Abstract
Expression of the oncogenic transcription factor MYC is disproportionately elevated in triple-negative breast cancer (TNBC) compared to estrogen, progesterone and human epidermal growth factor 2 receptor-positive (RP) breast tumors1,2. We and others have shown that MYC alters metabolism during tumorigenesis3,4. However, the role of MYC in TNBC metabolism remains largely unexplored. We hypothesized that MYC-dependent metabolic dysregulation is essential for MYC-overexpressing (MO) TNBC and may thus identify novel therapeutic targets for this clinically challenging subset of breast cancer. Using a targeted metabolomics approach, we identified fatty acid oxidation (FAO) intermediates as being dramatically upregulated in a MYC-driven model of TNBC. A lipid metabolism gene signature was identified in patients with TNBC from The Cancer Genome Atlas (TCGA) database and multiple other clinical datasets, implicating FAO as a dysregulated pathway critical for TNBC metabolism. We find that MO-TNBC displays increased bioenergetic reliance upon fatty acid oxidation (FAO), and that pharmacologic inhibition of FAO catastrophically decreases energy metabolism of MO-TNBC, blocks growth of a MYC-driven transgenic TNBC model and that of MO-TNBC patient-derived xenografts. Our results demonstrate that inhibition of FAO is a novel therapeutic strategy against MO-TNBC.
We hypothesized that MYC-dependent metabolic dysregulation is essential for MO-TNBC. To test this hypothesis, we investigated the metabolism of a conditional doxycycline-inducible transgenic model of MO-TNBC (MTB-TOM)5,6. We utilized mass spectrometry to compare the global metabolic profile of MTB-TOM tumors to naïve transgenic mammary glands in which MYC was not induced. Metabolite analysis revealed a number of dysregulated pathways in MTB-TOM tumors commonly associated with tumorigenesis including glycolysis, the tricarboxylic acid (TCA) cycle and fatty acid metabolism (Fig. 1a, Supplementary Table 1 and Fig. 1). Although fatty acid synthesis (FAS) is upregulated in many types of cancer, a MYC-dependent role for FAO in breast tumorigenesis has not been described4,7,8. Considering the proximity of primary breast tumors to the adipose-rich mammary gland, we chose to focus on the dysregulation of FAO in MO-TNBC.
Figure 1.
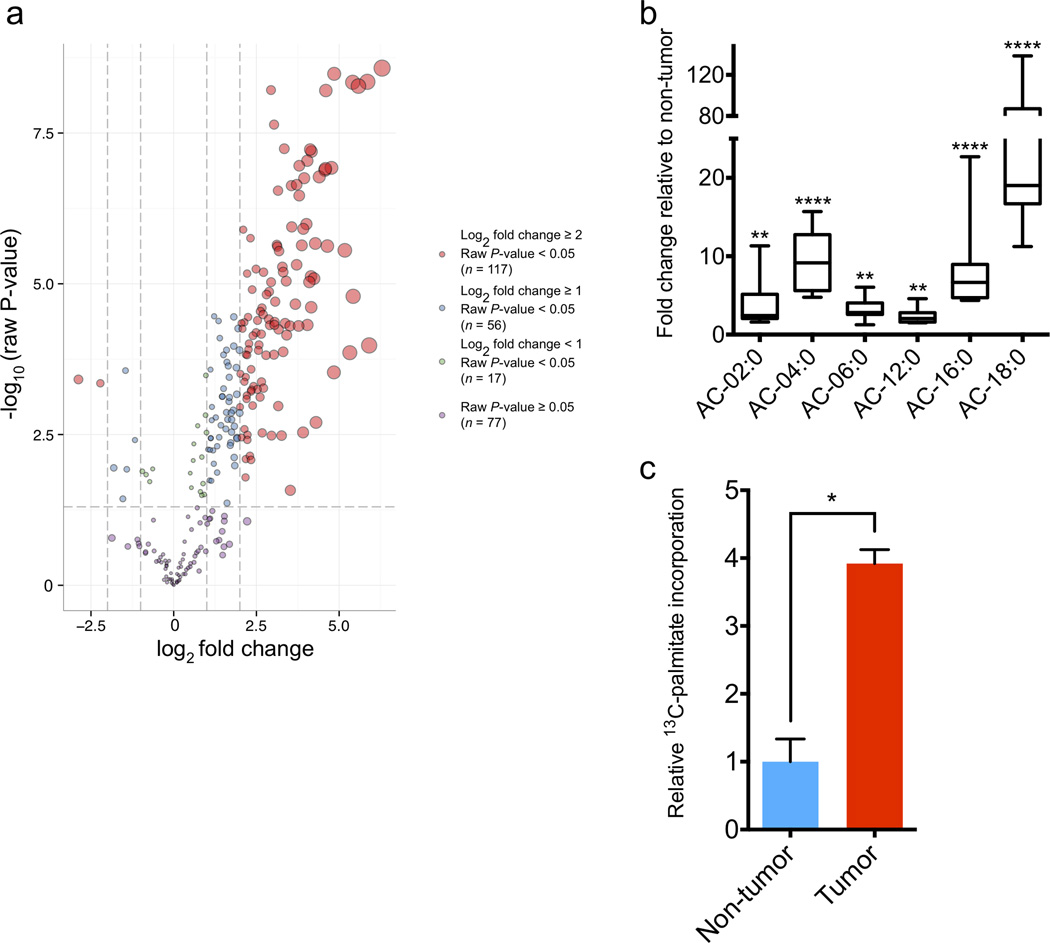
MTB-TOM tumors display dysregulated FAO. (a) Volcano plot of dysregulated metabolites in MTB-TOM tumors compared to non-tumor mammary glands. Values shown are log fold change of metabolites from seven tumors from five induced mice compared to those from five mammary glands from four uninduced mice. (b) Fold-change in AC levels in MTB-TOM tumors versus non-tumor mammary glands. Values are shown as min-to-max box plots of same mice as in (a). (c) Carbon flux analysis showing 13C-palmitoyl-carnitine production from 13C-palmitate in MTB-TOM orthotopic transplants compared to contralateral non-tumor mammary gland. A two-tailed unpaired t-test was used to compare non-tumor to tumor. Values shown are mean ± s.e.m. of four mice. All differential metabolite abundance analyses were performed using the limma R package (a, b). *P < 0.05, **P < 0.01, ****P < 0.0001
To investigate the role of MYC in TNBC FAO upregulation, we returned to our MTB-TOM metabolomic data (Supplementary Table 1). Acyl-carnitines (AC) are an essential intermediate and the first committed step of FAO9. To catabolize long-chain fatty acids such as palmitate to acetyl-CoA, a major fuel for biosynthetic and bioenergetic metabolism, acyl-CoAs must be converted to acyl-carnitines across the outer mitochondrial membrane by carnitine-palmitoyl transferase 1 (CPT1)9. Within our metabolomic data, we found that all six acyl-carnitine (AC) intermediates detected were significantly elevated in MTB-TOM tumors compared to control tissue (Fig. 1b). To further validate our steady-state findings in vivo, we assayed AC production in MTB-TOM tumors 4 hours (h) following intraperitoneal (IP) injection of 13C-palmitate into tumor-bearing mice. We found that tumor tissue had a four-fold increase in 13C-palmitoyl-carnitine production compared to control mammary gland (Fig. 1c). This experiment was not normalized by overall uptake, and thus, may reflect a relative increase in FA uptake, FA utilization via oxidation or both. Nevertheless, these data overall indicate that the first committed step of FAO is upregulated in MO-TNBC.
