomotypes only, these proportions did not vary significantly from those expected from random mating (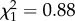 , p = 0.348). However, there were significant variations in reproductive success among PA males (
, p = 0.348). However, there were significant variations in reproductive success among PA males (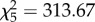 , p < 0.001), with a single individual fathering 86.5% (77–89%) of the offspring produced by this genomotype (male coded as e in table 1). When this individual was excluded from analysis, no differences were found among the remaining PA males (
, p < 0.001), with a single individual fathering 86.5% (77–89%) of the offspring produced by this genomotype (male coded as e in table 1). When this individual was excluded from analysis, no differences were found among the remaining PA males (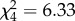 , p = 0.176), and the proportion of offspring fathered by the PPAA male became significantly higher than expected (
, p = 0.176), and the proportion of offspring fathered by the PPAA male became significantly higher than expected (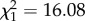 , p < 0.001). Most males reproduced with more than one female, but the PA male fathering the most YOYs crossed with more females than the remaining PA males (13 versus one to three females, respectively; table 1). The number of YOYs produced was independent of the length of PA and PPAA males (R2 = 0.27, F1,5 = 1.86, p = 0.231).
, p < 0.001). Most males reproduced with more than one female, but the PA male fathering the most YOYs crossed with more females than the remaining PA males (13 versus one to three females, respectively; table 1). The number of YOYs produced was independent of the length of PA and PPAA males (R2 = 0.27, F1,5 = 1.86, p = 0.231).
Table 1.
Paternity and maternity of the sub-sample (N = 100) of the offspring produced in the artificial pond, where females had free access to males with distinct genomotypes, expressed in number of YOYs per cross. Total crosses' count exclude YOYs with undetermined maternity. Note that no descendants from PAA, PPA, and PP males were found. Similarly, PP females produced no offspring. See text for further details. ND = undetermined maternity.
| PAA females |
total YOYs | total crosses | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | ND | |||||||
| males | PPAA | a | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 11 | 11 | 6 | 6 | |||||||||
| PA | b | 1 | 1 | 1 | 1 | 4 | 89 | 3 | 14 | |||||||||||||
| c | 3 | 1 | 1 | 5 | 2 | |||||||||||||||||
| d | 1 | 1 | 1 | |||||||||||||||||||
| e | 4 | 10 | 3 | 4 | 3 | 3 | 10 | 23 | 2 | 3 | 1 | 5 | 1 | 5 | 77 | 13 | ||||||
| f | 1 | 1 | 1 | |||||||||||||||||||
| g | 1 | 1 | 0 | |||||||||||||||||||
| total YOYs | 5 | 2 | 11 | 5 | 5 | 4 | 6 | 4 | 10 | 24 | 2 | 5 | 1 | 6 | 1 | 9 | 100 | |||||
| total crosses | 2 | 2 | 2 | 3 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | — | ||||||
The YOYs analysed for paternity were mothered by 15 out of the 23 PAA females in the pond. There was no variation in length between females with (6.5 ± 1.3 cm, 3.7–7.9) and without (6.0 ± 0.8 cm, 4.5–6.9) offspring (t21 = 0.92, p = 0.367). Moreover, the number of YOYs produced was independent of female length (R2 = 0.15, F1,21 = 3.60, p = 0.071), and most females with offspring (nine out of 15) reproduced with more than one male. Individual females mothered a proportion of offspring significantly different than expected if all of them had the same reproductive success (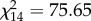 , p < 0.001), and their individual reproductive success was not independent of male genomotype (PA versus PPAA) (
, p < 0.001), and their individual reproductive success was not independent of male genomotype (PA versus PPAA) (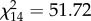 , p < 0.001). A similar result was found if only the offspring fathered by PA males was considered (
, p < 0.001). A similar result was found if only the offspring fathered by PA males was considered ( , p < 0.001), indicating that distinct PAA females had higher reproductive success with distinct PA males. However, this pattern was lost if the PA male that produced the most YOYs was excluded from analysis (
, p < 0.001), indicating that distinct PAA females had higher reproductive success with distinct PA males. However, this pattern was lost if the PA male that produced the most YOYs was excluded from analysis (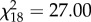 , p = 0.079). Moreover, in this case, females showed similar tendencies for crossing with PA and PPAA male genomotypes (
, p = 0.079). Moreover, in this case, females showed similar tendencies for crossing with PA and PPAA male genomotypes (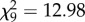 , p = 0.163).
