Abstract
Background
This meta-analysis aimed to assess available evidence on possible associations of interleukin-28B polymorphisms rs8099917 and rs12979860 with development of hepatitis virus-related hepatocellular carcinoma (HCC).
Methods
PubMed, EMBASE, Google Scholar, and the Chinese National Knowledge Infrastructure databases were systematically searched to identify relevant studies. Meta-analyses were performed to examine the association of interleukin-28B rs8099917 G/T and rs12979860 T/C polymorphisms with development of hepatitis virus-related HCC. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated.
Results
A total of ten studies involving 2,529 cases and 2,412 controls were included. The G-allele and GT genotype of rs8099917 were significantly associated with increased risk of hepatitis B virus (HBV)-related HCC (allelic model, OR 1.49, 95% CI 1.13–1.96, P=0.005; heterozygous model, OR 1.39, 95% CI 1.04–1.88, P=0.03). Conversely, the TT genotype was found to be significantly associated with lower risk of HBV-related HCC (dominant model, OR 0.68, 95% CI 0.51–0.91, P=0.01). Similar results were observed in the subgroup of Chinese patients and controls. In the pooled data set, the T-allele and TT genotype of rs12979860 showed a significant association with increased HCC risk (allelic model, OR 1.36, 95% CI 1.05–1.78, P=0.02; recessive model, OR 1.75, 95% CI 1.28–2.39, P=0.005; homozygous model, OR 1.99, 95% CI 1.41–2.80, P<0.001). Subgroup analysis based on ethnicity and etiology showed rs12979860 polymorphism to be significantly associated with HCC risk in Caucasians, especially hepatitis C virus-related HCC, according to all five genetic models. In contrast, only the TT genotype of rs12979860 was found to be significantly associated with increased risk of HBV-related HCC, especially in Asians.
Conclusion
The G-allele of rs8099917 may confer elevated risk of HBV-related HCC, while the wild-type TT genotype may protect against the disease. The T-allele of rs12979860 may increase the risk of HCC, in Caucasians, especially hepatitis C virus-related HCC. The TT genotype of rs12979860 may confer increased risk of HBV-related HCC, especially in Asians. These conclusions should be verified in large, well-designed studies.
Keywords: interleukin-28B, polymorphisms, hepatitis virus-related hepatocellular carcinoma, meta-analysis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related mortality worldwide.1 It is well known that infection with hepatitis B virus (HBV) or hepatitis C virus (HCV) is a major cause of chronic hepatitis, which can progress to HCC.2 Since only a fraction of infected patients develop HCC during their lifetime, genetic factors may play a role in tumor development.
Interferon (IFN) treatment is usually the preferred approach for eliminating HBV and HCV in chronic hepatitis patients and thereby inhibiting HCC progression.3,4 IFN-α, IFN-β, and IFN-λ are multifunctional cytokines responsible for transcription of subsets of genes, and they play a crucial role in antiviral immune responses involving activation of the JAKSTAT and MAPK pathways.5,6 IFN-λ shows both antiviral and antitumor activities in experimental models: it has been found to inhibit HBV and HCV replication,7 while activation of type III IFN group which consists of IFN-λ1, IFN-λ2, and IFN-λ3 induces apoptosis.8
Genome-wide association studies have shown that polymorphisms near the interleukin-28B (IL28B) gene on chromosome 19, which codes for IFN-λ3, are powerful predictors not only of sustained virologic response to combination therapy with Pegylated interferon (PEG-IFN) and ribavirin but also of spontaneous HCV clearance in patients with chronic hepatitis C infection.9,10 Some studies have also suggested that IL28B polymorphisms are related to serological response to PEGy-lated IFN and to HCV recovery in patients with chronic hepatitis B infection, and that these relationships to HCV recovery are the opposite of those observed with HBV recovery.11,12
Since IL28B polymorphisms affect responses to IFN, and since IFN therapy affects the prognosis of patients chronically infected with HBV or HCV, such polymorphisms may also affect the risk of developing HCC. Thus, several case–control studies have focused on the association of two common polymorphisms near the IL28B gene, rs8099917 and rs12979860, with onset of hepatitis virus-related HCC.13–22 These studies have not provided a clear answer because of limited sample size and heterogeneity in patient ethnicity and HCC etiology.
Therefore, we conducted the present meta-analysis of available data to gain a more reliable picture of associations of polymorphisms rs8099917 and rs12979860 with HCC risk. We also meta-analyzed the data according to whether the patients had HBV- or HCV-related HCC. To the best of our knowledge, this is the first meta-analysis to analyze association of the rs12979860 polymorphism with HCC risk in subgroups of patients with HBV- or HCV-related disease.
