Abstract
Antibiotics are routinely used to improve livestock health and growth. However, this practice may have unintended environmental impacts mediated by interactions among the wide range of micro- and macroorganisms found in agroecosystems. For example, antibiotics may alter microbial emissions of greenhouse gases by affecting livestock gut microbiota. Furthermore, antibiotics may affect the microbiota of non-target animals that rely on dung, such as dung beetles, and the ecosystem services they provide. To examine these interactions, we treated cattle with a commonly used broad-spectrum antibiotic and assessed downstream effects on microbiota in dung and dung beetles, greenhouse gas fluxes from dung, and beetle size, survival and reproduction. We found that antibiotic treatment restructured microbiota in dung beetles, which harboured a microbial community distinct from those in the dung they were consuming. The antibiotic effect on beetle microbiota was not associated with smaller size or lower numbers. Unexpectedly, antibiotic treatment raised methane fluxes from dung, possibly by altering the interactions between methanogenic archaea and bacteria in rumen and dung environments. Our findings that antibiotics restructure dung beetle microbiota and modify greenhouse gas emissions from dung indicate that antibiotic treatment may have unintended, cascading ecological effects that extend beyond the target animal.
Keywords: insect, microbiome, antimicrobial, methane, greenhouse gases
1. Introduction
Antibiotics are extensively used in agriculture to promote growth and to treat or prevent livestock disease [1–7]; yet they may have major consequences for human and environmental health [4,8,9]. For example, the use of antibiotics in agriculture can favour the evolution of antibiotic resistance among pathogens and the spread of antibiotic resistance genes to surrounding environments (e.g. [10–12]). In addition, this practice can have other ecosystem-level ramifications that are likely to be important, but less appreciated. In particular, antibiotic treatment could affect two distinct, but potentially interacting ecological processes: the removal and recycling of livestock dung by decomposer organisms, and the release of greenhouse gases from dung.
Livestock dung provides a source of nutrients, organic matter and microbes to pasture ecosystems [13–15]. Dung can also act as a source of pathogens [16] and emit significant quantities of greenhouse gases, including methane [17,18]. However, these effects can be modified by the diverse communities that inhabit, consume and/or interact with dung, of which dung beetles have been particularly well studied [19–23]. In general, dung beetles play a critical role in carbon and nitrogen cycling, and the maintenance of soil fertility in both natural and agricultural pasture ecosystems [24,25]. Veterinary pharmaceuticals can harm beetles and other downstream consumers of livestock dung, which may depress dung decomposition and reduce the diversity of dung-based communities. But while the effects of antiparasiticides have been relatively well studied in this context (e.g. [26–29]), the effects of antibiotics on dung beetles remain poorly known [30].
In contrast to antiparasiticides, which may directly act on the physiology of dung consumers (e.g. [31]), broad-spectrum antimicrobial compounds could have far-reaching, microbially mediated ecological effects. For example, as microbial symbionts (microbiota) are often critical to insect health and reproduction [32,33], antibiotics retained in dung could affect beetle performance by altering beetle microbiota. Furthermore, antibiotic-induced restructuring of livestock gut microbiota could change the nutritional, chemical and microbiological properties of dung, the diet of dung beetles.
Antibiotics could also directly or indirectly modulate greenhouse gas emissions from livestock dung. Antibiotics have been shown to alter the structure and activity of mammalian gut and faecal microbiota [34–36]. In livestock such as cattle, certain members of the gut microbiota are responsible for producing key greenhouse gases such as methane, nitrous oxide and carbon dioxide [37–39]. Despite interest in reducing emissions of these microbially mediated greenhouse gases, and the frequent use of antimicrobial compounds in agriculture, the relationship between the two remains unclear. Furthermore, the effects of antibiotics on greenhouse gas fluxes and on dung beetles could be linked. For example, dung beetles can reduce methane production from dung to an extent that could have impacts in some agricultural systems [40]. As methanogenesis is thought to be inhibited by oxygen entering the dung pat through beetle tunnels [20], it could be sensitive to the size and number of dung beetles. In turn, if beetle performance and fitness are influenced by antibiotic-sensitive microbiota, the effect of dung beetles on greenhouse gas emissions could depend upon the antibiotic treatment.
We hypothesized that antibiotics administered to cattle alter dung beetle microbiota and, as a consequence, depress beetle fitness. In addition, we hypothesized that antibiotics modulate gas emissions from dung and the extent to which beetles influence emissions. To experimentally test these hypotheses, we treated cattle with and without a broad-spectrum antibiotic, and assayed microbial communities in cow dung and in field-collected dung beetles (Aphodius fossor L.). We also tested the effect of antibiotics on beetle size and numbers, and on fluxes of multiple greenhouse gases (carbon dioxide, nitrous oxide and methane) from dung. Our findings demonstrate that the common practice of administering antibiotics to livestock [1–7] can have important, unintended impacts on dung biota and the biogeochemical processes they mediate in agroecosystems.