, p = 0.163).
(b). Offspring produced in directional crosses
More than 100 eggs were produced per mating pair, except in three crosses involving one PPAA and two PP males. Because we cannot be sure that females used in these crosses were fertile, they were discarded from analyses, reducing the sample size to 26 crosses. Overall, females laid an average of 1 026.0 eggs (±620.4, 139–2 324) and spawned 2.9 times (±1.6, 1–6), with batches including on average 329.3 eggs (±124.3, 102–595). There were no associations between female length and total number of eggs (R2 = 0.00, F1,13 = 0.01, p = 0.912), number of egg batches (R2 = 0.05, F1,13 = 0.67, p = 0.429), and average batch size (R2 = 0.00, F1,13 = 0.01, p = 0.929). Similarly, no associations were found between male length and total number of eggs (R2 = 0.14, F1,13 = 2.04, p = 0.177), number of egg batches (R2 = 0.15, F1,13 = 2.23, p = 0.159), and average batch size (R2 = 0.19, F1,13 = 3.15, p = 0.099).
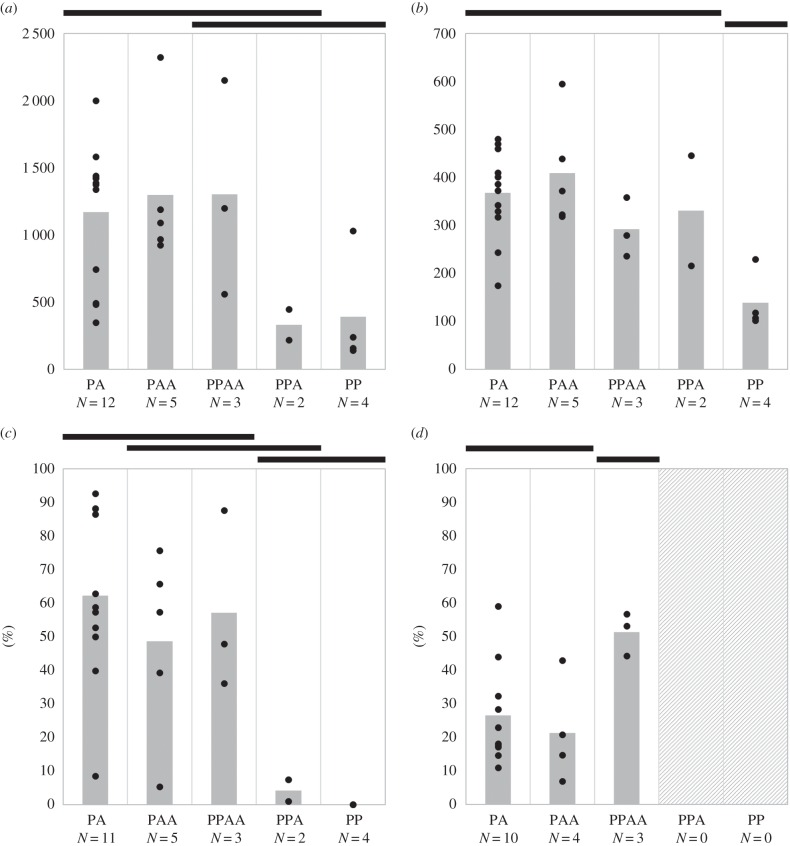
Egg allocation showed considerable variation among crosses involving different male genomotypes. The number of egg batches laid by females remained virtually the same (F4,21 = 2.39, p = 0.084), but there were significant variations in the total number of eggs (F4,21 = 4.10, p = 0.013), with females laying fewer eggs with PP than with PA and PAA males (figure 2a), but showing no significant difference between PPA and PPAA males and the remaining male genomotypes. Similarly, there were significant differences in the average batch size (F4,21 = 9.30, p < 0.001), with females laying fewer eggs per batch with PP males than with any other male genomotype (figure 2b).
Figure 2.
Variation in reproductive traits among directional crosses between PAA females and PA, PAA, PPAA, PPA, and PP males. Tukey HSD grouping (see text for more details) is shown above each graph, represented by black bars, and the total number of crosses analysed for each male genomotype is shown below the x-axis. (a) Total number of eggs, (b) batch size, (c) fertilization rate, and (d) larval survival. Dots represent mating pairs, and grey bars the average values obtained for each male genomotype.