To determine how FAO is altered in TNBC, we analyzed RNA-expression data from primary human tumors (TCGA, 771 patients). Of 336 genes associated with fatty acid metabolism by the Gene Ontology database (GO:0006631), we found that 244 (73%) were significantly dysregulated (FDR < 0.05) in TN versus RP tumors (Supplementary Table 2). We found that TN tumors displayed upregulation of many activators of FAO, including the master transcriptional regulator PGC1α, and downregulation of many activators of fatty acid synthesis (FAS), including FASN and ACACB (ACC2) (Supplementary Table 2). We applied the fatty acid metabolism signature identified from the TCGA samples (Fig. 2a) to four additional breast cancer clinical cohorts (2,119 total patients including TCGA) and confirmed that it was highly correlated with TN tumor samples (Supplementary Fig. 2 and Table 3)10–13. We confirmed upregulation of key FAO activators and downregulation of key fatty acid synthesis activators at the protein level in TN versus RP human breast cancer cell lines (Fig. 2b)4,14,15. Interestingly, while FASN and ACC2 were markedly downregulated in TN cell lines compared to RP cells, ACC1 is expressed in both TN and RP cell lines. To directly test if FAO was increased in MYChigh TNBC we measured 14C-oleic acid conversion to 14CO2. We found a ~50% increase in FAO in TN MYChigh cells compared to TN MYClow and RP cells (Fig. 2c). Taken together these data suggest FAO is upregulated in MYChigh TNBC, while FAS, if maintained, may occur preferentially through ACC1 activity rather than ACC2.
Figure 2.
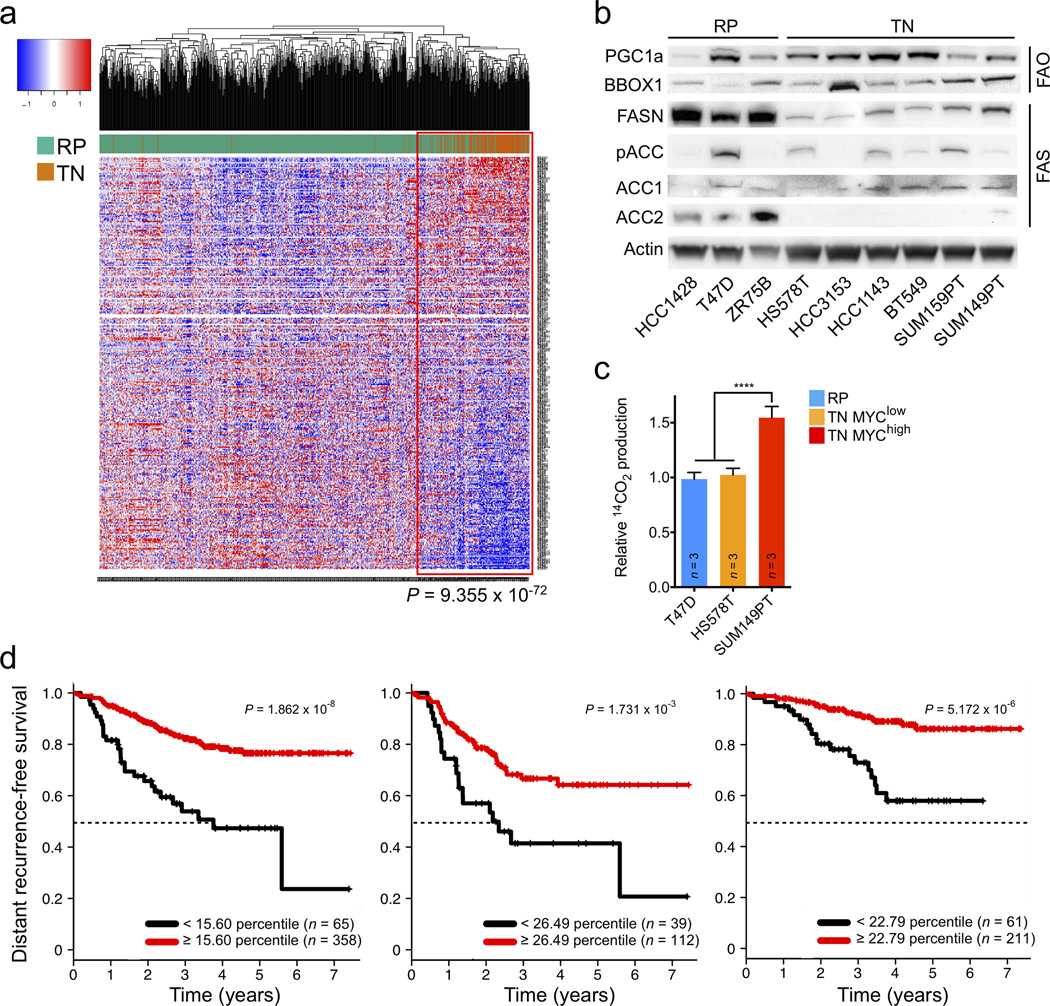
Human TNBC displays dysregulated FAO. (a) Hierarchical clustering of TCGA RNASeq samples of 771 breast cancer patients for 336 fatty acid metabolism genes in TN and RP tumors. Individual gene expression is row-normalized from −1 (blue) to 1 (red). A Fisher’s exact test was used to calculate indicated P-value demonstrating significant enrichment of TN tumors (116/123) in the clade outlined in red. (b) Immunoblot analysis showing expression levels of FAO activators PGC-1α and BBOX1, and fatty acid synthesis enzymes FASN, ACC1, ACC2 and pACC1/2 in a panel of TN and RP human cell lines. (f) Kaplan-Meier graphs of all (left), TN (middle) and RP (right) patients, dichotomized by ACACB (ACC2) mRNA expression at an optimal threshold, from a pooled neoadjuvant chemotherapy treated cohort. Samples with decreased ACACB expression are represented with black lines. Median survival time (MST) is indicated with black dotted lines. For all tumors ACACBlow MST = 3.76 years. For TN tumors ACACBlow MST = 2.18 years. MST not reached in any other group. A log-rank test was used to calculate P-values. All differential gene expression analyses were performed using the limma R package (a).