2. Material and methods
(a). Experimental set-up
Ten cows were randomly assigned to two treatments: five were given a standard 3-day course of tetracycline and five were left as controls. None of the cows had undergone antibiotic treatment within the previous six months, except for one cow in the antibiotic group, which had received a course of penicillin six weeks prior to the experiment (for the full history of each cow, see the electronic supplementary material, Methods). From each cow, we collected fresh dung on a single day, beginning 1 h after the last administration of antibiotics. That afternoon, we separated the dung from each cow into six 1 l pats and placed each pat in a mescosm (an open-bottom, mesh-covered plastic bucket) in the field. We also included four control mesocosms without dung to measure background fluxes of carbon dioxide (CO2), methane (CH4) and nitrous oxide (N2O) from the soil. More detail on experimental design is given in the electronic supplementary material, Methods.
To examine the effects of antibiotics on dung beetle microbiota, on the performance of beetles and on beetle-mediated effects on gas fluxes from cow pats, we focused on the dung beetle Aphodius fossor (L.). Aphodius fossor is a regionally widespread and locally common species [41], and its ecology and interactions with dung have been extensively studied (e.g. [20,21,42]). The beetles were collected in the field in early June from different localities and stored at 4°C until they were added to dung pats.
Dung beetles were added to four of the six dung pats produced by each cow (randomly chosen). Gas measurements and dung samples for microbial analysis were taken from two intact pats with beetles and two without beetles. The two additional pats with beetles were used for more invasive sampling of beetles for microbial characterization and to measure beetle reproduction and development. Based on beetle densities recorded in the field and on a previous study [42], we added 12 beetles to each pat, maintaining a sex ratio of 1 : 1.
(b). Gas flux measurements
Gas fluxes of CO2, N2O and CH4 were measured from dung mesocosms in the field at five time points over the course of the experiment (more information given in the electronic supplementary material). Net gas fluxes emitted from each dung pat were calculated as in Penttilä et al. [20].
(c). Dung and beetle sampling
All dung and beetle samples were preserved for subsequent characterization of microbiota using 95% ethanol, which is an effective storage medium for microbial community analysis [43]. On days 2, 11 and 23 of the experiment, dung samples of approx. 1 ml were taken and stored in ethanol (see the electronic supplementary material, Methods).
To test whether antibiotic treatment affected beetle microbiota, two parent beetles from each pat were preserved in ethanol on day 7 of the experiment. To measure antibiotic effects on beetle size, reproduction and survival, we sampled pats on day 43 (for half-grown larvae) and days 71 and 73 (for the next generation of adult beetles). Larvae recovered were weighed while fresh and the width of their head capsule was measured. Total offspring counts by the end of the experiment were used as an integrated measure of both adult reproduction and offspring survival.
(d). Molecular protocol and sequence data processing
To characterize the overall beetle-associated microbial community, whole adults were homogenized and DNA was extracted from approximately 100 mg of homogenate with the MoBio PowerSoil kit, following similar studies [43,44]. Approximately 100 mg subsamples of dung were used for DNA extraction with the same kit. Using barcoded primers, we PCR-amplified the V4 region of the bacterial and archaeal 16S rRNA gene, with the amplicons sequenced on an Illumina MiSeq platform as previously described [43,45]. Detail regarding sequence data processing and taxonomic identification is given in the electronic supplementary material, Methods.
(e). Statistical analysis
All statistical analyses were performed in R v. 3.2.2 [46], and plots were constructed with the ggplot2 package [47]. The vegan package [48] was used to analyse microbial community data. Following previous work (e.g. [43–45]), we used the Bray–Curtis dissimilarity metric to quantify variation in overall microbial community structure among samples. Non-metric multidimensional scaling plots were constructed to visualize Bray–Curtis dissimilarity in two dimensions. To avoid pseudoreplication of beetle data, dissimilarities (multivariate analyses) or performance variables (univariate analyses) were averaged between the beetles in each mesocosm; thus, multilevel statistical tests were conducted on the effects of antibiotic treatment and cow individual.
The effects of antibiotic treatment and cow individual on beetle and dung microbiota were tested using nested permutational multivariate ANOVA in the BiodiversityR package [49], with cow nested within treatment (999 permutations). Unless noted otherwise, below we report results from dung sampled on day 2 of the experiment, which we expected to most closely represent microbiota in the cow gut, but analyses of dung sampled on days 11 and 23 are shown in the electronic supplementary material. With each of the gases measured, we used ANOVA to assess effects of antibiotics and beetle presence on cumulative gas fluxes over the course of the experiment, as well as the interaction of the two factors. We analysed the proportion of methanogens in dung microbiota (all time points) with the same procedure, but with prior log transformation to meet the assumption of normality.
To assess antibiotic effects on the performance of larval offspring, we chose to focus on weight, which was highly correlated with head size (Spearman's rank correlation, ρ = 0.68, p = 0.005). Weight can be considered as a proxy for performance as it is correlated with adult fecundity in both Aphodius [50] and other insects (e.g. [51,52]). A nested ANOVA (cow individual within antibiotic treatment) was used to test for effects on larval weight; normality of model residuals was confirmed visually. The effect of antibiotic treatment on total counts of beetle offspring recovered from each dung pat was analysed by fitting a generalized linear mixed model with cow individual as a random effect, Poisson-distributed errors and a log link function.