Fertilization rate was estimated for only 25 crosses, given eggs produced in the only batch laid by a mating pair involving a PA male were infected by fungi and lost. Overall, the average fertilization rate was 44.35% (±33.10%, 0.00–92.67%), with no eggs being fertilized by PP males. The fertilization rate was independent of the length of females (R2 = 0.15, F1,13 = 2.22, p = 0.160) and males (R2 = 0.14, F1,13 = 2.14, p = 0.167).
The proportion of fertilized eggs varied significantly among crosses involving different male genomotypes (F4,20 = 6.88, p = 0.001), with fertilization being lower for PP than for PA, PAA, and PPAA males, and PPA males fertilizing fewer eggs than PA males (figure 2c). When crosses involving PP males were excluded from the analysis, the average fertilization rate increased to 52.80% (±29.09%, 1.00–92.67%), and there were no significant differences among crosses (F3,17 = 3.00, p = 0.060), meaning all other male genomotypes were equally successful at fertilizing eggs. However, considering the much lower fertilization rate observed for PPA males (figure 2c), the lack of significance was likely related to the small sample sizes.
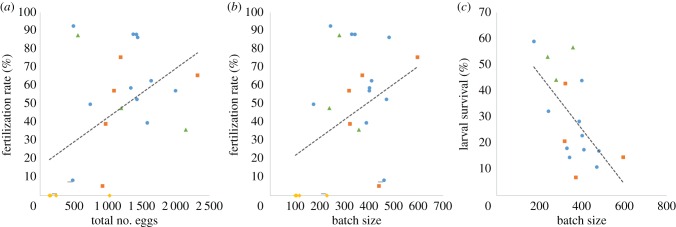
Considering all mating pairs, the fertilization rate tended to increase with the total number of eggs (R2 = 0.25, F1,23 = 7.81, p = 0.010; figure 3a) and batches (R2 = 0.19, F1,23 = 5.45, p = 0.029) laid by females, but showed only a nearly significant association with average batch size (R2 = 0.14, F1,23 = 3.80, p = 0.064) (figure 3b).
Figure 3.
Relationships between reproductive traits of PAA females over all crosses. (a) Variation in average fertilization rate with total number of eggs, (b) variation in average fertilization rate with average batch size, and (c) variation in larval survival with average batch size. Markers for male genomotypes are as follows: PA, circles; PAA, squares; PPA, dotted lines; PPAA, triangles; PP, diamonds. (Online version in colour.)
The larval survival rate was only assessed for crosses involving PA, PAA, and PPAA males, given no eggs were fertilized in batches produced in crosses involving PP males and the average fertilization rate was only 4.3% in those involving PPA males (figure 2c). For the same reason, two mating pairs involving PA and PAA males, with average fertilization rates of 8.5% and 5.3%, respectively, were also excluded from the analysis. For the remaining 17 mating pairs, fertilization rates of the analysed batches ranged between 53.0 and 100.0%, and larval survival rate averaged 29.69% (±17.0%, 7.0–59.0%).
Larval survival varied significantly among crosses (F2,14 = 4.43, p = 0.032), being higher in those involving PPAA than PA and PAA males (figure 2d). Regardless of parental male genomotype, larval survival significantly decreased with average batch size (R2 = 0.39, F1,15 = 9.44, p = 0.008; figure 3c), but showed no association with the total number of eggs (R2 = 0.06, F1,15 = 0.99, p = 0.336) and batches (R2 = 0.05, F1,15 = 0.76, p = 0.397) and fertilization rate (R2 = 0.20, F1,15 = 3.76, p = 0.072). Moreover, larval survival was independent of the length of females (R2 = 0.32, F1,7 = 3.26, p = 0.114) and males (R2 = 0.23, F1,7 = 2.11, p = 0.190).
4. Discussion
Despite the high diversity of genomotypes and reproductive modes in the S. alburnoides complex, central and southern populations are generally dominated by PAA females. The results of our study suggest that this may reflect, at least partially, the influence of mate selection and reproductive success of male genomotypes. Indeed, the genomotype composition of the offspring produced by PAA females having free access to mates differed from the expected if mating preferences and reproductive success were similar among male genomotypes. Moreover, egg allocation and fertilization were superior when PAA females mated with PA and PPAA males, with which they produce PAA offspring, in comparison to PP males, with which they produce PA offspring. Furthermore, survival seemed to be higher in offspring fathered by PPAA males.