TN is the most aggressive subtype of breast cancer and is characterized by poor clinical outcome1,2. To determine if FAO gene expression is associated with prognosis, we performed univariate analysis of the 336 fatty acid metabolism genes on a patient cohort with long-term distant recurrence-free survival data (Supplementary Table 4)11. We found that decreased ACACB (ACC2), a critical FAS gene, that produces malonyl-CoA to directly inhibit CPT1 and therefore FAO, was associated with worse prognosis in all patients, as well as within a TN cohort (Fig. 2d)9. While diminished ACACB was also associated with worse outcome for patients with RP tumors, median distant recurrence-free survival was not reached in this cohort (Fig. 2d). These data suggest that decreased ACC2 and thus increased FAO directly contribute to the aggressiveness of breast tumors, with worst outcome found amongst the TN cohort.
FAO is the primary bioenergetic pathway in many non-tumor tissues9. We therefore investigated if this pathway is essential for energy production in MO-TNBC. We used etomoxir, a clinically tested specific inhibitor of CPT116, to determine the effects of FAO inhibition on ATP or NAD(P)H production. We first examined nine human breast cancer cell lines (six TN, three RP) with various levels of MYC expression15. Etomoxir treatment dramatically inhibited ATP production in TNBC cell lines expressing high MYC, while low MYC TN and RP cell lines were significantly less affected (Fig. 3a,f). Analysis of proliferation and apoptosis revealed that etomoxir-treated MYChigh cells displayed decreased proliferation without a decrease in viability (Supplementary Fig. 3).
Figure 3.
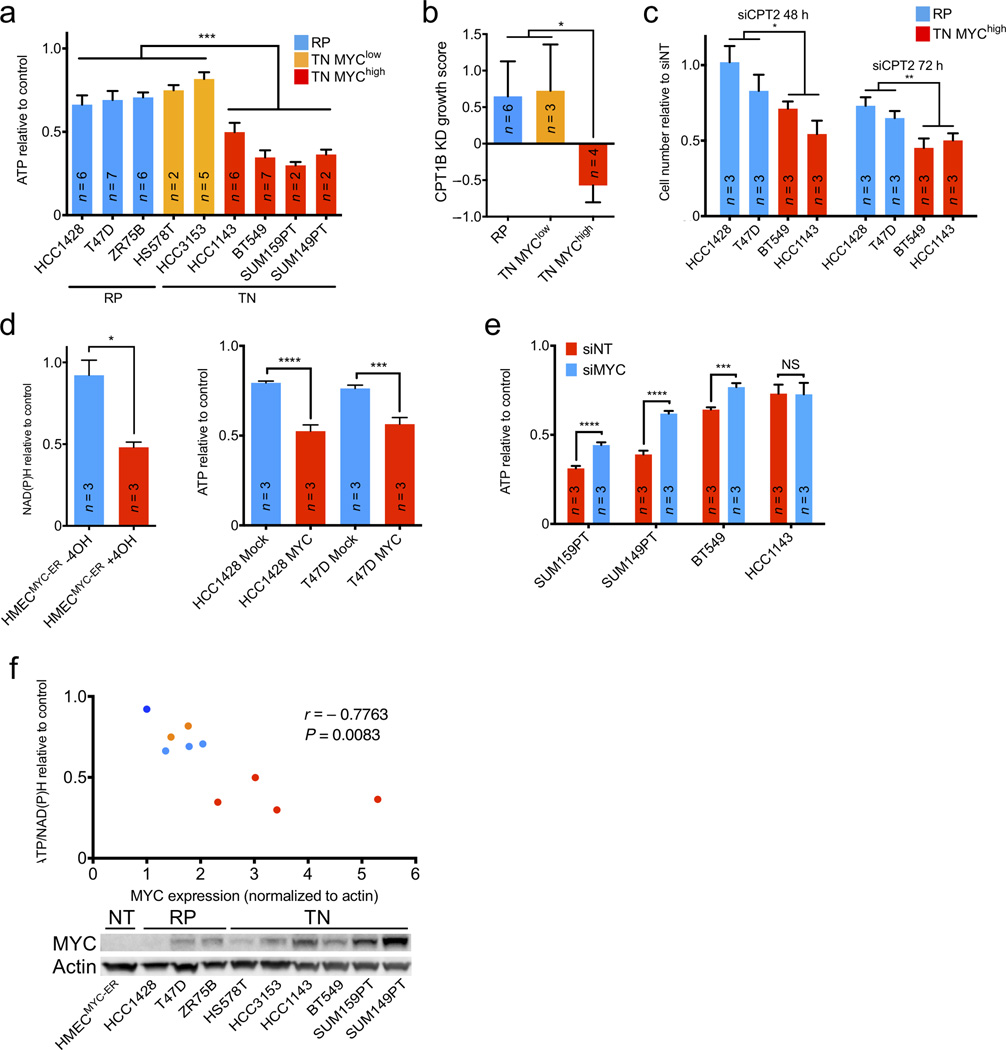
FAO inhibition shows MYC-dependent bioenergetic effects in vitro. (a) ATP response of TN MYChigh versus TN MYClow and RP cells untreated or treated with 200 µM etomoxir for 48 h. (b) Growth response of an independent panel of TN MYChigh versus TN MYClow and RP cells treated with a pool of shRNAs targeting CPT1. (c) Proliferation response of TN versus RP cells to siRNA-mediated CPT2 knockdown after 48 and 72 h. (d) Fatty acid oxidation in TN MYChigh versus TN MYClow and RP cells. Relative 14CO2 production was normalized per cell line to total protein levels. (e) Left, NAD(P)H response of HMECMYC-ER with or without MYC activation for 48 h, then untreated or treated with 200 µM etomoxir for 24 h. Right, ATP response of RP cells with or without MYC overexpression untreated or treated with 200 µM etomoxir for 48 h. (f) ATP response of TN MYChigh cells with or without siRNA-mediated MYC knockdown untreated or treated with 200 µM etomoxir for 48 h. (g) Above, correlation of MYC protein expression and mean ATP/NAD(P)H response to etomoxir of TN and RP cells in (a) and HMECMYC-ER cells without MYC in (b). Pearson correlation and two-tailed t-test were used to generate correlation coefficient and associated P-value. Coloring same as (a) except HMECMYC-ER (dark blue). Below, immunoblot analysis showing MYC protein levels in indicated cell lines. A two-tailed unpaired t-test was used to compare experimental groups (a–f). Values shown are mean ± s.e.m. from triplicate samples (a, d–f), indicated number of cell lines (b), or three biological replicates (c). Number of biological replicates is indicated (a, d–f). *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
To validate the requirement for FAO in MO-TNBC with an orthogonal approach, we extracted data specific to the knockdown of CPT1 (isoform B) from the Project Achilles dataset17. We focused specifically on an independent set of breast cancer cell lines and categorized them as RP, TN MYClow and TN MYChigh as in Fig. 3a. We found that the growth of TN MYChigh cell lines was significantly more sensitive to CPT1B knockdown than that of RP or TN MYClow cells (Fig 3b). Additionally, we utilized siRNA to deplete CPT2, a different essential FAO enzyme9. CPT2 knockdown markedly decreased proliferation of MYChigh TN cell lines, but had a significantly reduced effect on the growth of RP lines (Fig. 3c and Supplementary Fig. 4). Thus, small molecule inhibition and knockdown of CPT1, as well as knockdown of CPT2 suggest FAO is an essential metabolic pathway in MO-TNBC.