Methods
Search strategy
PubMed, EMBASE, Google Scholar, and the Chinese National Knowledge Infrastructure (CNKI) databases were systematically searched to identify clinical and experimental case–control studies of possible associations of IL28B polymorphisms rs8099917 or rs12979860 with risk of hepatitis virus-related HCC published through January 15, 2016. No language restrictions were imposed. The following search terms were used: rs8099917; rs12979860; these two terms in combination with polymorphism, polymorphisms, SNP, variant, variants, variation, genotype, genetic or mutation; and all the earlier terms in combination with hepatocellular carcinoma and liver cancer. Reference lists in identified articles and reviews were also searched manually to identify additional eligible studies.
Inclusion criteria
To be included in our review and meta-analysis, studies should 1) have used a case–control design to assess the association of rs8099917 or rs12979860 polymorphisms with HCC risk in humans, 2) be available as full-text articles and report sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI), and 3) report genotype frequencies. If multiple publications were identified from the same research group and apparently involved the same population, only the most recent study was used in the meta-analysis.
Data extraction
Two authors (YZ and SLZ) independently extracted the following data from included studies: first author’s family name, year of publication, country of origin, testing methods, P for Hardy–Weinberg equilibrium in controls, type of control population, numbers of cases and controls, and frequencies of genotypes in cases and controls. Discrepancies were resolved by consensus. Only those studies that met the predetermined inclusion criteria were included.
Statistical analysis
Following previous approaches,23–25 we calculated the unadjusted OR with 95% CI from the genotype frequencies in cases and controls in order to assess the strength of association of rs8099917 and rs12979860 polymorphisms with HCC risk. The significance of pooled ORs was determined using the Z test, with P<0.05 defined as the significance threshold. Meta-analysis was conducted using a fixed-effect model when P was >0.10 for the Q test, indicating lack of heterogeneity among studies; otherwise, a random-effect model was used. All statistical tests were performed using Review Manager 5.2 (Cochrane Collaboration, London, UK).
Publication bias was assessed using Begg’s funnel plot and Egger’s weighted regression, generated using Stata 12.0 (Stata Corp, College Station, TX, USA). P<0.05 was considered statistically significant.
Results
Description of studies
Figure 1 illustrates the search and selection criteria for studies in the meta-analysis. A total of 368 potentially relevant publications up to January 15, 2016 were systematically identified in the PubMed, EMBASE, Google Scholar, and CNKI databases. Of these, we excluded 349 based on review of the titles and abstracts. The study by He et al was excluded because it was a meta-analysis,26 and three studies were excluded because they did not have a case–control design.27–29 The article by Shi et al was excluded because it did not report precise genotypes,30 and the article by Lee et al was excluded because it did not report usable data.31 Three articles investigating the association between IL28B polymorphism and the combination of HCC and liver cirrhosis were excluded.32–34
Figure 1.
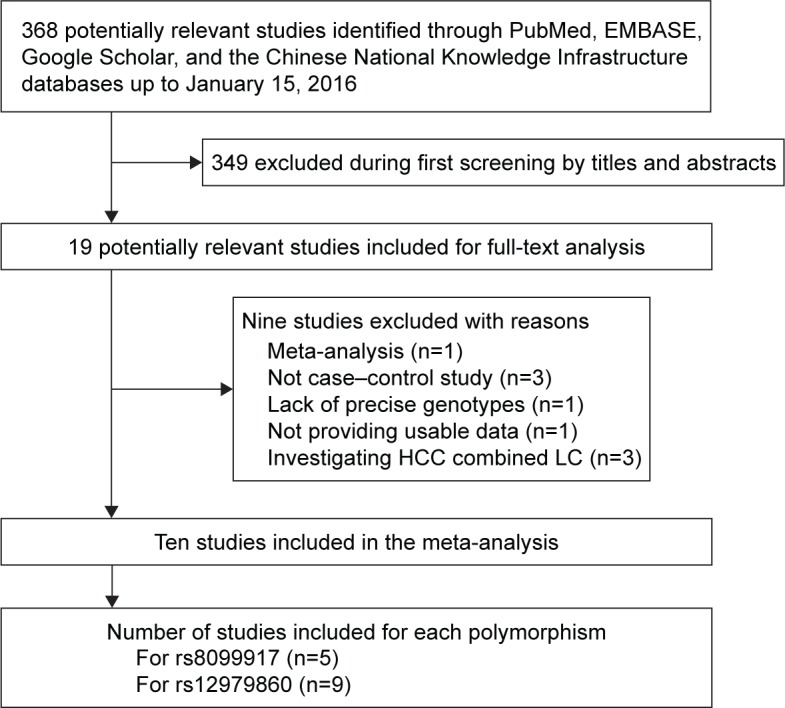
Flowchart of study inclusion.
Abbreviations: HCC, hepatocellular carcinoma; LC, liver cirrhosis.