3. Results and discussion
(a). Microbial community structure in beetles
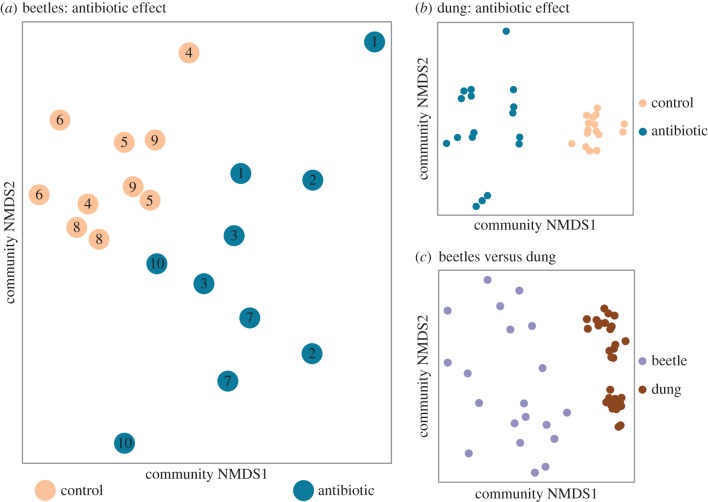
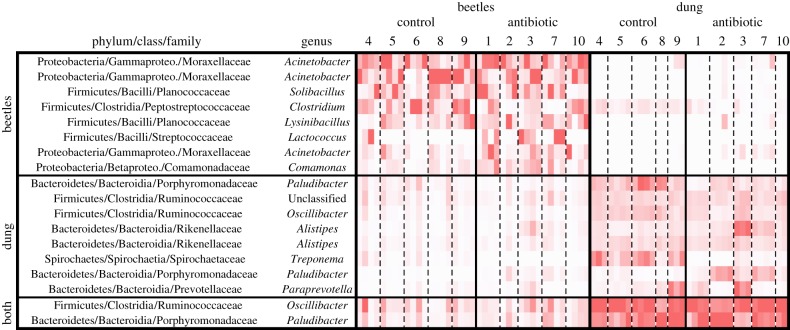
Dung beetles are extraordinarily diverse [53], and both scientifically [54] and ecologically important [25]; yet only one previous study has used DNA sequence-based methods to characterize their microbiota (Onthophagus taurus [55]). Despite feeding on a microbe-rich substrate, we found that A. fossor adults transform dung microbiota during digestion or host symbionts not present in their diet. Overall, beetles and dung had compositionally distinct microbiota (F1,51 = 17.32, p = 0.001; figures 1c and 2), regardless of the dung sampling time point (electronic supplementary material, figures S1a,b), and beetles contained lower microbial diversity than their diet (F1,51 = 45.12, p < 0.0001; electronic supplementary material, figure S2). While the dung communities were dominated by Bacteroidia, Clostridia and Spirochaetes, the dung beetle microbiota were dominated by Gammaproteobacteria and Bacilli. Less than 25% of the operational taxonomic units (OTUs) that were relatively abundant in beetle communities (overall proportion greater than or equal to 1%) were also abundant in dung, implying that the dominant beetle microbes are either vertically transmitted or rare in dung. Indeed, the genus Acinetobacter was highly abundant in A. fossor (figure 2) and also found in O. taurus fed sterilized dung [55], suggesting that Acinetobacter could be vertically transmitted symbionts common among dung beetles.
Figure 1.
Microbial communities diverge upon antibiotic treatment and cluster according to sample habitat (beetle versus dung). Non-metric multidimensional scaling ordinations of microbiota in (a) parental dung beetles, where cow individual is indicated by numbers and treatment by colour, (b) dung samples only and (c) beetles versus dung. The ordinations visually represent Bray–Curtis dissimilarities among samples in two dimensions. (Online version in colour.)
Figure 2.
Variability in dominant microbial OTUs between individuals, sample habitats and treatments. The heatmap shows OTU relative abundance in each sample (columns) where darker shading indicates a higher proportion; the range of values is scaled separately for each sample. Numbers above columns denote cow individuals. For clarity, only the top 10 OTUs for each sample type are shown (two OTUs were in the top 10 of both beetles and dung). All OTUs shown are bacteria, as archaea were less abundant overall. (Online version in colour.)
(b). Effects of antibiotics on dung and dung beetle microbiota
As expected from previous work [34–36,56], we found a clear effect of antibiotic treatment on dung microbiota (F1,23 = 4.84, p = 0.01; figure 1b; electronic supplementary material, figure S2), and this effect persisted even 23 days after defecation and sample collection (electronic supplementary material, figure S1c,d). As in other animal taxa that maintain individual-specific microbiota (e.g. [56,57]), we also found that dung microbial communities clustered by cow individual (F8,23 = 4.87, p = 0.001; figure 2). Antibiotic treatment administered to cattle also affected dung beetle microbiota (F1,10 = 2.39, p = 0.007; figure 1a; electronic supplementary material, figure S2), and mirroring the patterns found for dung, the microbial communities in beetles also clustered by cow individual (F8,10 = 1.38, p = 0.001; figure 1a).
