Abstract
MicroRNAs (miRNAs) are negative regulators of gene expression, and miRNA deregulation is found in various tumors. We previously reported that suppression of adenine nucleotide translocase 2 (ANT2) by short hairpin RNA (shRNA) inhibits hepatocellular carcinoma (HCC) development by rescuing miR-636 expression. However, the tumor-suppressive mechanisms of ANT2 shRNA are still poorly understood in HCC. Here, we hypothesized that miRNAs that are specifically downregulated by ANT2 shRNA might function as oncomiRs, and we investigated the roles of ANT2 shRNA-regulated miRNAs in the pathogenesis of HCC. Our data show that miR-19a and miR-96, whose expression is regulated by ANT2 suppression, were markedly upregulated in HCC cell lines and clinical samples. Ectopic expression of miR-19a and miR-96 dramatically induced the proliferation and colony formation of hepatoma cells in vitro, whereas inhibition of miR-19a and miR-96 reduced these effects. To investigate the in vivo function, we implanted miR-96-overexpressing HepG2 cells in a xenograft model and demonstrated that the increase in miR-96 promoted tumor growth. We also found that miR-19a and miR-96 inhibited expression of tissue inhibitor of metalloproteinase-2. Taken together, our results suggest that ANT2-regulated miR-19a and miR-96 play an important role in promoting the proliferation of human HCC cells, and the knockdown of ANT2 directly downregulates miR-19a and miR-96, ultimately resulting in the suppression of tumor growth.
Introduction
Among RNAs with regulatory functions, microRNAs (miRNAs) have been extensively studied for their potential roles in cancer diagnosis and as therapeutic targets. The miRNA molecules are transcribed from noncoding genes, and the transcribed products stabilize into a duplex structure that activates the RNA interference machinery and downregulates gene expression posttranscriptionally.1 The miRNAs play a crucial role in both physiological and pathological processes, and >50% of the known miRNAs participate in human tumorigenesis by regulating gene expression.2, 3 Their deregulation is observed in many types of cancer,4, 5 and it is negatively associated with the clinical outcomes in cancer patients.6, 7 Recent studies have reported that alterations in miRNA expression are frequently observed in hepatocellular carcinomas (HCCs).8, 9 Indeed, deregulation of miR-199-3p was observed in 40/40 patients, and its low expression correlated with poor survival of HCC patients.10 In addition, one study showed that miR-519d was upregulated in ∼50% of HCCs.11
Adenine nucleotide translocase (ANT) is abundant in the inner mitochondrial membrane. ANT plays an important role in cellular energy metabolism by catalyzing the exchange of mitochondrial adenosine triphosphate for cytosolic adenosine diphosphate, thus influencing mitochondrial bioenergetics.12 In addition, it is involved in the formation of the mitochondrial permeability transition pore complex that interacts with Bcl2 family proteins, contributing to mitochondrial-mediated apoptosis.13 Human ANT has four isoforms (ANT1, ANT2, ANT3 and ANT4), and the relative expression of these isoforms is dependent on developmental stage, proliferation status and cell or tissue types. Among these isoforms, ANT2 is specifically expressed in proliferative and undifferentiated cells.13 ANT2 imports glycolytically derived adenosine triphosphate into the mitochondria, maintaining the mitochondrial membrane potential and ensuring cell survival. Previously, we showed that ANT2 is overexpressed in a wide variety of tumors.14 Because the expression of ANT2 is closely associated with the mitochondrial bioenergetics of tumors, ANT2 could be a promising target for cancer therapy. In fact, we have demonstrated that ANT2 depletion by short hairpin RNA (shRNA) significantly inhibits tumorigenesis in breast cancer15 and HCC.16, 17
An essential step in cancer invasion and metastasis is the breakdown of the extracellular matrix by proteinases, such as the matrix metalloproteinases (MMPs).18, 19 MMPs are capable of degrading many types of extracellular matrix proteins and are abundant in advanced tumors. Therefore, MMPs are thought to be involved in the regulation of cancer progression, invasion and metastasis. Under normal physiological conditions, the regulation of the MMP family is dependent on gene expression, proenzyme activation and interactions with specific inhibitors, such as tissue inhibitors of metalloproteinases (TIMPs).20, 21 All four TIMP members—TIMP-1, 2, 3, and 4—interact with MMPs in a stoichiometric (1:1 molar) manner and regulate the function and activation of MMPs. In addition, phosphatidylinositol-3-kinase (PI3K)/Akt activation is involved in the regulation of the MMP family, suggesting that selective targeting of the PI3K/Akt signaling pathway may effectively block potentially invasive cancer cells.22
Based on our previous observation that ANT2 suppression resulted in inhibition of HCC,17 we hypothesized that ANT2 shRNA regulates a specific subset of miRNAs that are crucial for HCC development, and investigated the functions of ANT2 shRNA-regulated miRNAs in the pathogenesis of HCC.