Although sample sizes used in our study may be regarded as small, this was a logistical limitation that reflected the scarcity of some genomotypes in the wild (e.g. PPA). Moreover, there are severe legal restrictions regarding the capture of S. pyrenaicus (PP), which lists as ‘Endangered’ [32]. Although small sample sizes together with the absence of balanced experimental design could have limited our results to some extent, we are confident that the patterns now perceived provide valid insights into the reproductive dynamics of the S. alburnoides complex, which should deserve further research (see below).
Results of free-access crosses revealed that mating choices and offspring viability are unlikely to be similar among male genomotypes and individual mating pairs. Although the experimental population included males with five genomotypes (PA, PAA, PPA, PPAA, and also PP), only PA and PPAA males produced offspring with PAA females. Moreover, a single PA male fathered 77% of all the offspring, suggesting that the offspring produced by some mating pairs may be much more successful than that produced by other pairs of the same genomotypes. Ignoring this ‘individual’ effect may lead to misleading conclusions about population structuring in studies focusing on overall genomotype patterns and should, thus, be prevented in further studies.
Patterns of individual variation in offspring production perceived in free-access crosses were likely expressed pre- and post-zygotically. Indeed, PAA females differed not only in egg production in relation to male genomotype, but also in fertilization rate and offspring survival, suggesting that natural selection is probably in action in both phases. This is consistent with previous studies showing that S. alburnoides females are choosy and favour certain male genomotypes, independently of their frequency or density [13,14].
Overall, PP males appeared to be the least favourite of PAA females. The differential allocation hypothesis predicts that choosy females invest more reproductive resources towards high-quality than low-quality males, drawing a positive relationship between energetic investment and reproductive success [33,34], and meaning that differential allocation is directly influenced by mate choice. In fishes with external fertilization and displaying no parental care, differential allocation is often expressed in the number of eggs laid by choosy females [35–38]. In directional crosses, PAA females laid fewer eggs with PP males than with the remaining. This may, at least partially, explain the lack of PA offspring when females had free access to males. In these circumstances, it appears that S. alburnoides females may invest more in hybrid ‘conspecific’ males, belonging to the allopolyploid complex itself.
Multiple factors may be involved in shaping mate choice. PAA females may identify some bad quality indicators in PP males and allocate their eggs accordingly. Indeed, in directional crosses, the total number of eggs was positively associated with fertilization success, and PP males seemed unable to fertilize the eggs of PPA females. Moreover, egg allocation was independent of male length, suggesting that size plays a minor role in defining male quality for females. However, it is also possible that male mate choice is also at play. PP males may be less available to mate with hybrid females and invest less in each spawning event than other more willing males, for example, by displaying less vigorous courtships or releasing insufficient amounts of sperm to fertilize the eggs. Note that PP males were significantly bigger than S. alburnoides male genomotypes and, thus, theoretically able to produce more sperm. Although both male and female mate choices could influence each other, this is unlikely to be the case here. Indeed, PP males were the second favourites of PAA females in previous affiliation trials [14], suggesting there was probably a low interest of PP males in mating with PAA females and not the opposite. Differential fertilization rates among male genomotypes have been reported for other polyploid fish [39–41], and, in certain fish species, non-spermiating males were found to exhibit courtship behaviours and induce spawning in mature females [42]. Although we cannot exclude the hypothesis that the null fertilization found for PP males in directional crosses was due to the lack of adequate substrate for preparing spawning pits [19], this seems unlikely, because, in the artificial pond, PP males did not produce any offspring, neither with PP nor PAA females, despite adequate substrate being available. Note that S. alburnoides and S. pyrenaicus specimens display external fertilization and probably share, to some extent, the spawning habitats in the wild, which may result in accidental intercrosses by sperm dispersal, that possibly contribute to the maintenance of some PA fish frequency.