FAO has been shown to be important during matrix detachment and under nutrient-deprived conditions18,19. We asked whether TN and MYC status affected responses to FAO inhibition when tumor cells are not attached or deprived of glucose. We utilized ultra-low adhesion plates and grew breast cancer cell lines as spheres to model matrix detachment. While TN MYChigh, TN MYClow and RP cells were all able to form viable spheres upon matrix detachment, the TN MYChigh cells were significantly more sensitive to FAO inhibition (Supplementary Fig. 5). Next, we tested the additive effect of FAO inhibition and glucose deprivation. Both TN MYChigh and TN MYClow cells display increased sensitivity to glucose deprivation, consistent with a well-established dependence on glycolysis for TNBC growth. Depleting glucose and inhibiting FAO results in additive ATP depletion in TN but not in RP cells (Supplementary Fig. 6). This is consistent with previous findings that FAO is important for breast cancer cell survival and NADPH production when glycolysis is limited20. Although FAO is important during matrix detachment and nutrient-deprivation, data presented in this study suggest TN MYChigh cells are uniquely more reliant upon FAO in both contexts.
To confirm the bioenergetic reliance upon FAO in MO-TNBC is a MYC-dependent phenotype, we examined if conditional MYC expression could alter sensitivity to FAO inhibition. We employed two distinct approaches: non-tumor human mammary epithelial cells expressing a four-hydroxytamoxifen (TAM)-inducible MYC-ER fusion protein (HMECMYC-ER) and two RP lines stably transduced with a MYC overexpression construct or control vector1,15 (Supplementary Fig. 7). We found that TAM-induced HMECMYC-ER and MYC-transduced RP cells were significantly more sensitive to etomoxir than uninduced HMECMYC-ER and vector-transduced RP cells, respectively (Fig. 3d). Furthermore, siRNA-mediated knockdown of MYC significantly rescued etomoxir sensitivity in three of four MYChigh TNBC cell lines (Fig. 3e and Supplementary Fig. 8). Finally, MYC protein expression was significantly correlated with bioenergetic sensitivity to FAO inhibition across our panel of human breast cell lines (Fig. 3f). While MYC expression is not tightly correlated with FAO or FAS protein expression (Figs. 2b and 3g), it is well correlated with FAO inhibition sensitivity and FAO activity (Figs. 2c and 3g). These data suggest MYC expression is both necessary and sufficient to induce a bioenergetic reliance upon FAO in breast epithelial cells.
Given that our initial observation of dysregulated FAO was made in MYC-driven breast tumors, we sought to test the effects of FAO inhibition on MO-TNBC metabolism in vivo. We administered 60 mg per kg of body weight (mg/kg) etomoxir or vehicle by IP injection at t = 0 h and 24 h to a cohort of mice bearing orthotopic HCI-002 MYChigh TNBC patient-derived xenografts (PDX) (Fig. 4a)21. Metabolite analysis of tumors harvested at 26 h revealed a significant reduction in long-chain AC levels as well as a reduction in TCA cycle intermediates, especially α-ketoglutarate, in drug-treated tumors compared to vehicle-treated tumors (Fig. 4b and Supplementary Table 5). Phosphorylation of AMPK (pAMPK) is a well-established marker of bioenergetic stress. We found that etomoxir-treated tumors had a significant increase in pAMPK compared to control-treated tumors (Fig. 4c). Further, we conducted a second metabolomic study comparing MTB-TOM tumors treated with 20 mg/kg for 14 days (d) to untreated tumors. Our analysis of tumors treated for 14 d corroborated our 26 h data as etomoxir-treated tumors displayed a significant reduction in the majority of TCA cycle intermediates, as well as ATP, and a significant increase in AMP/ATP, ADP/ATP, UMP/UTP and UDP/UTP ratios (Supplementary Fig. 9 and Table 6). These data indicate that treatment with an FAO inhibitor decreases bioenergetic metabolism in vivo in both transgenic and PDX models of TNBC.
Figure 4.