Antibiotics could alter beetle microbiota by multiple mechanisms. For example, antibiotic-induced changes in dung microbiota (figure 1b) could change the nutritional quality of the beetles' diet, or provide an altered inoculum to the beetle community. Alternatively, antibiotic residues present in dung and consumed by beetles may act on beetle gut microbiota. Concern has been raised over the persistence of antiparasiticides in the environment, where—as eukaryote-specific pharmaceuticals—they are directly toxic to animals [30,31,58]. Our data suggest that even antibacterials such as tetracycline may have similar far-reaching effects, by altering the microbiota of non-target animals.
(c). Beetle size and numbers
Despite clear antibiotic effects on beetle microbiota (figure 1a), and the often critical role of microbiota in insect biology [32,33], antibiotics did not influence dung beetle size, reproduction or survival. Larval weight was unaffected by antibiotic treatment (F1,13 = 0.018, p = 0.90), and the total number of beetle offspring recovered from dung pats by the end of the experiment was similar between treatments (Poisson GLMM, z = −0.35, p = 0.73). An average of 4.2 ± 1.5 versus 3.0 ± 0.71 offspring were collected from dung pats of control and antibiotic treatments, respectively (mean ± s.e.m., n = 10 per treatment). As not all members of the beetle microbiota responded to antibiotics (figure 2), one possible explanation is that the antibiotic effect on overall microbiota was driven by commensal and not mutualistic species. For example, Acinetobacter abundance was not sensitive to antibiotic treatment (F1,10 = 0.35, p = 0.57; figure 2), and tetracycline resistance has been documented among Acinetobacter in other environments [59,60]. Alternatively, microbial symbionts may simply not be important to the nutrition or development of dung beetles [61]. However, given the wide variety of processes that can be influenced by microbiota (e.g. [62–64]), the antibiotic-induced microbial restructuring we observed could affect unmeasured aspects of dung beetle biology.
(d). Greenhouse gas emissions from dung
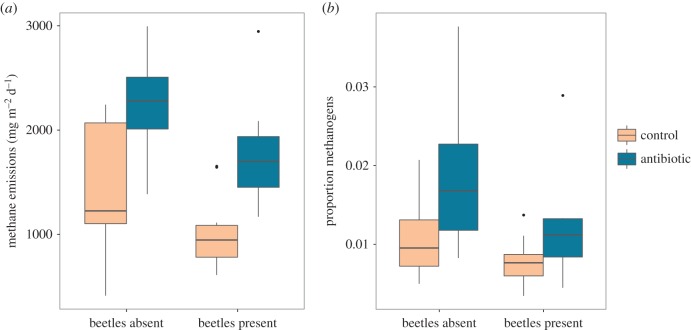
The presence of beetles decreased methane fluxes from dung (F1,36 = 7.49, p < 0.01; figure 3a), an effect reported previously and probably due to oxygenation of the pat caused by beetle tunnels [20,40]. By contrast, antibiotic treatment consistently increased methane emissions (F1,36 = 22.21, p < 0.0001; figure 3a), which are probably related to changes in dung microbiota (figure 1b). Contrary to our expectations, the two effects were unrelated (interaction F1,36 = 0.004, p = 0.95), suggesting that beetle tunnelling activity is not affected by antibiotic modification of dung (and its impact on the beetle microbiome).
Figure 3.
Dung methanogenesis is raised by antibiotic treatment and lowered by the presence of dung beetles, a pattern matched by methanogen abundance in dung microbiota. (a) Boxplot showing the factors affecting total methane emissions quantified from dung pats. (b) Boxplot showing the proportion of methanogenic archaea in DNA sequence libraries from the same dung samples. Bold horizontal lines indicate medians, box limits show first and third quartiles, whiskers extend to the most extreme values within 1.5× the inter-quartile range, and dots show outlying data points. (Online version in colour.)
Carbon dioxide emissions were similar between treatments (F1,36 = 0.001, p = 0.98), and there was no effect of beetle presence (F1,36 = 0.02, p = 0.88) nor an interaction between the two factors (F1,36 = 0.22, p = 0.64), suggesting that antibiotics do not affect overall microbial decomposition rates. This result indicates that the antibiotic effect on methane production is not simply due to an increase in overall microbial activity. By contrast, fluxes of nitrous oxide, another potent greenhouse gas, were influenced by beetles in an antibiotic-dependent manner (electronic supplementary material, figure S3). Specifically, the presence of beetles raised nitrous oxide emissions (F1,36 = 17.31, p < 0.001), in agreement with a previous report [20] (but see [40]). In support of our original hypothesis, antibiotic treatment appeared to weaken this beetle-mediated increase (interaction F1,36 = 5.85, p = 0.02). There was no main effect of antibiotics alone (F1,36 = 1.99, p = 0.17), despite clear antibiotic-induced changes to dung microbiota (figure 1b). It remains to be determined whether antibiotics modulate the effect of beetle presence on nitrous oxide specifically through their impact on beetle microbiota (figure 1a).