Materials and methods
Cell culture and transfection
The hepatocellular carcinoma cell lines HepG2 and Hep3B cells were purchased from the Korean Cell Line Bank (KCLB, Seoul, Korea). These cells were then cultured in Dulbecco's modified Eagle's medium (WelGENE, Seoul, Korea) supplemented with 10% fetal bovine serum, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco, Grand Island, NY, USA) in a humidified 5% CO2/95% air atmosphere at 37 °C.
For transfection, cells were plated in a 6-well plate (2 × 105 cells per well) or 100 mm dish (2 × 106 cells per dish) and then transfected with the appropriate plasmid(s) in combination with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). pSilencer 3.1-H1 puro ANT2 or scrambled small interfering RNA vectors were transfected into the cells. Transfected cells were then cultured for 4 h, and the culture media were replaced with fresh media supplemented with 10% fetal bovine serum. The cells were harvested 24–48 h after transfection. Negative mimics, miR-19a and miR-96 mimics, negative inhibitors and miR-19a and miR-96 inhibitors were transfected into the cells using the same method.
Synthesis of miR mimics and miR inhibitors
The miR-19a and miR-96 mimics and inhibitors, as well as a negative control of miRNA mimics (negative mimics) or inhibitors (negative inhibitors), were synthesized by Genolution (Seoul, Korea) and GenePharma (Shanghai, China) with the following sequences: miR-19a mimic (5′-AGUUUUGCAUAGUUGCACUACA-3′) and inhibitor (5′-UCAGUUUUGCAUAGAUUUGCACA-3′: 2′OMe modification), miR-96 mimic (5′-UUUGGCACUAGCACAUUUUUGCU-3′) and inhibitor (5′-AGCAAAAAUGUGCUAGUGCCAAA-3′: 2′OMe modification) and negative mimic/inhibitor (5′-UUGUACUACACAAAAGUACUG-3′).
Quantitative real-time PCR
Quantitative real-time PCR was used to confirm microarray data and measure the miRNA impact on putative targets. Total RNA was extracted from cell lines using TRIzol reagent (Invitrogen) or 10-mm thick formalin-fixed paraffin-embedded tissue sections using a RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA). Quantitative real-time PCR was carried out with a Mir-X miRNA First-Strand Synthesis Kit and a SYBR Green Real-time PCR Master Mix (TaKaRa, Otsu, Japan) according to the manufacturer's instructions. The housekeeping gene U6 was used for standardization of the initial miRNA content of a sample. Relative changes in gene expression were calculated by the formula previously described.17
Western blot analysis
Cell lysates were separated on 10% SDS–polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA) and then detected with the respective antibodies indicated below. Immunoblots were visualized using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Uppsala, Sweden). Anti-Akt and anti-pAkt were obtained from Cell Signaling Technology (Beverly, MA, USA), and anti-TIMP-2 and anti-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell proliferation assay
For the determination of cell growth, cells were seeded at 1 × 104 per well in 96-well plates and incubated under the indicated conditions. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed as previously described.23 The MTT-derived formazan generated by cells was quantified. After the addition of 20 μl of MTT (5 mg ml−1) to each well and a 4-h incubation at 37 °C, MTT crystals were dissolved with dimethyl sulfoxide without discarding the cell supernatants, and the absorbance (570 nm) was measured. All experiments were repeated a minimum of 3 times in 96-well plates.
Soft agar colony formation assay
Transfected cells were mixed in 0.3% Noble agar (in RPMI-1640 medium supplemented with 10% fetal bovine serum) and plated at 1000 cells per well in 6-well plates containing a solidified bottom layer (0.6% Noble agar in the same growth medium). After 14 days, colonies were stained with 0.1% crystal violet (Sigma, St Louis, MO, USA) and photographed.