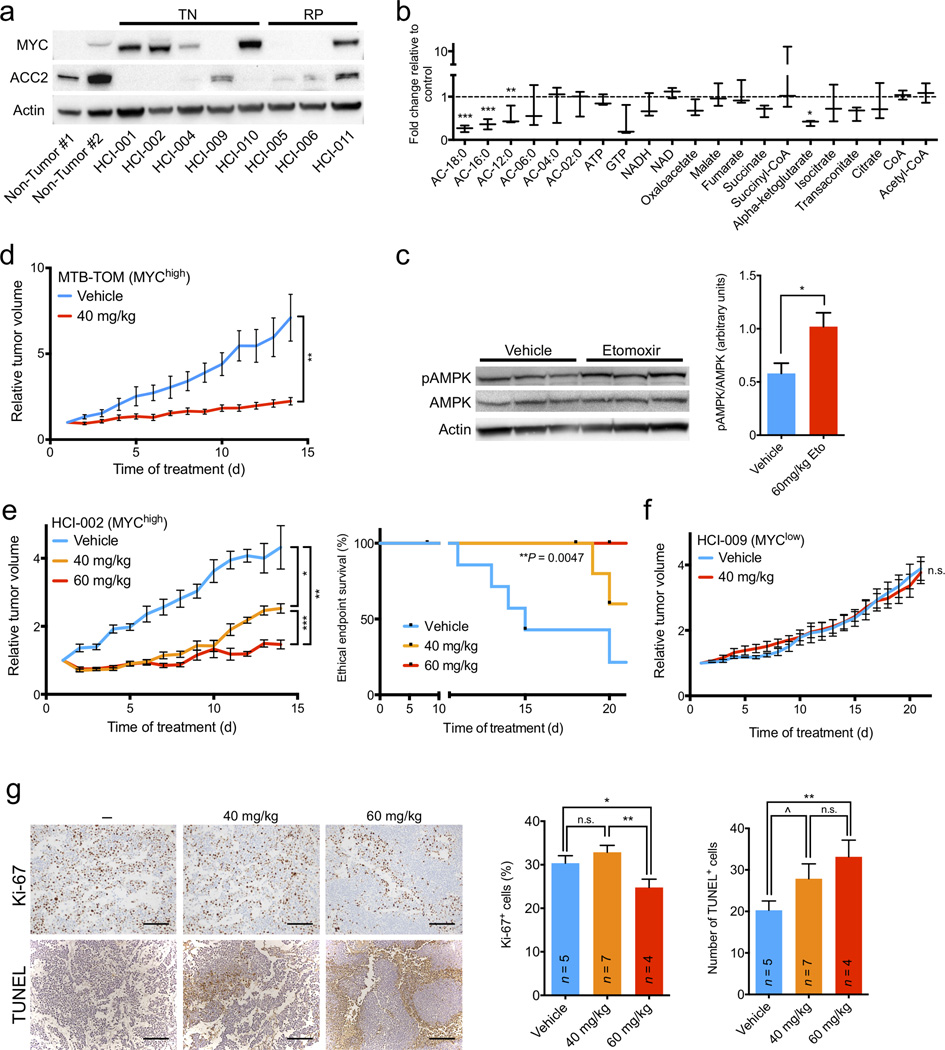
FAO inhibition shows MYC-dependent bioenergetic and growth effects in vivo. (a) Immunoblot analysis of indicated protein expression in TN and RP patient-derived xenografts and human non-tumor reduction mammoplasty tissues. (b) Fold change in metabolite levels in etomoxir-treated xenografts versus vehicle-treated tumors. Values are shown as min-to-max box plots from three mice in each group. (c) Etomoxir- and vehicle-treated tumors were examined by immunoblotting for indicated protein expression. pAMPK/AMPK ratio was normalized to β-actin. (d) FVB/N mice with orthotopic MTB-TOM tumor allografts were treated with vehicle or etomoxir (40 mg/kg) daily for 14 d. Growth plots are shown. (e) Left, NOD/SCID mice with orthotopic HCI-002 xenografts were treated with vehicle or etomoxir (40 or 60 mg/kg) daily for 21 d. Growth plots are shown. Statistical analysis was performed using a log-rank test. (f) NOD/SCID mice with orthotopic HCI-009 xenografts were treated with vehicle or etomoxir (40 mg/kg) daily for 21 d. Growth plots are shown. (g) Left, representative Ki-67 and TUNEL staining of untreated, 40 or 60 mg/kg etomoxir-treated HCI-002 tumors from mice euthanized at the end of the study. Right, quantification of percent Ki-67 positive cells per field and number of TUNEL positive cells per field. Number of mice analyzed in each treatment is indicated. Scale bar indicates 200 µm. All differential metabolite abundance analyses were performed using the limma R package. A two-tailed unpaired t-test was used to compare experimental groups (c–g). Values shown are mean ± s.e.m. from three individual mice (c), six mice in the control group and seven mice in the experimental group (d), seven mice in the control and 40 mg/kg etomoxir groups and five mice in the 60 mg/kg group (e), three mice in each group (f), or three high-powered (20×) fields from two separate areas of each tumor (g). ^P ≤ 0.10, *P ≤ 0.05, **P < 0.01, ***P < 0.001
The observed reduction in bioenergetic metabolism prompted us to analyze the effects of prolonged FAO inhibition on the growth of MO-TNBC tumors. We performed orthotopic transfer of MTB-TOM or HCI-002 PDX tumors into the mammary fat pad of FVB/N or NOD/SCID mice, respectively. We administered 40 mg/kg etomoxir or vehicle by IP injection daily for 14 d to MTB-TOM allograft-bearing mice. We administered 40 or 60 mg/kg etomoxir or vehicle by IP injection daily for 21 d to HCI-002 PDX-bearing mice. Etomoxir treatment resulted in significant attenuation of tumor growth in both models, and a significant extension of the time to ethical endpoint in the PDX model (Fig. 4d,e). In contrast, we observed no significant attenuation in tumor growth of the HCI-009 MYClow TNBC PDX tumors treated with 40 mg/kg etomoxir (Fig. 4a,f). The MYClow TNBC PDX tumors (HCI-009) were found to have moderate ACC2 expression indicating that FAO may be attenuated in these tumors (Fig 4a), explaining their resistance to etomoxir. Thus, both MYChigh transgenic and PDX models, but not a MYClow PDX model, demonstrate in vivo efficacy of etomoxir treatment and low ACC2 expression may serve as a biomarker of sensitivity.
Ki-67 is a well-established marker of proliferation. We found that PDX tumors treated with 60 mg/kg etomoxir had significantly decreased Ki-67 staining (Fig. 4g). We next sought to determine if cell death was increased in PDX tumors treated with etomoxir. We found a dose-dependent increase in TUNEL staining in etomoxir-treated tumors compared to controls (Fig. 4g). Although etomoxir treatment of cultured MO-TNBC cell lines had a marked effect on cell proliferation without appreciable changes in cell death (Supplementary Fig. 3), in vivo FAO inhibition resulted in a concurrent decrease in proliferation and an increase in apoptosis of MO-TNBC (Fig. 4g). These results are consistent with FAO playing a more critical role for in vivo tumor development.
Elevated MYC expression was recently discovered to be a defining factor of TNBC1,2. The present study is amongst the first to investigate the role of MYC in TNBC metabolism in vivo. Here we show that MO-TNBC upregulates FAO, that TNBC is sensitive to FAO inhibition in a MYC-dependent manner, and that FAO inhibition abrogates growth of distinct models of MO-TNBC in vitro and in vivo. This work supports a critical role for FAO in TNBC7,8, and identifies a dependency on MYC as a marker for this phenotype. Inhibition of FAO demands further investigation as a therapeutic strategy for MO-TNBC.