(e). Implications and potential mechanism of methane effect
The large (1.8-fold) increase in methane emissions from the dung of cattle treated with antibiotics (figure 3a) has not been documented, despite the considerable literature on methane production from agricultural systems (e.g. [17,37–39,65,66]), and the long-standing and increasing administration of antibiotics to livestock. Previous studies have found either a short-lived decrease [65], or no effect following antibiotic treatment [66,67]; to the best of our knowledge, this is the first report of antibiotics increasing methane emissions. While dung emissions of methane are typically lower than those released from belching [68], they still comprise a substantial proportion of total agricultural methanogenesis in pasture systems [40]. Moreover, as the effects of antibiotics apparently derive from microbial interactions within the gut (explained below), they probably extend to gas emissions from enteric fermentation as well. Hence, we suggest that future research should be focused on antibiotic effects on methane emissions from belching.
The pattern of methane emissions (figure 3a) was qualitatively similar to that of the relative abundance of methanogens in the same dung pats (figure 3b). As with gas fluxes, antibiotics raised the proportional abundance of methanogens (F1,35 = 8.72, p = 0.006), and there was no interaction of antibiotics with beetle presence (F1,35 = 0.26, p = 0.62). We suggest that these patterns may be explained by competitive dynamics among gut microbiota. In the gut of ruminants and other mammals, methanogenic archaea may compete with bacteria for hydrogen, which is often scarce [69,70]. Tetracycline and some other broad-spectrum antibiotics are generally less effective against archaea, including methanogens isolated from mammalian digestive tracts [71,72]. Therefore, we propose that by specifically suppressing bacteria in the gut and subsequently in dung, antibiotic treatment enables methanogens to outcompete bacteria for hydrogen, increasing their concomitant methane output.
4. Conclusion
The routine practice of administering antibiotics to livestock [1–7] can have unexpected consequences for dung biota and greenhouse gas emissions from agriculture. First, antibiotics altered the composition of microbial communities associated with dung beetles, an ecologically important group of insects in many environments. This finding highlights a unique feature of antibacterial pharmaceuticals: even if not directly toxic to non-target animals, they may have a range of unanticipated effects by altering the microbiota of both livestock and wildlife. Second, we provide the first demonstration that antibiotics can increase dung emissions of methane, a potent greenhouse gas. Our findings call for analyses at larger scales (e.g. [40]) that take other factors into account, such as the relative importance of dung versus belching in gas emissions, and the global extent and purpose of antibiotic use in livestock production. Improved monitoring and estimates of agricultural antibiotic use will be necessary to identify whether antibiotics may impact the overall contribution of livestock production to global warming. Finally, further research into this effect will require unravelling the ecological interactions between microbes in the gut of livestock and their susceptibilities to antibiotic disturbance.
Supplementary Material
Acknowledgements
We acknowledge E. Beaury for assistance processing samples, J. Henley for operating the Illumina MiSeq, E. Tripp for use of the bead shaker and E. Hakanen for assisting with the experiment. The Director of the Viikki Experimental Farm, Miika Kahelin, kindly provided cattle logistics. We thank members of N. Fierer and M. D. Bowers' lab groups for valuable discussions, and two anonymous reviewers for comments that improved the manuscript.
Ethics
In accordance with the Finnish Act on Animal Experimentation (62/2006), the Animal Experiment Board in Finland (ELLA) approved the procedures used in this study.
Data accessibility
DNA sequences and all metadata are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.6bs01.
Authors' contributions
T.J.H., N.F., T.R. and E.S. conceived the original idea. T.J.H. helped design the study, conducted molecular laboratory work, analysed data, and drafted the manuscript with additional input from N.F. and T.R. N.F. participated in study design and data analysis, and provided equipment and facilities for sequencing. B.H., T.R., H.V., J.T. and E.S. helped design the experiment, and J.T. administered the antibiotic treatments and handled the cows. B.H. and H.V. implemented all phases of the field experiment, including response measurements and beetle sampling. A.S. directed gas flux measurements and their interpretation. All authors contributed to the final version of the manuscript and gave approval for publication.
Competing interests
We have no competing interests.
Funding
T.J.H. was supported by the US National Science Foundation (NSF) Graduate Research Fellowship Program under grant no. 1144083. N.F. was supported by NSF (grant no. DEB-0953331). B.H., E.S., H.V. and T.R. were supported by a grant from the Academy of Finland (grant no. 138346 to T.R.).
References
- 1.Mellon MG, Benbrook C, Benbrook KL. 2001. Hogging it: estimates of antimicrobial abuse in livestock. Cambridge, MA: Union of Concerned Scientists Publications. [Google Scholar]
- 2.McEwen SA, Fedorka-Cray PJ. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34, S93–106. ( 10.1086/340246) [DOI] [PubMed] [Google Scholar]
- 3.Sarmah AK, Meyer MT, Boxall AB. 2006. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. ( 10.1016/j.chemosphere.2006.03.026) [DOI] [PubMed] [Google Scholar]
- 4.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc. Natl Acad. Sci. USA 112, 5649–5654. ( 10.1073/pnas.1503141112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S. Food and Drug Administration. 2015. Summary report on antimicrobials sold or distributed for use in food-producing animals. See http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM476258.pdf.