In the end, ten studies13–22 were included in the meta-analysis (Table 1). Five studies13–17 were conducted in Asian populations (1,041 cases and 907 controls) and evaluated the association between rs8099917 polymorphism and HCC risk. Nine studies,14–22 involving 1,503 cases and 1,505 controls, evaluated the association between rs12979860 polymorphism and HCC risk. The distribution of genotypes in controls was consistent with Hardy–Weinberg equilibrium (P>0.05) in all but two studies.
Table 1.
Characteristics of studies in the meta-analysis
| Author | Year | Country | Methods | P (HWE) | Type of control group | Cases/controls | Case genotype | Control genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs8099917 (1,041 cases and 907 controls) | TT | GT | GG | TT | GT | GG | ||||||
| Jiao et al13 | 2011 | People’s Republic of China | PCR | 0.602 | Healthy | 99/144 | 87 | 12 | 0 | 132 | 12 | 0 |
| Ren et al14 | 2012 | People’s Republic of China | TaqMan | 0.001 | Healthy | 154/47 | 127 | 21 | 6 | 44 | 2 | 1 |
| Chen et al15 | 2012 | People’s Republic of China | Real-time PCR | 0.757 | Healthy | 406/244 | 356 | 48 | 2 | 218 | 25 | 1 |
| Wang et al16 | 2012 | People’s Republic of China | PCR | 0.458 | Healthy | 299/308 | 255 | 41 | 3 | 283 | 25 | 0 |
| Kimkong et al17 | 2015 | Thailand | PCR-RFLP | 0.291 | Patients with self-limiting HBV infection | 83/164 | 72 | 10 | 1 | 139 | 25 | 0 |
| rs12979860 (1,503 cases and 1,505 controls) | CC | CT | TT | CC | CT | TT | ||||||
| Fabris et al18 | 2011 | Italy | PCR-RFLP | 0.386 | Patients with liver cirrhosis | 85/171 | 23 | 50 | 12 | 79 | 78 | 14 |
| Agundez et al19 | 2012 | Spain | Real-time PCR | 0.383 | Patients with CHC virus infection | 134/97 | 52 | 66 | 16 | 37 | 49 | 11 |
| Eurich et al20 | 2012 | Germany | TaqMan | 0.219 | Patients with CHC virus infection | 61/106 | 13 | 31 | 17 | 35 | 57 | 14 |
| Ren et al14 | 2012 | People’s Republic of China | TaqMan | 0.761 | Healthy | 154/47 | 115 | 29 | 10 | 43 | 4 | 0 |
| Chen et al15 | 2012 | People’s Republic of China | Real-time PCR | 0.369 | Healthy | 406/244 | 348 | 53 | 5 | 213 | 29 | 2 |
| Wang et al16 | 2012 | People’s Republic of China | PCR | <0.001 | Healthy | 298/290 | 97 | 198 | 3 | 80 | 210 | 0 |
| Akkiz et al21 | 2014 | Turkey | PCR-RFLP | 0.732 | Healthy | 187/208 | 80 | 83 | 24 | 92 | 91 | 25 |
| De Re et al22 | 2014 | Italy | TaqMan | 0.076 | Healthy | 95/178 | 28 | 49 | 18 | 75 | 89 | 14 |
| Kimkong et al17 | 2015 | Thailand | PCR-RFLP | 0.312 | Patients with self-limiting HBV infection | 83/164 | 72 | 10 | 1 | 140 | 24 | 0 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; HBV, hepatitis B virus; CHC, chronic hepatitis C.
Meta-analysis of the association between rs8099917 polymorphism and HCC risk
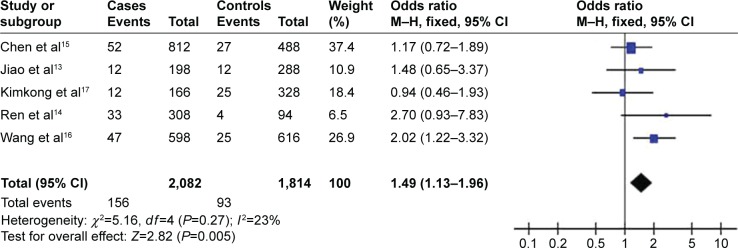
Meta-analyses of the possible association between rs8099917 polymorphism and risk of HCC are summarized in Table 1; the overall results are summarized in Table 2. All 1,041 cases in the five studies examining rs8099917 polymorphism13–17 had HBV-related HCC. Fixed-effect meta-analysis of the total population showed the rs8099917 polymorphism to be significantly associated with increased risk of HBV-related HCC according to the allelic model (OR 1.49, 95% CI 1.13–1.96, P=0.005; Figure 2) and heterozygous model (OR 1.39, 95% CI 1.04–1.88, P=0.03). These results suggest that the G-allele of rs8099917 may increase susceptibility to HBV-related HCC. In contrast, fixed-effect meta-analysis using the dominant model suggested a protective role of the wild-type TT genotype of rs8099917 in patients with HBV-related HCC (OR 0.68, 95% CI 0.51–0.91, P=0.01).