Tumorigenicity assays in nude mice
Animal experiments were carried out according to the guidelines of the institutional animal care and use committee of Seoul National University. The experiments were performed after receiving approval from the institutional biosafety committee (SNUIB-P110525-1) and institutional animal care and use committee (SNU-110527-1). The miR-96- and negative control (NC)-transfected HepG2 cells (1 × 107) were suspended in 100 μl phosphate-buffered saline and then injected subcutaneously into either side of the posterior flank of the same female BALB/c athymic nude mouse at 5 to 6 weeks of age. Tumor sizes were measured twice a week and calculated with the following formula: (length × width2) × 0.5.
Statistical analysis
Statistical analyses were performed primarily with Student's t-test with SigmaPlot 2001 software (Systat Software, Chicago, IL, USA). For the analysis of animal experiments, two-way analysis of variance was used with Prism 3.0 software (GraphPad Software, La Jolla, CA, USA). P-values of <0.05 were regarded as statistically significant.
Results
Upregulation of miR-19a and miR-96 in human HCC samples
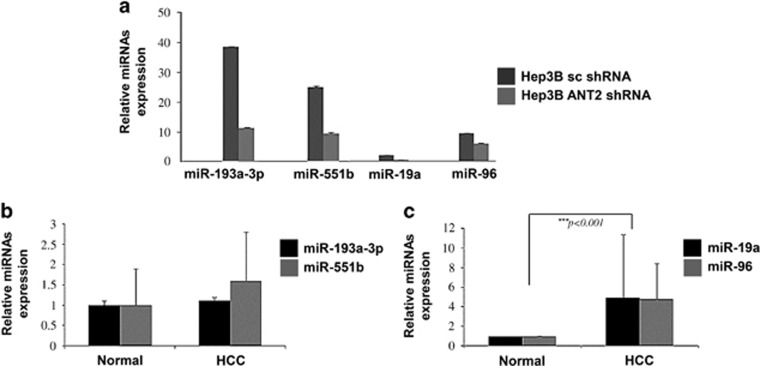
In our previous study, we performed miRNA microarray screenings to identify ANT2 shRNA-regulated miRNAs in Hep3B cells and reported that 14 miRNAs were downregulated by ANT2 inhibition.17 To validate the 14 candidate miRNAs, quantitative real-time PCR was carried out, and only four miRNAs (miR-193a-3p, miR-551b, miR-19a and miR-96) exhibited significant differences between ANT2 shRNA and scramble transfection in Hep3B cells (Figure 1a). To determine whether these 4 miRNAs are biologically relevant in HCC, we evaluated the expression of the 4 miRNAs in 30 HCC tissue pairs. Surprisingly, expression of miR-193a-3p and miR-551b, which showed marked downregulation by ANT2 inhibition in Hep3B cells, was not significantly different in HCC tissues compared with matched normal liver tissues (Figure 1b). However, miR-19a and miR-96 were highly expressed in HCC tissues, implying that miR-19a and miR-96 might be associated with HCC development (Figure 1c).
Figure 1.
miR-19a and miR-96 are upregulated in human hepatocellular carcinoma (HCC) samples. (a) After 24 h of transfection with scramble and adenine nucleotide translocase 2 (ANT2) short hairpin RNA (shRNA) in Hep3B cells, total RNA was extracted and subjected to quantitative real-time PCR (RT-qPCR) using primers for miR-193a-3p, miR-551b, miR-19a, miR-96 or U6 (internal control). The mean±s.d. of triplicate samples is shown. (b, c) RT-qPCR of HCC tumor tissue pairs was carried out (n=30). The mean±s.d. is shown, and data were evaluated using Student's t-test.