Offspring survival may also play an important role in shaping the genomotype composition of S. alburnoides populations. Overall, offspring survival was higher in smaller egg batches, probably reflecting the influence of egg size on larval survival. Although no data on egg size was gathered for PAA females herein, females producing larger eggs are generally constrained to lay fewer eggs than females producing smaller ones [28]. Ultimately, this may lead to a higher survival of the offspring produced by the former females, because larvae hatching from larger eggs tend to be larger and more capable of resisting starvation and other environmental constraints [28].
Offspring produced in directional crosses involving PPAA males showed the highest survival rate. This was consistent with PAA females favouring PPAA males in affiliation trials [14] and with the results of paternity analysis of the offspring produced in the artificial pond. Excluding the PA male progenitor that produced the most offspring, the PPAA male fathered a higher proportion of offspring than all the other PA males. Taken together, these results suggest that a ‘good genes' type of mate choice seems to be occurring in S. alburnoides, with females showing a preference towards the male genomotype with which they produce higher quality offspring (i.e. with higher survival). Similar mate choice trends have been documented for other organisms (e.g. [43,44]), including cyprinids with external fertilization and other freshwater fishes (e.g. [36,45]). Moreover, it is possible that S. alburnoides mate choice may also be upheld by a heterozygosity-based component [46]. Indeed, among all male genomotypes, PPAA tetraploids are the only male hybrids undergoing regular meiosis (producing PA sperm), thus contributing to a higher genetic variability of the offspring, which ultimately may contribute to its higher survival rate.
In conclusion, multiple mechanisms may be involved in shaping the genomotype composition of natural S. alburnoides populations. Besides the variation in mate choice, egg allocation, fertilization rate, and offspring survival among genomotypes, individual variation in reproductive success within genomotypes may also be important. The production of the PAA genomotype seems to be favoured by higher egg allocation, fertilization rate and offspring survival in crosses involving PAA females and PA or PPAA males, whereas the production of the PA genomotype seems to be hampered by lower allocation and fertilization of eggs produced by mating pairs involving PP males. Therefore, it appears that natural selection acting early on spawning and larval developmental stages may strongly contribute to structure the genomotype composition of populations. These findings open a path for future research on the ecology and evolutionary biology of S. alburnoides, namely, on the actual relevance of the sympatric Squalius bisexual species in the reproductive dynamics, and on the way the breeding net among genomotypes may lead to the tetraploidization of populations observed in some northern rivers [2] and, consequently, to a possible event of hybrid speciation. Altogether, these findings add substantially to knowledge on the influence of reproductive dynamics in driving allopolyploid populations through their multiplicity of possible evolutionary pathways.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Professor M. M. Coelho's team for providing the AMT intron sequence that comprises the microsatellite and ICNF for authorizing fish sampling and use in experimental trials. Finally, we thank two anonymous reviewers for their insightful comments.
Ethics
Field and laboratorial procedures followed the ethical guidelines [47] and conformed to the Portuguese legislation regarding animal capture, manipulation, and experimentation for scientific purposes. Fish sampling was conducted with the permission of the Instituto da Conservação da Natureza e das Florestas (permits 140/2012/CAPT and 239/2013/CAPT). Because studied taxa are listed as threatened [32], sample sizes were small to avoid natural stock depletion. All efforts were made to minimize accidental deaths and stress on fish throughout the study. At the end of the study, parental fish were in good condition and returned to the Ocreza River, whereas larvae and YOYs were kept in captivity for further research.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
Conception and design: M.M.S., L.V., and M.J.C.P. Acquisition of data: M.M.S. and S.C. Analysis and interpretation of data: M.M.S., S.C., M.F.M., and M.J.C.P. Drafting the article: M.M.S. Revising the article critically: M.F.M., S.C., L.V., and M.J.C.P. The final approval of the version to be published: all authors.
Competing interests
We have no competing interests.
Funding
This study was financed by Portuguese National Funds, through FCT—Fundação para a Ciência e a Tecnologia, within the projects UID/BIA/00329/2013 and PEstOE/BIA/UI0329/2014 and the grant SFRH/BD/65154/2009.