Table 2.
Overall meta-analysis of the association of rs8099917GT and rs12979860C/T polymorphisms with risk of hepatocellular carcinoma
| Polymorphism | Allelic model
|
Recessive model
|
Dominant model
|
Homozygous model
|
Heterozygous model
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | Z (P-value) | I2 (%) | OR | 95% CI | Z (P-value) | I2(%) | OR | 95% CI | Z (P-value) | I2 (%) | OR | 95% CI | Z (P-value) | I2 (%) | OR | 95% CI | Z (P-value) | I2 (%) | |
| rs8099917 | G versus T | GG versus GT + | TT | TT versus GT | + GG | GG versus TT | GT versus TT | |||||||||||||
| Overall | 1.49 | 1.13–1.96 | 2.82 (0.005) | 23 | 2.76 | 0.78–9.80 | 1.57 (0.12) | 0 | 0.68 | 0.51–0.91 | 2.56 (0.01) | 24 | 2.91 | 0.82–10.30 | 1.65 (0.10) | 0 | 1.41 | 1.04–1.89 | 2.24 (0.02) | 23 |
| Ethnicity | ||||||||||||||||||||
| Chinese | 1.61 | 1.19–2.18 | 3.08 (0.002) | 13 | 3.61 | 0.78–16.67 | 1.65 (0.10) | 0 | 0.61 | 0.43–0.87 | 2.70 (0.007) | 31 | 3.84 | 0.83–17.71 | 1.72 (0.09) | 0 | 1.54 | 1.07–2.22 | 2.32 (0.02) | 35 |
| rs12979860 | T versus C | TT versus CT + | CC | CC versus CT | + TT | TT versus CC | CT versus CC | |||||||||||||
| Overall | 1.36 | 1.05–1.78 | 2.29 (0.02) | 74 | 1.75 | 1.28–2.39 | 3.48 (0.005) | 6 | 0.77 | 0.58–1.03 | 1.76 (0.08) | 59 | 1.99 | 1.41–2.80 | 3.95 (<0.001) | 26 | 1.12 | 0.94–1.34 | 1.28 (0.20) | 43 |
| Ethnicity | ||||||||||||||||||||
| Caucasian | 1.47 | 1.05–2.06 | 2.27 (0.02) | 75 | 1.62 | 1.16–2.25 | 2.87 (0.004) | 34 | 0.69 | 0.49–0.97 | 2.14 (0.03) | 49 | 1.84 | 1.29–2.64 | 3.34 (<0.001) | 55 | 1.30 | 1.02–1.66 | 2.13 (0.03) | 28 |
| Asian | 1.20 | 0.78–1.86 | 0.84 (0.40) | 64 | 3.51 | 1.09–11.23 | 2.11 (0.03) | 0 | 0.90 | 0.56–1.44 | 0.44 (0.66) | 60 | 3.54 | 1.11–11.34 | 2.13 (0.03) | 0 | 0.95 | 0.73–1.23 | 0.41 (0.69) | 43 |
| Chinese | 1.32 | 0.75–2.34 | 0.97 (0.33) | 0 | 3.28 | 0.95–11.36 | 1.88 (0.06) | 0 | 0.81 | 0.44–1.52 | 0.65 (0.52) | 73 | 3.33 | 0.96–11.53 | 1.90 (0.06) | 0 | 1.09 | 0.65–1.84 | 0.33 (0.74) | 61 |
| Etiology | ||||||||||||||||||||
| HCV-related HCC | 1.44 | 1.03–2.10 | 2.15 (0.03) | 66 | 1.82 | 1.04–3.20 | 2.09 (0.04) | 52 | 0.64 | 0.42–0.99 | 1.99 (0.05) | 55 | 2.26 | 1.04–4.92 | 2.05 (0.04) | 68 | 1.37 | 1.03–1.84 | 2.15 (0.03) | 30 |
| HBV-related HCC | 1.16 | 0.86–1.57 | 0.99 (0.32) | 56 | 1.80 | 1.03–3.15 | 2.08 (0.04) | 0 | 0.90 | 0.63–1.28 | 0.59 (0.55) | 52 | 1.97 | 1.09–3.56 | 2.26 (0.02) | 0 | 0.99 | 0.79–1.25 | 0.07 (0.94) | 33 |
Abbreviations: OR, odds ratio; CI, confidence interval; HCV, hepatitis C virus; HCC, hepatocellular carcinoma; HBV, hepatitis B virus.
Figure 2.
Forest plot describing the association between rs8099917 polymorphism and hepatocellular carcinoma risk (G-allele versus T-allele).
Abbreviations: CI, confidence interval; M–H, Mantel–Haensze.