ONLINE METHODS
TB-TOM tumor generation
All protocols described in this and other sections regarding animal studies were approved by the UCSF Institutional Animal Care and Use Committee. MTB-TOM (MMTV-rtTA/TetO-MYC) were generated as previously described5. Mice were bred and maintained off doxycycline. At 12–15 weeks of age female mice were put on doxycycline (200 mg/kg doxy chow, Bio-Serv) to induce MYC expression and tumorigenesis. Mice were monitored daily for tumor growth by inspection and caliper measurement in two dimensions. Mice were sacrificed as per ethical guidelines (tumors reaching 2 cm in any single dimension) and tumor(s) or mammary gland(s) were flash frozen in liquid nitrogen.
Metabolomics
For U-13C-palmitate flux analyses, the labeled palmitate (Cambridge Isotope Laboratories Inc, CLM-409-0.5) or unlabeled palmitate (Sigma) diluted in PEG40 (Spectrum) via sonication was administered via intraperitoneal injection at 100 mg/kg. 4 h after injection, animals were sacrificed and tumors were flash frozen.
Nonpolar metabolites were extracted using Dounce homogenizers in 4 mL of 2:1:1 chloroform:methanol:PBS spiked with 10 nmol of internal standards (C12:0 dodecylglycerol and pentadecanoic acid, Sigma). Separation of organic and aqueous layers was achieved via centrifugation at 1000g for 5 minutes at 4 °C. Secondary extraction of the aqueous layer was performed using 0.1% formic acid followed by addition of 2 mL chloroform and further centrifugation. After combining the organic extractions, the lipid-containing mixture was dried under N2 and dissolved in 120 uL chloroform. Single-reaction monitoring (SRM) LC-MS/MS on an Agilent 6400 series QQQ using 10 uL of sample was achieved using a reverse-phase C5 column (Phenomenex, Luna 50mm × 4.6mm, 5um particle diameter). Mobile phases: Buffer A composed of 95:5 water:methanol and buffer B 60:35:5 2-propanol:methanol:water. Solvent modifiers were 0.1% formic acid with 5 mM ammonium formate or 0.1% ammonium hydroxide for positive and negative ionization modes, respectively.
For polar metabolites, frozen tissue was homogenized using a TissueLyser in 300 µl of 40:40:20 acetonitrile:methanol:water with the addition of 1 nM final concentration of D3-15N serine as an internal extraction standard (Cambridge Isotopes Laboratories Inc, DNLM-6863). 10 µl of cleared supernatant (via centrifugation at 15,000 rpm, 10 minutes, at 4 °C) was used for SRM-LC-MS/MS using a normal-phase Luna NH2 column (Phenomenex). Mobile phases: Buffer A composed of 100% acetonitrile, buffer B composed of 95:5 water:acetonitrile. Solvent modifiers were 0.1% formic acid or 0.2% ammonium hydroxide with 50 mM ammonium acetate for positive and negative ionization modes, respectively.
All metabolites were analyzed using the MassHunter software package (Agilent Technologies) by quantifying the transition from parent precursor mass to product ions for each individual metabolite.
Gene expression analysis
The TCGA breast invasive carcinoma dataset was sourced from data generated by the TCGA Research Network: http://cancergenome.nih.edu/, made available on the University of California, Santa Cruz (UCSC) Cancer Browser. Series matrix files for ISPY1 (Accession: GSE22226) and a neoadjuvant chemotherapy-treated cohort (Accession: GSE25066) were downloaded from GEO and processed using the GEOquery R package10,11,22. A chemotherapy-naïve and an aggressively treated early-stage cohort were obtained from the UCSC Cancer Browser12,13. Multiple probes corresponding to the same gene were collapsed using the “MaxMean” method in the WGCNA R package23,24.
A fatty acid metabolism gene set was compiled using genes containing the Gene Ontology term, GO:0006631 (http://geneontology.org). An average expression profile (centroid) of these genes was calculated for the triple-negative subset of samples within the TCGA breast invasive carcinoma dataset. Similarities between this centroid and gene expression profiles of samples from the four independent clinical cohorts were then quantified using a Pearson correlation metric.
Heatmaps and clustering analyses were performed using the gplots (http://cran.r-project.org/web/packages/gplots/index.html) and cluster (http://cran.r-project.org/web/packages/cluster/index.html) R packages respectively.
To generate Kaplan Meier plots, samples were grouped by receptor status, and dichotomized by ACC2 expression at an optimal threshold, yielding groups with the most significant difference in distant recurrence-free survival based on the log-rank test. Kaplan Meier plots were then generated for the respective groups using the survival (http://cran.r-project.org/web/packages/survival/index.html) R package.
Univariate cox proportional hazards regression analysis was performed using the survival R package to assess the correlation of fatty acid metabolism gene expression to distant recurrence-free survival in the pooled neoadjuvant chemotherapy-treated cohort11. This was conducted for all tumors, TN tumors and RP tumors.
Cell lines and propagation
A panel of established TN and RP human breast cancer cell lines and their culture conditions have been previously described15. Primary human mammary epithelial cells (HMECs) were derived from histologically normal breast tissues and cultured as previously described25. The cells were infected with lentivirus encoding p16 shRNA and further infected with pBabe-MycER virus and named B1389-shp16-MycER (HMECMYC-ER)26,27. While expressing p16 shRNA delays senescence, the cells are not immortalized and undergo spontaneous senescence when continuously cultured. Therefore, cells were not used beyond twelve passages past derivation. HMECMYC-ER cells were treated with four-hydroxytamoxifen (TAM) at 500 nM to induce MYC activation. RP (HCC1428 and T47D) cells stably overexpressing MYC have been previously described1. No cell line used in this paper is listed in the database of commonly misidentified cell lines maintained by ICLAC (http://iclac.org/databases/cross-contaminations/). All lines were found negative for mycoplasma contamination.
Immunoblot analysis
Proteins were extracted using RIPA buffer: 50 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.5% Na-deoxycholate, 1% Triton X-100, 0.1% SDS, 2 mM EDTA, and proteinase (Roche) plus phosphatase (Roche) inhibitor cocktails. Protein extracts were resolved using 4–12% SDS-PAGE gels (Life Technologies) and transferred to nitrocellulose membranes (Life Technologies). Membranes were probed with primary antibodies overnight on 4 °C shaker, then incubated with horseradish-peroxidase conjugated secondary antibodies and signals were visualized with ECL (Bio Rad). The primary antibodies used are as follows: β-Actin (Actin) (sc-47778 HRP, Santa Cruz, 1:10,000), PGC1α (ab54481, Abcam, 1:500), BBOX1 (WH0008424M1, Sigma Aldrich, 1:500), CPT2 (ab71435, Abcam, 1:500), FASN (SAB1403807, Sigma Aldrich, 1:1000), pACC1/2 (11818, Cell Signaling, 1:1000), ACC1 (4190, Cell Signaling, 1:1000), ACC2 (8578, Cell Signaling, 1:1000), AMPK (2532, Cell Signaling, 1:1000), pAMPK (2535, Cell Signaling, 1:1000) and c-MYC (MYC) (ab32072, Abcam, 1:1000).