References
- 1.Mallet J. 2007. Hybrid speciation. Nature 446, 279–283. ( 10.1038/nature05706) [DOI] [PubMed] [Google Scholar]
- 2.Cunha C, Doadrio I, Coelho MM. 2008. Speciation towards tetraploidization after intermediate processes of non-sexual reproduction. Phil. Trans. R. Soc. B 363, 2921–2929. ( 10.1098/rstb.2008.0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nolte A, Tautz D. 2010. Understanding the onset of hybrid speciation. Trends Genet. 26, 54–58. ( 10.1016/j.tig.2009.12.001) [DOI] [PubMed] [Google Scholar]
- 4.Collares-Pereira MJ, Matos I, Morgado-Santos M, Coelho MM. 2013. Natural pathways towards polyploidy in animals: the Squalius alburnoides fish complex as a model system to study genome size and genome reorganization in polyploids. Cytogenet. Genome Res. 140, 97–116. ( 10.1159/000351729) [DOI] [PubMed] [Google Scholar]
- 5.Abt G, Reyer HU. 1993. Mate choice and fitness in a hybrid frog—Rana esculenta females prefer Rana lessonae males over their own. Behav. Ecol. Sociobiol. 32, 221–228. ( 10.1007/BF00166511) [DOI] [Google Scholar]
- 6.Christiansen DG, Fog K, Pedersen BV, Boomsma JJ. 2005. Reproduction and hybrid load in all-hybrid populations of Rana esculenta water frogs in Denmark. Evolution 59, 1348–1361. ( 10.1111/j.0014-3820.2005.tb01784.x) [DOI] [PubMed] [Google Scholar]
- 7.Christiansen DG. 2009. Gamete types, sex determination and stable equilibria of all-hybrid populations of diploid and triploid edible frogs (Pelophylax esculentus). BMC Evol. Biol. 9, 135 ( 10.1186/1471-2148-9-135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christiansen DG, Reyer H-U. 2009. From clonal to sexual hybrids: genetic recombination via triploids in all-hybrid populations of water frogs. Evolution 63, 1754–1768. ( 10.1111/j.1558-5646.2009.00673.x) [DOI] [PubMed] [Google Scholar]
- 9.Lamatsch DK, Stöck M. 2009. Sperm-dependent parthenogenesis and hybridogenesis in teleost fishes. In Lost sex (eds Schön I, Martens K, van Dijk P). pp. 399–432. New York, NJ: Springer. [Google Scholar]
- 10.Janko K, Kotusz J, De Gelas K, Slechtová V, Opoldusová Z, Drozd P, Choleva L, Popiołek M, Baláž M. 2012. Dynamic formation of asexual diploid and polyploid lineages: multilocus analysis of Cobitis reveals the mechanisms maintaining the diversity of clones. PLoS ONE 7, e45384 ( 10.1371/journal.pone.0045384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruvost NBM, Mikulíček P, Choleva L, Reyer H-U. 2015. Contrasting reproductive strategies of triploid hybrid males in vertebrate mating systems. J. Evol. Biol. 28, 189–204. ( 10.1111/jeb.12556) [DOI] [PubMed] [Google Scholar]
- 12.Gregory TR, Mable BK. 2005. Polyploidy in animals. In The evolution of the genome (ed. Gregory TR.). pp. 427–517. Princeton, NJ: Dreibelbis, Dana. [Google Scholar]
- 13.Sousa-Santos C, Collares-Pereira MJ, Almada VC. 2006. Reproductive success of nuclear nonhybrid males of Squalius alburnoides hybridogenetic complex (Teleostei, Cyprinidae): an example of interplay between female choice and ecological pressures? Acta Ethol. 9, 31–36. ( 10.1007/s10211-006-0012-8) [DOI] [Google Scholar]
- 14.Morgado-Santos M, Pereira HM, Vicente L, Collares-Pereira MJ. 2015. Mate choice drives evolutionary stability in a hybrid complex. PLoS ONE 10, e0132760 ( 10.1371/journal.pone.0132760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgado-Santos M, Matos I, Vicente L, Collares-Pereira MJ. 2010. Scaleprinting: individual identification based on scale patterns. J. Fish Biol. 76, 1228–1232. ( 10.1111/j.1095-8649.2010.02591.x) [DOI] [PubMed] [Google Scholar]