Similar results, with even higher statistical significance, were observed when we meta-analyzed the subgroup of studies involving Chinese populations (958 cases and 743 controls): the OR and 95% CI were 1.61 and 1.19–2.18 for the allelic model (P=0.002), 1.54 and 1.07–2.22 for the heterozygous model (P=0.02), and 0.61 and 0.43–0.87 for the dominant model (P=0.07), respectively. This suggests an association between rs8099917 polymorphism and HBV-related HCC risk in Chinese population. Meta-analysis results for the rs8099917 polymorphism in Chinese population are summarized in Table 2.
Meta-analysis of the association between rs12979860 polymorphism and HCC risk
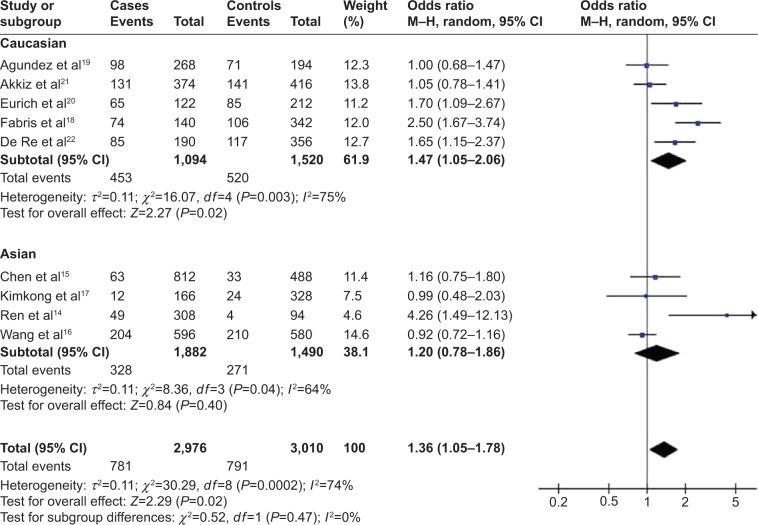
Meta-analyses of the possible association between rs12979860 polymorphism and HCC risk are summarized in Table 1, and overall results are summarized in Table 2. In the total population of 1,503 cases and 1,505 controls, rs12979860 polymorphism showed a significant relationship with HCC in the allelic model (OR 1.36, 95% CI 1.05–1.78, P=0.02; Figure 3), recessive model (OR 1.75, 95% CI 1.28–2.39, P=0.005), and homozygous model (OR 1.99, 95% CI 1.41–2.80, P<0.001).
Figure 3.
Forest plot describing the association between rs12979860 polymorphism and hepatocellular carcinoma risk (T-allele versus C-allele).
Abbreviation: CI, confidence interval.
In the Caucasian subgroup of 562 cases and 760 controls, rs12979860 polymorphism was associated with HCC risk in all five genetic models: allelic, OR 1.47, 95% CI 1.05–2.06, P=0.02 (Figure 3); recessive, OR 1.62, 95% CI 1.16–2.25, P=0.004; dominant, OR 0.69, 95% CI 0.49–0.97, P=0.03; homozygous, OR 1.84, 95% CI 1.29–2.64, P<0.001; and heterozygous, OR 1.30, 95% CI 1.02–1.66, P=0.03. These results suggest that the T-allele of rs12979860 may be a genetic risk factor for HCC in Caucasians. In contrast, in the Asian subgroup of 941 cases and 745 controls, rs12979860 polymorphism was significantly related to HCC risk only in the recessive model (OR 3.51, 95% CI 1.09–11.23, P=0.03) and homozygous model (OR 3.54, 95% CI 1.11–11.34, P=0.03). These results suggest that the TT genotype of rs12979860 may be a genetic risk factor for HCC in Asians. Meta-analyses of association of rs12979860 polymorphism with HCC risk in different ethnic subgroups are summarized in Table 2.
A subset of studies that clearly reported whether their cases had HBV- or HCV-related HCC were meta-analyzed in order to examine whether the association between rs12979860 polymorphism and HCC risk depends on etiology. These included two studies on Caucasians involving 272 cases with HBV- or HCV-related disease,18,21 a study by Fabris et al involving 35 HCV-related and 50 undefined cases,18 and a study by Akkiz et al involving 24 HCV-related, 110 HBV-related, and 49 undefined cases.21 Meta-analysis of data for each type of HCC showed that the rs12979860 polymorphism was associated with risk of HCV-related HCC according to four genetic models (Tables 2 and 3): allelic, OR 1.44, 95% CI 1.03–2.10, P=0.03; recessive, OR 1.82, 95% CI 1.04–3.20, P=0.04; homozygous, OR 2.26, 95% CI 1.04–4.92, P=0.04; and heterozygous, OR 1.37, 95% CI 1.03–1.84, P=0.03. These results suggest that the T-allele of rs12979860 may increase risk of HCV-related HCC. In contrast, rs12979860 polymorphism was associated with risk of HBV-related HCC in the recessive model (OR 1.80, 95% CI 1.03–3.15, P=0.04) and homozygous model (OR 1.97, 95% CI 1.09–3.56, P=0.02). These results suggest that the TT genotype of rs12979860 may increase risk of HBV-related disease.