- 6.European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption. 2015. Sales of veterinary antimicrobial agents in 26 EU/EEA countries in 2013. See http://www.ema.europa.eu/docs/en_GB/document_library/Report/2015/10/WC500195687.pdf.
- 7.Krishnasamy V, Otte J, Silbergeld E. 2015. Antimicrobial use in Chinese swine and broiler poultry production. Antimicrob. Resist. Infect. Control. 4, 17 ( 10.1186/s13756-015-0050-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustafson RH, Bowen RE. 1997. Antibiotic use in animal agriculture. J. Appl. Microbiol. 83, 531–541. ( 10.1046/j.1365-2672.1997.00280.x) [DOI] [PubMed] [Google Scholar]
- 9.Kumar K, Gupta SC, Chander Y, Singh AK. 2005. Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87, 1–54. ( 10.1016/S0065-2113(05)87001-4) [DOI] [Google Scholar]
- 10.Schwarz S, Kehrenberg C, Walsh TR. 2001. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17, 431–437. ( 10.1016/S0924-8579(01)00297-7) [DOI] [PubMed] [Google Scholar]
- 11.Teuber M. 2001. Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 4, 493–499. ( 10.1016/S1369-5274(00)00241-1) [DOI] [PubMed] [Google Scholar]
- 12.Aarestrup FM. 2005. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 96, 271–281. ( 10.1111/j.1742-7843.2005.pto960401.x) [DOI] [PubMed] [Google Scholar]
- 13.Eghball B, Wienhold BJ, Gilley JE, Eigenberg RA. 2002. Mineralization of manure nutrients. J. Soil Water Conserv. 57, 470–473. [Google Scholar]
- 14.Aarons SR, O'Connor CR, Hosseini HM, Gourley CJP. 2009. Dung pads increase pasture production, soil nutrients and microbial biomass carbon in grazed dairy systems. Nutr. Cycl. Agroecosystems 84, 81–92. ( 10.1007/s10705-008-9228-5) [DOI] [Google Scholar]
- 15.Yoshitake S, Soutome H, Koizumi H. 2014. Deposition and decomposition of cattle dung and its impact on soil properties and plant growth in a cool-temperate pasture. Ecol. Res. 29, 673–684. ( 10.1007/s11284-014-1153-2) [DOI] [Google Scholar]
- 16.Mawdsley JL, Bardgett RD, Merry RJ, Pain BF, Theodorou MK. 1995. Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 2, 1–15. ( 10.1016/0929-1393(94)00039-A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis SC, Lovell RD, Panayides R. 1995. Patterns of methane emission from excreta of grazing animals. Soil Biol. Biochem. 27, 1581–1588. ( 10.1016/0038-0717(95)00092-S) [DOI] [Google Scholar]
- 18.Saggar S, Bolan NS, Bhandral R, Hedley CB, Luo J. 2004. A review of emissions of methane, ammonia, and nitrous oxide from animal excreta deposition and farm effluent application in grazed pastures. New Zeal. J. Agric. Res. 47, 513–544. ( 10.1080/00288233.2004.9513618) [DOI] [Google Scholar]
- 19.Yokoyama K, Kai H, Koga T, Aibe T. 1991. Nitrogen mineralization and microbial populations in cow dung, dung balls and underlying soil affected by paracoprid dung beetles. Soil Biol. Biochem. 23, 649–653. ( 10.1016/0038-0717(91)90078-X) [DOI] [Google Scholar]
- 20.Penttilä A, Slade EM, Simojoki A, Riutta T, Minkkinen K, Roslin T. 2013. Quantifying beetle-mediated effects on gas fluxes from dung pats. PLoS ONE 8, e71454 ( 10.1371/journal.pone.0071454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slade EM, Roslin T, Santalahti M, Bell T. 2015. Disentangling the ‘brown world’ faecal–detritus interaction web: dung beetle effects on soil microbial properties. Oikos 125, 629–635. ( 10.1111/oik.02640) [DOI] [Google Scholar]
- 22.Nichols E, Gomez A. 2014. Dung beetles and fecal helminth transmission: patterns, mechanisms and questions. Parasitology 141, 614–623. ( 10.1017/S0031182013002011) [DOI] [PubMed] [Google Scholar]
- 23.Manning P, Slade EM, Beynon SA, Lewis OT. 2016. Functionally rich dung beetle assemblages are required to provide multiple ecosystem services. Agric. Ecosyst. Environ. 218, 87–94. ( 10.1016/j.agee.2015.11.007) [DOI] [Google Scholar]
- 24.Bang HS, Lee JH, Kwon OS, Na YE, Jang YS, Kim WH. 2005. Effects of paracoprid dung beetles (Coleoptera: Scarabaeidae) on the growth of pasture herbage and on the underlying soil. Appl. Soil Ecol. 29, 165–171. ( 10.1016/j.apsoil.2004.11.001) [DOI] [Google Scholar]
- 25.Nichols E, Spector S, Louzada J, Larsen T, Amezquita S, Favila ME. 2008. Ecological functions and ecosystem services provided by Scarabaeinae dung beetles. Biol. Conserv. 141, 1461–1474. ( 10.1016/j.biocon.2008.04.011) [DOI] [Google Scholar]