- 16.Lamatsch DK, Steinlein C, Schmid M, Schartl M. 2000. Noninvasive determination of genome size and ploidy level in fishes by flow cytometry: detection of triploid Poecilia formosa. Cytometry 39, 91–95. ( 10.1002/(SICI)1097-0320(20000201)39:2%3C91::AID-CYTO1%3E3.0.CO;2-4) [DOI] [PubMed] [Google Scholar]
- 17.Sousa-Santos C, Robalo JI, Collares-Pereira MJ, Almada VC. 2005. Heterozygous indels as useful tools in the reconstruction of DNA sequences and in the assessment of ploidy level and genomic constitution of hybrid organisms. DNA Seq. 16, 462–467. ( 10.1080/10425170500356065) [DOI] [PubMed] [Google Scholar]
- 18.Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215 ( 10.1093/nar/16.3.1215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa-Santos C, Robalo J, Almada V. 2014. Spawning behavior of a threatened Iberian cyprinid and its implications for conservation. Acta Ethol. 17, 99–106. ( 10.1007/s10211-014-0185-5) [DOI] [Google Scholar]
- 20.Martelo J, Grossman GD, Porto M, Magalhães MF. 2014. Habitat patchiness affects distribution and microhabitat use of endangered Mira chub Squalius torgalensis (Actinopterygii, Cypriniformes). Hydrobiologia 732, 93–109. ( 10.1007/s10750-014-1850-4) [DOI] [Google Scholar]
- 21.Pires DF, Pires AM, Collares-Pereira MJ, Magalhães MF. 2010. Variation in fish assemblages across dry-season pools in a Mediterranean stream: effects of pool morphology, physicochemical factors and spatial context. Ecol. Freshwater Fish 19, 74–86. ( 10.1111/j.1600-0633.2009.00391.x) [DOI] [Google Scholar]
- 22.Guerrero RD, Shelton WL. 1974. An aceto-carmine squash method for sexing juvenile fishes. Prog. Fish Cult. 36, 56 ( 10.1577/1548-8659(1974)36%5B56:AASMFS%5D2.0.CO;2) [DOI] [Google Scholar]
- 23.Turner TF, Dowling TE, Broughton RE, Gold JR. 2004. Variable microsatellite markers amplify across divergent lineages of cyprinid fishes (subfamily Leuciscinae). Conserv. Genet. 5, 279–281. ( 10.1023/B:COGE.0000029998.11426.ab) [DOI] [Google Scholar]
- 24.Muenzel FM, Sanetra M, Salzburger W, Meyer M. 2007. Microsatellites from the vairone Leuciscus souffia (Pisces: Cyprinidae) and their application to closely related species. Mol. Ecol. Resour. 7, 1048–1050. ( 10.1111/j.1471-8286.2007.01772.x) [DOI] [Google Scholar]
- 25.Vyskočilová M, Šimková A, Martin J-F. 2007. Isolation and characterization of microsatellites in Leuciscus cephalus (Cypriniformes, Cyprinidae) and cross-species amplification within the family Cyprinidae. Mol. Ecol. Res. 7, 1150–1154. ( 10.1111/j.1471-8286.2007.01813.x); [DOI] [Google Scholar]
- 26.Schuelke M. 2000. An economic method for fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234. ( 10.1038/72708) [DOI] [PubMed] [Google Scholar]
- 27.Robalo JI, Almada VC, Levy A, Doadrio I. 2007. Re-examination and phylogeny of the genus Chondrostoma based on mitochondrial and nuclear data and the definition of 5 new genera. Mol. Phylogenet. Evol. 42, 362–372. ( 10.1016/j.ympev.2006.07.003) [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro F, Cowx IG, Tiago P, Filipe AF, da Costa LM, Collares-Pereira MJ. 2003. Growth and reproductive traits of diploid and triploid forms of the Squalius alburnoides cyprinid complex in a tributary of the Guadiana River, Portugal. Arch. Hydrobiol. 156, 471–484. ( 10.1127/0003-9136/2003/0156-0471) [DOI] [Google Scholar]
- 29.StatSoft, Inc. 2013. Electronic statistics textbook. Tulsa, OK: StatSoft; (http://www.statsoft.com/textbook/) (accessed 17 January 2015). [Google Scholar]
- 30.Zar JH. 2010. Biostatistical analysis, 5th edn Upper Saddle River, NJ: Prentice Hall. [Google Scholar]
- 31.Morgado-Santos, et al. In preparation.