ATP and NAD(P)H quantification
To determine ATP effects of etomoxir treatment, tumor cells were seeded in 96-well plates at 5,000–7,000 cells per well and cultured in the presence of 0 or 200 µM etomoxir (Sigma-Aldrich) for 48 h, with triplicate samples for each condition. Relative ATP concentration was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). To determine NAD(P)H effects of etomoxir treatment, HMECMYC-ER cells were seeded in 96-well plates at 2,000 cells per well and cultured in the presence of 0 or 500 nM TAM for 48 h, then 0 or 200 µM etomoxir for 24 h, with six samples for each condition. Relative NAD(P)H concentration was determined using CellTiter-Glo 96 AQueous One Solution Cell Proliferation Assay (Promega).
Fatty acid oxidation assay
To determine relative fatty acid oxidation pathway activity, tumor cells were seeded in 24-well plates at 100,000 cells per well in triplicate. Cells were incubated in serum-free medium for 2 h and then incubated back in standard medium for 1 h. 14C-oleic acid (Moravek Biochemicals, MC406) at 0.1 µCi/µL was added, 2×2 cm squares of Whatman paper were taped over wells, and cells were incubated for 3 h at 37 °C. After adding 200 µL 3 M NaOH directly to each square of Whatman paper, 100 µL 70% perchloric acid was added to each well and CO2 was captured at room temperature for 1 h. Whatman paper was dried at room temperature and placed in scintillation vial with 5 mL scintillation fluid. 14C radioactivity was measured by liquid scintillation counter and normalized to protein concentration.
RNAi knockdown
MYC (L-003282-02-0005), CPT2 (M-008574-01-005) and non-targeting (D-001810-10-20) siRNAs were purchased from GE Dharmacon (SMARTpool, 4 siRNAs per gene) and used to transfect cells with 30 pmol using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) according to the manufacturer’s instruction. Cells were incubated with siRNA for 72 h. For siMYC studies, media was changed at 24 h and 0 and 200 µM etomoxir was added for 48 h before ATP levels were quantified as described above. For siCPT2 studies, relative number of cells was determined at 72 h as described below.
The CPT1 knockdown data was extracted from the Project Achilles dataset17. The viability score represents the ATARiS solution, which is the computationally derived score that quantifies the gain or loss of growth specific to knockdown of the gene from pooled shRNA screens.
Proliferation and viability assays
To determine proliferative effects of etomoxir treatment, cells were seeded in 6-well plates at 100,000–150,000 cells per well and cultured in the presence of 0 or 200 µM etomoxir. Cells were harvested at 24, 48 and 72 h. Cell counts were determined using the Countess Automated Cell Counter (Life Technologies) according to the manufacturer’s instruction, and cell viability was assessed by performing the flow cytometry-based Guava ViaCount viability assay (Millipore) according to the manufacturer’s instruction. To determine proliferative effects of siCPT2 treatment, cells were seeded in 6-well plates at 100,000–150,000 cells per well and transfected with siNT or siCPT2 as described above. Cells were harvested at 24, 48 and 72 h and cell counts were determined using the Countess Automated Cell Counter (Life Technologies) according to the manufacturer’s instruction. Three independent biological replicates were performed for each time-point and condition.
Matrix detachment assays
To determine the effects of matrix detachment, cells were seeded in ultra-low adhesion round bottom 96-well plates at 1,000 cells per well. Sphere formation was judged by brightfield imaging 4 d after seeding, and then spheres were subsequently culture in 0 or 200 µM etomoxir for 48 h, with triplicate samples for each condition. Relative ATP concentration was determined as above.
Glucose starvation assays
To determine the effects of glucose starvation, cells were seeded in 96-well plates at 5,000–7,000 cells per well and cultured in replete medium, glucose-depleted medium, 200 µM etomoxir or glucose-depleted medium and 200 µM etomoxir for 24 h, with triplicate samples for each condition. Relative ATP concentration was determined as above.
Orthotopic allograft and xenograft studies
4 week-old WT FVB/N and immunocompromised NOD/SCID female mice were purchased from Taconic Biosciences. The derivation of the HCI-002 and HCI-009 TN patient-derived xenograft has been previously described21. Viably frozen MTB-TOM, HCI-002 and HCI-009 tumor samples were transplanted into the cleared mammary fat pads of FVB/N and NOD/SCID mice, respectively. Tumor growth was monitored daily by caliper measurement in two dimensions. Researchers were not blinded to treatment group.
For the 13C-palmitate experiment, the tumors were allowed to reach ~1.5 cm3 and then mice were randomized into experimental groups. Mice received 100 mg/kg 12C- or 13C-palmitate delivered by IP injection at t = 0 h. The palmitate was prepared by sonication into PEG-40 (Spectrum). Mice were euthanized at t = 4 h and tumors and contralateral non-tumor mammary glands were flash frozen in liquid nitrogen.
For the HCI-002 acute etomoxir experiment (Fig. 2g), tumors were allowed to reach ~1.5 cm3 and then mice were randomized into experimental groups. Mice received vehicle or 60 mg/kg etomoxir delivered by IP injection at t = 0 and 24 h. Mice were euthanized at t = 26 h and tumors were flash frozen in liquid nitrogen.
For the remaining in vivo studies, tumors reached ~1 cm3 when mice were randomized into experimental groups and drug treatment was initiated. For the MTB-TOM studies (Fig. 2h, Supplementary Fig. 12 and Supplementary Table 6), mice received vehicle or 20 or 40 mg/kg etomoxir delivered by IP injection daily for 14 d. Tumor growth was monitored daily by caliper measurement. Mice were euthanized after 14 d of treatment or if tumors reached 2cm in any dimension and tumors were flash frozen in liquid nitrogen. For the HCI-002 and HCI-009 studies (Fig. 2i,j), mice received vehicle, 40 or 60 mg/kg etomoxir delivered by IP injection daily for 21 d. Tumor growth was monitored daily by caliper measurement. Mice were euthanized after 21 d of treatment or if tumors reached 2 cm in any dimension and for the HCI-002 study two pieces of tumor from separate locations were fixed in 4% paraformaldehyde (n = 5 for vehicle, n = 7 for 40 mg/kg, n = 4 for 60 mg/kg). Remaining tumor was flash frozen in liquid nitrogen.