- 26.Wall R, Strong L. 1987. Environmental consequences of treating cattle with the antiparasitic drug ivermectin. Nature 327, 418–421. ( 10.1038/327418a0) [DOI] [PubMed] [Google Scholar]
- 27.Madsen M, Nielsen BO, Holter P, Pedersen OC, Jespersen JB, Jensen K-MV, Nansen P, Gronvold J. 1990. Treating cattle with ivermectin: effects on the fauna and decomposition of dung pats. J. Appl. Ecol. 27, 1–15. ( 10.2307/2403564) [DOI] [Google Scholar]
- 28.Floate KD, Wardhaugh KG, Boxall ABA, Sherratt TN. 2005. Fecal residues of veterinary parasiticides: nontarget effects in the pasture environment. Annu. Rev. Entomol. 50, 153–179. ( 10.1146/annurev.ento.50.071803.130341) [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Cogollo LC, Rodríguez-Vivas RI, Delfín-González H, Reyes-Novelo E, Ojeda-Chi MM. 2015. Lethal and sublethal effects of ivermectin on Onthophagus landolti (Coleoptera: Scarabaeidae). Environ. Entomol. 44, 1634–1640. ( 10.1093/ee/nvv139) [DOI] [PubMed] [Google Scholar]
- 30.Schmitt H, Römbke J. 2008. The ecotoxicological effects of pharmaceuticals (antibiotics and antiparasiticides) in the terrestrial environment: a review. In Pharmaceuticals in the environment (ed. Kümmerer K.), pp. 285–303. Berlin, Germany: Springer. [Google Scholar]
- 31.Verdú JR, Cortez V, Ortiz AJ, González-Rodríguez E, Martinez-Pinna J, Lumaret J-P, Lobo JM, Numa C, Sánchez-Piñero F. 2015. Low doses of ivermectin cause sensory and locomotor disorders in dung beetles. Sci. Rep. 5, 13912 ( 10.1038/srep13912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel P, Moran NA. 2013. The gut microbiota of insects: diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. ( 10.1111/1574-6976.12025) [DOI] [PubMed] [Google Scholar]
- 33.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. ( 10.1146/annurev-ento-010814-020822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Looft T, et al. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl Acad. Sci. USA 109, 1691–1696. ( 10.1073/pnas.1120238109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Looft T, Allen HK. 2012. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. ( 10.4161/gmic.21288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50. ( 10.1016/j.cell.2012.10.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss AR, Jouany J, Newbold J. 2000. Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49, 231–253. ( 10.1051/animres:2000119) [DOI] [Google Scholar]
- 38.Kebreab E, Clark K, Wagner-Riddle C, France J. 2006. Methane and nitrous oxide emissions from Canadian animal agriculture: a review. Can. J. Anim. Sci. 86, 135–158. ( 10.4141/A05-010) [DOI] [Google Scholar]
- 39.Martin C, Morgavi DP, Doreau M. 2010. Methane mitigation in ruminants: from microbe to the farm scale. Animal 4, 351–365. ( 10.1017/S1751731109990620) [DOI] [PubMed] [Google Scholar]
- 40.Slade EM, Riutta T, Roslin T, Tuomisto HL. 2016. The role of dung beetles in reducing greenhouse gas emissions from cattle farming. Sci. Rep. 6, 18140 ( 10.1038/srep18140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roslin T. 2001. Spatial population structure in a patchily distributed beetle. Mol. Ecol. 10, 823–837. ( 10.1046/j.1365-294X.2001.01235.x) [DOI] [PubMed] [Google Scholar]
- 42.Vessby K. 2001. Habitat and weather affect reproduction and size of the dung beetle Aphodius fossor. Ecol. Entomol. 26, 430–435. ( 10.1046/j.1365-2311.2001.00331.x) [DOI] [Google Scholar]
- 43.Hammer TJ, Dickerson JC, Fierer N. 2015. Evidence-based recommendations on storing and handling specimens for analyses of insect microbiota. PeerJ 3, e1190 ( 10.7717/peerj.1190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammer TJ, McMillan WO, Fierer N. 2014. Metamorphosis of a butterfly-associated bacterial community. PLoS ONE 9, e86995 ( 10.1371/journal.pone.0086995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez KS, et al. 2014. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc. R. Soc. B 281, 20141988 ( 10.1098/rspb.2014.1988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 47.Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York, NY: Springer. [Google Scholar]
- 48.Oksanen J, et al. 2015. vegan: Community Ecology Package. R package version 2.3-0. See http://CRAN.R-project.org/package=vegan .
- 49.Kindt R, Coe R. 1995. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. Nairobi, Kenya: World Agroforestry Centre.