Table 3.
Genotype frequencies of the rs12979860 polymorphism according to etiology of hepatocellular carcinoma
| Author | Year | Country | Method | P (HWE) | Type of control | Cases/controls | Case genotype | Control genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCV-related HCC (374 cases and 760 controls) | CC | CT | TT | CC | CT | TT | ||||||
| Fabris et al18 | 2011 | Italy | PCR-RFLP | 0.386 | Patients with liver cirrhosis | 35/171 | 6 | 21 | 8 | 79 | 78 | 14 |
| Agundez et al19 | 2012 | Spain | PCR-RFLP | 0.383 | Patients with CHC virus infection | 134/97 | 52 | 66 | 16 | 37 | 49 | 11 |
| Eurich et al20 | 2012 | Germany | PCR-RFLP | 0.219 | Patients with CHC virus infection | 61/106 | 13 | 31 | 17 | 35 | 57 | 14 |
| Akkiz et al21 | 2014 | Turkey | PCR-RFLP | 0.732 | Healthy | 49/208 | 21 | 24 | 4 | 92 | 91 | 25 |
| De Re et al22 | 2014 | Italy | TaqMan | 0.076 | Healthy | 95/178 | 28 | 49 | 18 | 75 | 89 | 14 |
| HBV-related HCC (1,051 cases and 953 controls) | CC | CT | TT | CC | CT | TT | ||||||
| Ren et al14 | 2012 | People’s Republic of China | TaqMan | 0.006 | Healthy | 154/47 | 115 | 29 | 10 | 43 | 4 | 0 |
| Chen et al15 | 2012 | People’s Republic of China | Real-time PCR | 0.369 | Healthy | 406/244 | 356 | 48 | 2 | 218 | 25 | 1 |
| Wang et al16 | 2012 | People’s Republic of China | MALDI-TOF MS | <0.001 | Healthy | 298/290 | 97 | 198 | 3 | 80 | 210 | 0 |
| Akkiz et al21 | 2014 | Turkey | PCR-RFLP | 0.732 | Healthy | 110/208 | 43 | 50 | 17 | 92 | 91 | 25 |
| Kimkong et al17 | 2015 | Thailand | PCR-RFLP | 0.312 | Patients with self-limiting HBV infection | 83/164 | 72 | 10 | 1 | 140 | 24 | 0 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; HCV, hepatitis C virus; HCC, hepatocellular carcinoma; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; CHC, chronic hepatitis C; HBV, hepatitis B virus; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
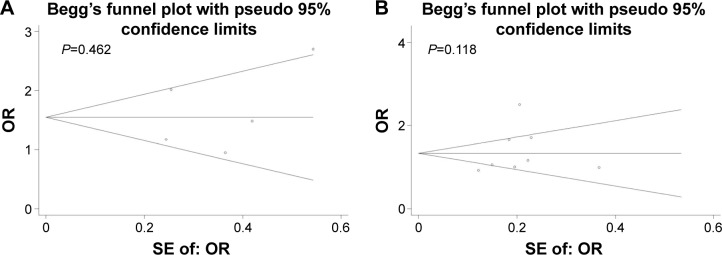
Publication bias
Begg’s funnel plot and Egger’s test were performed to detect potential publication bias in the meta-analysis. No obvious asymmetry was observed in Begg’s funnel plots for rs8099917 (Figure 4A) or rs12979860 (Figure 4B) according to the allelic model. Similarly, Egger’s tests showed no significant risk of publication bias in the allelic model for rs8099917 (G-allele versus T-allele: t=0.51, P=0.647) or rs12979860 (T-allele versus C-allele: t=2.07, P=0.077).
Figure 4.
Begg’s funnel plots according to allelic models.
Notes: Begg’s funnel plots to detect publication bias in analysis of the association of (A) rs8099917 polymorphism and hepatocellular carcinoma risk according to the allelic model (G-allele versus T-allele), or the association of (B) rs12979860 polymorphism and hepatocellular carcinoma risk according to the allelic model (T-allele versus C-allele).
Abbreviations: OR, odds ratio; SE, standard error.
Discussion
Few meta-analyses have focused on the association between rs8099917 polymorphism and risk of developing HCC, and individual studies have failed to reach a consensus.13–17 Most of those studies suggest that rs8099917 polymorphism does not affect risk of HBV-related HCC,13–15,17 but the present meta-analysis suggests that the G-allele of rs8099917 may be associated with increased risk, and that the wild-type TT genotype may protect against the disease. Similar results were observed specifically in Chinese population. It is worth noting that all five previous studies,13–17 involving 1,041 cases and 907 controls, were conducted on Asian populations in which all cases had HBV-related HCC.