- 32.Cabral MJ et al. (eds). 2005. Livro Vermelho dos Vertebrados de Portugal. Lisboa, Portugal: Instituto da Conservação da Natureza. [Google Scholar]
- 33.Burley N. 1988. The differential-allocation hypothesis—an experimental test. Am. Nat. 132, 611–628. ( 10.1086/284877) [DOI] [Google Scholar]
- 34.Sheldon BC. 2000. Differential allocation: tests, mechanisms and implications. Trends Ecol. Evol. 15, 397–402. ( 10.1016/S0169-5347(00)01953-4) [DOI] [PubMed] [Google Scholar]
- 35.Skinner AMJ, Watt PJ. 2007. Strategic egg allocation in the zebra fish, Danio rerio. Behav. Ecol. 18, 905–909. ( 10.1093/beheco/arm059) [DOI] [Google Scholar]
- 36.Casalini M, Agbali M, Reichard M, Konecna M, Bryjova A, Smith C. 2009. Male dominance, female mate choice, and intersexual conflict in the rose bitterling (Rhodeus ocellatus). Evolution 63, 366–376. ( 10.1111/j.1558-5646.2008.00555.x) [DOI] [PubMed] [Google Scholar]
- 37.Evans JP, Box TM, Brooshooft P, Tatter JR, Fitzpatrick JL. 2010. Females increase egg deposition in favor of large males in the rainbowfish, Melanotaenia australis. Behav. Ecol. 21, 465–469. ( 10.1093/beheco/arq006) [DOI] [Google Scholar]
- 38.Uusi-Heikkila S, Boeckenhoff L, Wolter C, Arlinghaus R. 2012. Differential allocation by female zebrafish (Danio rerio) to different-sized males—an example in a fish species lacking parental care. PLoS ONE 7, e48317 ( 10.1371/journal.pone.0048317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linhart O, Rodina M, Flajshans M, Mavrodiev N, Nebesarova J, Gela D, Kocour M. 2006. Studies on sperm of diploid and triploid tench, Tinca tinca (L.). Aquacult. Int. 14, 9–25. ( 10.1007/s10499-005-9010-5) [DOI] [Google Scholar]
- 40.Fujimoto T, Yasui GS, Yoshikawa H, Yamaha E, Arai K. 2008. Genetic and reproductive potential of spermatozoa of diploid and triploid males obtained from interspecific hybridization of Misgurnus anguillicaudatus female with M. mizolepis male. J. Appl. Ichthyol. 24, 430–437. ( 10.1111/j.1439-0426.2008.01131.x) [DOI] [Google Scholar]
- 41.Peruzzi S, Rudolfsen G, Primicerio R, Frantzen M, Kauric G. 2009. Milt characteristics of diploid and triploid Atlantic cod (Gadus morhua L.). Aquacult. Res. 40, 1160–1169. ( 10.1111/j.1365-2109.2009.02212.x) [DOI] [Google Scholar]
- 42.Iguchi K, Ito F. 1994. Occurrence of cross-mating in ayu: amphidromous×land-locked forms, and diploid x triploid. Fish.Sci. 60, 653–655. [Google Scholar]
- 43.Kotiaho JS, Puurtinen M. 2007. Mate choice for indirect genetic benefits: scrutiny of the current paradigm. Funct. Ecol. 21, 638–644. ( 10.1111/j.1365-2435.2007.01286.x) [DOI] [Google Scholar]
- 44.Hettyey A, Hegyi G, Puurtinen M, Hoi H, Torok J, Penn DJ. 2010. Mate choice for genetic benefits: time to put the pieces together. Ethology 116, 1–9. ( 10.1111/j.1439-0310.2009.01704.x)21132114 [DOI] [Google Scholar]
- 45.Watt PJ, Shohet AJ, Renshaw K. 2001. Female choice for good genes and sex-biased broods in guppies. J. Fish Biol. 59, 843–850. ( 10.1111/j.1095-8649.2001.tb00155.x) [DOI] [Google Scholar]
- 46.Brown JL. 1997. A theory of mate choice based on heterozygosity. Behav. Ecol. 8, 60–65. ( 10.1093/beheco/8.1.60) [DOI] [Google Scholar]
- 47.ASAB. 2013. Guidelines for the treatment of animals in behavioural research and teaching. Anim. Behav. 85, 287–295. ( 10.1016/S0003-3472(12)00550-7) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.