- 50.Hirschberger P. 1999. Larval population density affects female weight and fecundity in the dung beetle Aphodius ater. Ecol. Entomol. 24, 316–322. ( 10.1046/j.1365-2311.1999.00205.x) [DOI] [Google Scholar]
- 51.Tammaru T, Kaitaniemi P, Ruohomäki K. 1996. Realized fecundity in Epirrita autumnata (Lepidoptera: Geometridae): relation to body size and consequences to population dynamics. Oikos 77, 407–416. ( 10.2307/3545931) [DOI] [Google Scholar]
- 52.Honek A. 1993. Intraspecific variation in body size and fecundity in insects: a general relationship. Oikos 66, 483–492. ( 10.2307/3544943) [DOI] [Google Scholar]
- 53.Hanski I, Cambefort Y (eds). 1991. Dung beetle ecology. Princeton, NJ: Princeton University Press. [Google Scholar]
- 54.Simmons LW, Ridsdill-Smith TJ (eds). 2011. Ecology and evolution of dung beetles. New York, NY: Wiley-Blackwell. [Google Scholar]
- 55.Estes AM, Hearn DJ, Snell-Rood EC, Feindler M, Feeser K, Abebe T, Dunning Hotopp JC, Moczek AP. 2013. Brood ball-mediated transmission of microbiome members in the dung beetle, Onthophagus taurus (Coleoptera: Scarabaeidae). PLoS ONE 8, e79061 ( 10.1371/journal.pone.0079061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grønvold A-MR, Mao Y, L'Abée-Lund TM, Sørum H, Sivertsen T, Yannarell AC, Mackie RI. 2011. Fecal microbiota of calves in the clinical setting: effect of penicillin treatment. Vet. Microbiol. 153, 354–360. ( 10.1016/j.vetmic.2011.05.040) [DOI] [PubMed] [Google Scholar]
- 57.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. ( 10.1126/science.1177486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puniamoorthy N, Schäfer MA, Römbke J, Meier R, Blanckenhorn WU. 2014. Ivermectin sensitivity is an ancient trait affecting all ecdysozoa but shows phylogenetic clustering among sepsid flies. Evol. Appl. 7, 548–554. ( 10.1111/eva.12152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henwood CJ, Gatward T, Warner M, James D, Stockdale MW, Spence RP, Towner KJ, Livermore DM, Woodford N. 2002. Antibiotic resistance among clinical isolates of Acinetobacter in the UK, and in vitro evaluation of tigecycline (GAR-936). J. Antimicrob. Chemother. 49, 479–487. ( 10.1093/jac/49.3.479) [DOI] [PubMed] [Google Scholar]
- 60.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953. ( 10.1128/AAC.01388-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrne MJ, Watkins B, Bouwer G. 2013. Do dung beetle larvae need microbial symbionts from their parents to feed on dung? Ecol. Entomol. 38, 250–257. ( 10.1111/een.12011) [DOI] [Google Scholar]
- 62.Brownlie JC, Johnson KN. 2009. Symbiont-mediated protection in insect hosts. Trends Microbiol. 17, 348–354. ( 10.1016/j.tim.2009.05.005) [DOI] [PubMed] [Google Scholar]
- 63.Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543. ( 10.1111/j.1365-2311.2011.01318.x) [DOI] [Google Scholar]
- 64.Lizé A, McKay R, Lewis Z. 2013. Gut microbiota and kin recognition. Trends Ecol. Evol. 28, 325–326. ( 10.1016/j.tree.2012.10.013) [DOI] [PubMed] [Google Scholar]
- 65.Johnson KA, Johnson DE. 1995. Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. [DOI] [PubMed] [Google Scholar]
- 66.Shibata M, Terada F. 2010. Factors affecting methane production and mitigation in ruminants. Anim. Sci. J. 81, 2–10. ( 10.1111/j.1740-0929.2009.00687.x) [DOI] [PubMed] [Google Scholar]
- 67.Hashimoto AG, Varel VH, Chen YR. 1981. Ultimate methane yield from beef cattle manure: effect of temperature, ration constituents, antibiotics and manure age. Agric. Wastes 3, 241–256. ( 10.1016/0141-4607(81)90011-1) [DOI] [Google Scholar]
- 68.Tubiello FN, Salvatore M, Rossi S, Ferrara A, Fitton N, Smith P. 2013. The FAOSTAT database of greenhouse gas emissions from agriculture. Environ. Res. Lett. 8, 015009 ( 10.1088/1748-9326/8/1/015009) [DOI] [Google Scholar]
- 69.Strocchi A, Furne J, Ellis C, Levitt MD. 1994. Methanogens outcompete sulphate reducing bacteria for H2 in the human colon. Gut 35, 1098–1101. ( 10.1136/gut.35.8.1098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morvan B, Bonnemoy F, Fonty G, Gouet P. 1996. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr. Microbiol. 32, 129–133. ( 10.1007/s002849900023) [DOI] [PubMed] [Google Scholar]
- 71.Dridi B, Fardeau M-L, Ollivier B, Raoult D, Drancourt M. 2011. The antimicrobial resistance pattern of cultured human methanogens reflects the unique phylogenetic position of archaea. J. Antimicrob. Chemother. 66, 2038–2044. ( 10.1093/jac/dkr251) [DOI] [PubMed] [Google Scholar]
- 72.Khelaifia S, Drancourt M. 2012. Susceptibility of archaea to antimicrobial agents: applications to clinical microbiology. Clin. Microbiol. Infect. 18, 841–848. ( 10.1111/j.1469-0691.2012.03913.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences and all metadata are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.6bs01.