Two previous meta-analyses were published reporting that rs12979860 polymorphism is a genetic risk factor for hepatitis virus-related HCC.26,35 These results should be interpreted with caution, since the meta-analyses included 118 cases from a case–control study in which the patients appeared to have had HCC and liver cirrhosis and to have been in end-stage liver disease.32 This may make the meta-analysis results less accurate because genetic factors affect risk of HCC and liver cirrhosis differently. In addition, these meta-analyses did not include four recent case–control studies.15,17,21,22 The present meta-analysis not only included these four studies but also excluded the study by El-Awady et al.32
Our meta-analysis of all included patients suggests that the T-allele of rs12979860 significantly increases risk of HCC. The T-allele also seems to increase risk of HCC in the subset of Caucasians, especially HCV-related HCC. Although we failed to detect a significant association between the T-allele and risk of HBV-related HCC in the allelic model, the TT genotype of rs12979860 was associated with elevated risk of HBV-related disease across all patients and in the subgroup of Asians. Therefore, our meta-analysis provides new evidence of the important role of IL28B rs12979860 polymorphism in hepatitis virus-related carcinogenesis.
The findings of this systematic review are limited by the designs of the included studies. First, the controls in four case–control studies were not healthy individuals, which increases the risk of heterogeneity because not all controls were chosen according to the same well-defined criteria. Second, the P-value associated with Hardy–Weinberg equilibrium was <0.05 in two included studies14,16 (Table 1), suggesting that these study populations were not representative of the broader target population. Third, the results may be affected by additional confounding factors, such as tumor status, sex, or age, but most studies either did not report these baseline data or aggregated the data in different ways, preventing their inclusion in the meta-analysis. Fourth, the studies used various assay methods to characterize the polymorphisms; these methods differ in sensitivity and specificity, which may affect the reliability of the results. Lastly, the fact that all five studies of the rs8099917 polymorphism in our meta-analysis13–17 examined Asian populations highlights the need for large, well-designed studies in Caucasian and African populations to verify our findings.
Conclusion
The G-allele of rs8099917 may be associated with elevated risk of HBV-related HCC, while the wild-type TT genotype of rs8099917 may protect against this disease. The T-allele of rs12979860 may increase risk of HCC in Caucasians, especially HCV-related HCC. The TT genotype of rs12979860 may increase risk of HBV-related HCC, especially in Asians. Further detailed and well-designed studies involving larger, multiethnic samples are needed to clarify the influence of those two polymorphisms on HCC risk. Such studies should also explore gene–gene and gene–environment interactions that may mediate an association of rs809991 and rs12979860 with risk of developing
Acknowledgments
This research was supported by the National Science and Technology Major Special Project (2012ZX10002010001009), the Self-Raised Scientific Research Fund of the Ministry of Health of Guangxi Province (Z2015621 and Z2014241), and the Guangxi University of Science and Technology Research Fund (KY2015LX056).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard MJ, Navas-Martin S. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett. 2011;305:123–143. doi: 10.1016/j.canlet.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S, Yu M, Lee C, et al. Interferon therapy in HBeAg positive chronic hepatitis reduces progression to cirrhosis and hepatocellular carcinoma. J Hepatol. 2007;46:45–52. doi: 10.1016/j.jhep.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 4.Bruno S, Stroffolini T, Colombo M, et al. Sustained virological response to interferon-α is associated with improved outcome in HCV-related cirrhosis: a retrospective study. Hepatology. 2007;45:579–587. doi: 10.1002/hep.21492. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Liu X, Zhou Y, et al. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009;86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 7.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumoutier L, Tounsi A, Michiels T, et al. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem. 2004;279:32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 9.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Jilg N, Shao R, et al. IL28B inhibits hepatitis C virus replication through the JAK–STAT pathway. J Hepatol. 2011;55:289–298. doi: 10.1016/j.jhep.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonneveld MJ, Wong VWS, Woltman AM, et al. Polymorphisms near IL28B and serologic response to peginterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology. 2012;142:513–520. doi: 10.1053/j.gastro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Kim SU, Song KJ, Chang HY, et al. Association between IL28B polymorphisms and spontaneous clearance of hepatitis B virus infection. PLoS One. 2013;8:e69166. doi: 10.1371/journal.pone.0069166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao XL, Gao YT, Jing L, et al. Studies on the relationship between polymorphism of IL-28B rs8099917 and the outcome of HBV infection. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:1143–1147. Chinese. [PubMed] [Google Scholar]