Immunohistochemical analysis
PFA-fixed tumor samples were paraffin-embedded and a tissue microarray (TMA) of HCI-002 tumors following described treatment conditions was created using two 2 mm punch cores per each tumor. Immunohistochemical staining of the TMA for Ki-67 was performed using MIB-1 antibody clone (1:50 dilution; DAKO, Carpinteria, CA, USA) following 20 minutes antigen retrieval with epitope retrieval solution 2 (Leica Biosystems, Buffalo Grove, IL, USA). Images were scored as the percentage of Ki-67 positive tumor cell nuclei per total tumor cell nuclei in each captured field using Immunoratio software (http://jvsmicroscope.uta.fi/immunoratio/). TUNEL staining was performed using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer’s instruction. Images were scored as the total number of TUNEL positive cells per captured field. All quantification was performed blinded to treatment group; TMA was created with independent labeling system.
Statistical analysis
Prism software was used to generate and analyze Pearson correlations (Fig. 2f) and the survival plot (Fig. 2h). Correlation P-values were generated using a two-sided t-test. Clade enrichment P-value was generated using a Fisher’s exact test. Survival plot P-value was generated using a log-rank test. All differential metabolite abundance and gene expression analyses were performed using the limma R package.
Differential metabolite abundance between MTB-TOM tumors and non-tumor mammary glands was performed using the limma R package28. Metabolites that were significantly different between these groups at a false discovery rate of 0.05 were extracted for downstream analyses. Pathway enrichment within this set of metabolites was quantified using the maxmean method within the piano R package29, based on annotations from the Kyoto Encyclopedia of Genes and Genomes (KEGG): http://www.genome.jp/kegg/. Significantly enriched pathways were identified at a P-value cutoff of 0.05.
For all other comparisons, unpaired two-sided t-tests were used (GraphPad). No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during in vivo experiments. The investigators were blinded during immunohistochemical analysis. All in vivo studies were randomized as tumors reached predetermined volume on a per experiment basis as described above. For each dataset, the data meet the assumptions of the statistical test used as determined by distribution and variance.
Sample size for all experiments (in vitro and in vivo) was not chosen with consideration of adequate power to detect a pre-specified effect size. For in vitro studies, all completed experiments are reported. For in vivo studies, number of indicated mice represents total number of mice treated and processed for each experiment. No samples were fully processed for metabolomic analysis or otherwise and then excluded. For the etomoxir treatment studies, mice were euthanized at ethical endpoint (~2 cm) or at study endpoint unless they surpassed predetermined UCSF Institutional Animal Care and Use Committee quality of life guidelines. No mice that completed the studies were excluded from analyses. In the prolonged PDX etomoxir study (Fig. 2h), two mice from the vehicle group and one mouse from the 60 mg/kg experimental group were found dead of unknown causes, and these tumors were not included in further analyses.
Code availability
Publically available datasets were acquired as noted. Our annotations of the datasets are available (https://bitbucket.org/jeevb/brca). All code used for this project has been deposited to Github (https://github.com/snjvb/fam_study).
Supplementary Material
Acknowledgments
This work was supported in part by grants from the US Department of Defense – Congressionally Directed Medical Research Programs’ Era of Hope Scholar Award W81XWH-12-1-0272 (A.G.), the US National Institutes of Health R01 CA170447 (A.G.), T32 DK007418 Diabetes, Endocrinology & Metabolism Training Grant (R.C.), T32 CA108462 Molecular and Cellular Mechanisms of Cancer Training Grant (A.Y.Z.) and the Atwater Foundation. The authors thank A. Welm for guidance in the use of patient-derived xenografts, D. Lowe and A. Beardsley for technical guidance and helpful discussion, and K.A. Fontaine for helpful discussion and comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
R.C. designed and conducted all of the experiments except the initial MTB-TOM metabolomic and HMECMYC-ER studies, and wrote the manuscript. A.Y.Z. performed the in vivo studies and provided valuable discussion and intellectual input. R.A.K. performed the mass spectrometry and metabolomic analyses. S.B. performed bioinformatic analyses of the data from gene expression and metabolomic analyses. C.M. performed in vitro proliferation and viability studies, matrix detachment studies and assisted in TUNEL quantification. B.A. performed the HMECMYC-ER studies and provided valuable discussion. H.E. performed orthotopic tumor transplants. S.K. supervised the FAO activity analyses. A.T. provided CPT1B knockdown data, interpretation and valuable discussion. G.K. constructed the HCI-002 etomoxir study tissue-microarray and performed Ki-67 staining and quantification. D.K.N. supervised the mass spectrometry and metabolomic analyses, and provided valuable discussion. A.G. supervised all studies, and provided valuable discussion and intellectual input. All authors edited the manuscript.
The authors declare no competing financial interests.
MAIN TEXT REFERENCES
- 1.Horiuchi D, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J. Exp. Med. 2012;209:679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu S, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahlström T, Henriksson MA. Impact of MYC in regulation of tumor cell metabolism. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbagrm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 5.D’Cruz CM, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat. Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 6.Pfefferle AD, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14:R125. doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carracedo A, et al. A metabolic prosurvival role for PML in breast cancer. J. Clin. Invest. 2012;122:3088–3100. doi: 10.1172/JCI62129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaugg K, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnefont JP, et al. Carnitine palmitoyltransferases 1 and 2: Biochemical, molecular and medical aspects. Mol. Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Esserman LJ, et al. Chemotherapy response and recurrence-free survival in Neoadjuvant breast cancer depends on biomarker profiles: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res. Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatzis C, et al. A Genomic Predictor of Response and Survival Following Taxane-Anthracycline Chemotherapy for Invasive Breast Cancer. JAMA J. Am. Med. Assoc. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yau C, et al. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12:R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Morris EM, et al. PGC-1 overexpression results in increased hepatic fatty acid oxidation with reduced triacylglycerol accumulation and secretion. AJP Gastrointest. Liver Physiol. 2012;303:G979–G992. doi: 10.1152/ajpgi.00169.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holubarsch CJF, et al. A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (etomoxir for the recovery of glucose oxidation) stud. Clin. Sci. 2007;113:205. doi: 10.1042/CS20060307. [DOI] [PubMed] [Google Scholar]
- 17.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon S, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensaad K, et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 21.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature Medicine. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 22.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay R, et al. Promotion of variant human mammary epithelial cell outgrowth by ionizing radiation: an agent-based model supported by in vitro studies. Breast Cancer Res. 2010;12:R11. doi: 10.1186/bcr2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 27.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth GK. limma: Linear Models for Microarray Data. Bioinforma. Comput. Biol. Solut. Using R Bioconductor. 2005:397–420. [Google Scholar]
- 29.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.