- 14.Ren S, Lu J, Du X, et al. Genetic variation in IL28B is associated with the development of hepatitis B-related hepatocellular carcinoma. Cancer Immunol Immun. 2012;61:1433–1439. doi: 10.1007/s00262-012-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Wang L, Li Y, et al. Association analysis between SNPs in IL-28B gene and the progress of hepatitis B infection in Han Chinese. PLoS One. 2012;7:e50787. doi: 10.1371/journal.pone.0050787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhang HH, Chen YH, et al. Correlation between interleukin-28B genetic polymorphisms and primary hepatocellular carcinoma. Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46:527–532. Chinese. [PubMed] [Google Scholar]
- 17.Kimkong I, Chankaew J, Kunanopparat A, et al. Gene polymorphisms of interleukin 28B and the risk to chronic hepatitis B virus infection in Thai. Tissue Antigens. 2015;85:177–181. doi: 10.1111/tan.12517. [DOI] [PubMed] [Google Scholar]
- 18.Fabris C, Falleti E, Cussigh A, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54:716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Agundez JA, Garcia-Martin E, Maestro ML, et al. Relation of IL28B gene polymorphism with biochemical and histological features in hepatitis C virus-induced liver disease. PLoS One. 2012;7:e37998. doi: 10.1371/journal.pone.0037998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eurich D, Boas-Knoop S, Bahra M, et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation. 2012;93:644–649. doi: 10.1097/TP.0b013e318244f774. [DOI] [PubMed] [Google Scholar]
- 21.Akkiz H, Kuran S, Akgöllü E, et al. The role of interleukin 28B gene polymorphism in Turkish patients with hepatocellular carcinoma. Ann Hepatol. 2014;13:788–795. [PubMed] [Google Scholar]
- 22.De Re V, Gragnani L, Fognani E, et al. Impact of immunogenetic IL28B polymorphism on natural outcome of HCV infection. Biomed Res Int. 2014;2014:710642. doi: 10.1155/2014/710642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong JH, You XM, Gong WF, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7:e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong JH, Xiang BD, Ma L, et al. Meta-analysis of microsomal epoxide hydrolase gene polymorphism and risk of hepatocellular carcinoma. PLoS One. 2013;8:e57064. doi: 10.1371/journal.pone.0057064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong JH, Zhang ZM, Li LQ. mEH Tyr113His polymorphism and the risk of ovarian cancer development. J Ovarian Res. 2010;6:40. doi: 10.1186/1757-2215-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Yu G, Li Z, et al. Influence of interleukin-28B polymorphism on progression to hepatitis virus-induced hepatocellular carcinoma. Tumor Biol. 2014;35:8757–8763. doi: 10.1007/s13277-014-2142-3. [DOI] [PubMed] [Google Scholar]
- 27.Sticchi L, Di Biagio A, Rappazzo E, et al. Rs12979860 and rs8099917 single nucleotide polymorphisms of interleukin-28B gene: simultaneous genotyping in Caucasian patients infected with hepatitis C virus. J Prev Med Hyg. 2015;54:83–86. [PMC free article] [PubMed] [Google Scholar]
- 28.Hodo Y, Honda M, Tanaka A, et al. Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin Cancer Res. 2013;19:1827–1837. doi: 10.1158/1078-0432.CCR-12-1641. [DOI] [PubMed] [Google Scholar]
- 29.Asahina Y, Tsuchiya K, Nishimura T, et al. Genetic variation near interleukin 28B and the risk of hepatocellular carcinoma in patients with chronic hepatitis C. J Gastroenterol. 2014;49:1152–1162. doi: 10.1007/s00535-013-0858-2. [DOI] [PubMed] [Google Scholar]
- 30.Shi X, Chi X, Pan Y, et al. IL28B is associated with outcomes of chronic HBV infection. Yonsei Med J. 2015;56:625–633. doi: 10.3349/ymj.2015.56.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DH, Cho Y, Seo JY, et al. Polymorphisms near interleukin 28B gene are not associated with hepatitis B virus clearance, hepatitis B e antigen clearance and hepatocellular carcinoma occurrence. Intervirology. 2013;56:84–90. doi: 10.1159/000342526. [DOI] [PubMed] [Google Scholar]
- 32.El-Awady MK, Mostafa L, Tabll AA, et al. Association of IL28B SNP with progression of Egyptian HCV genotype 4 patients to end stage liver disease. Hepat Mon. 2012;12:271. doi: 10.5812/hepatmon.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al Qahtani AA, Al Anazi MR, Abdo AA, et al. Genetic variation in interleukin 28B and correlation with chronic hepatitis B virus infection in Saudi Arabian patients. Liver Int. 2014;34:e208–e216. doi: 10.1111/liv.12347. [DOI] [PubMed] [Google Scholar]
- 34.Kil H, Jeong S, Kim J, et al. Role of interleukin-28B genetic polymorphisms in Korean patients with hepatitis C virus infection. Gut Liver. 2014;8:70–78. doi: 10.5009/gnl.2014.8.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suo GJ, Zhao ZX. Association of the interleukin-28B gene polymorphism with development of hepatitis virus-related hepatocellular carcinoma and liver cirrhosis: a meta-analysis. Genet Mol Res. 2013;12:3708–3717. doi: 10.4238/2013.September.19.1. [DOI] [PubMed] [Google Scholar]