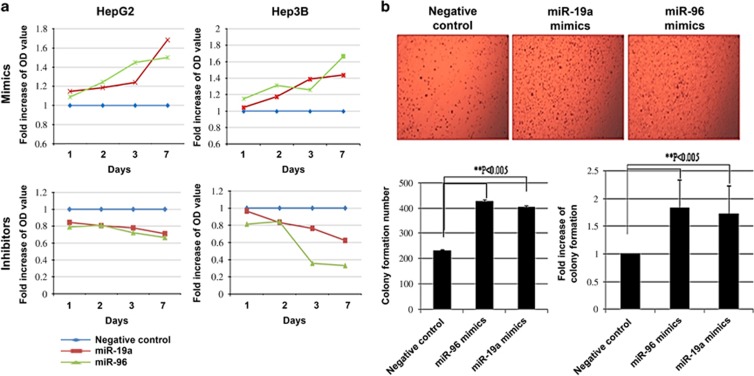
Upregulation of miR-19a and miR-96 enhances proliferation and tumorigenicity of HCC cells
To investigate the biological role of miR-19a and miR-96 in the development and progression of HCC, we transfected HepG2 and Hep3B cells with miR-19a and miR-96 mimic oligonucleotides and then examined the effect on cellular proliferation. Using the MTT assay, we observed that overexpression of miR-19a and miR-96 dramatically increased the growth rate of both HCC cells (HepG2 and Hep3B) compared with those of negative control (NC)-transfected cells (Figure 2a). In contrast, transfection with anti-miR-19a and anti-miR-96 impaired proliferation of HCC cells, suggesting that miR-19a and miR-96 may function as oncomiRs. Moreover, overexpression of miR-19a and miR-96 enhanced anchorage-independent growth of HepG2 cells in soft agar (Figure 2b). The colony formation number of miR-96- and miR-19a-treated cells increased ∼2- and 1.8-fold, respectively. These results suggest that upregulation of miR-19a and miR-96 could augment the tumorigenicity of HCC cells in vitro.
Figure 2.
miR-19a and miR-96 enhance proliferation and tumorigenicity of hepatocellular carcinoma (HCC) in vitro. (a) HepG2 and Hep3B liver cancer cells were transfected with miR-19a mimics, miR-96 mimics, miR-19a inhibitors and miR-96 inhibitors. Cell viability (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)) assays were performed 1, 2, 3 and 7 days after transfection. (b) Soft agar colony formation assays were conducted after HepG2 cells were transfected with miR-19a mimics, miR-96 mimics, miR-19a inhibitors and miR-96 inhibitors. Three independent experiments were performed. The mean±s.d. is shown, and data were analyzed using Student's t-test.
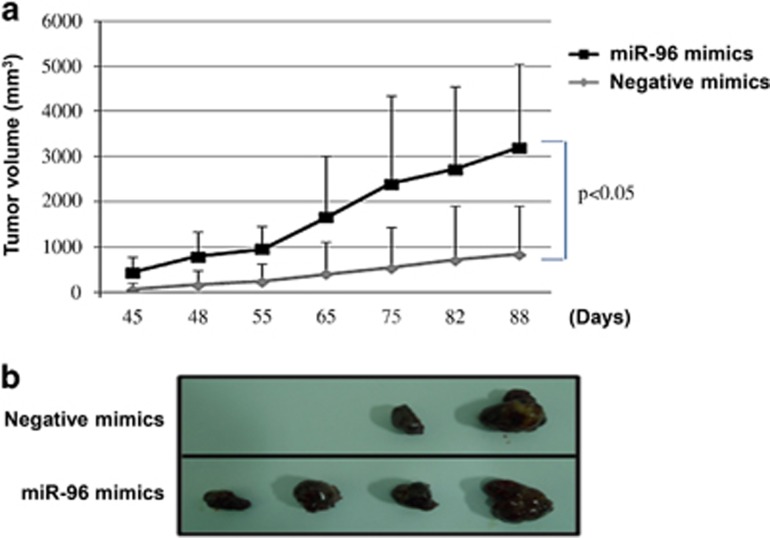
Overexpression of miR-96 contributes to progression of HCC
To further confirm these findings, an in vivo model was used. It has been reported that miR-96 is involved in hepatitis B virus (HBV)-associated HCC.24 However, the role of miR-96 in HBV-negative HCC remains largely unknown. To address this question, NC- and miR-96-transfected HepG2 cells (HBV negative) were subcutaneously injected into either side of the posterior flank of the same nude mice, respectively. The experimental group consisted of four nude mice, and 1 × 106 cells were injected into each posterior flank of the mice. At 6 weeks after injection, tumor growth on the flanks injected with miR-96-transfected HepG2 cells was observed and compared with the growth on flanks injected with NC-transfected HepG2 cells (Figures 3a and b). The miR-96-transfected HepG2 cells produced tumors (mean size of 2363±1846 mm3) in all mice (Figure 3a), whereas NC-transfected HepG2 generated only a few tumors (mean size of 835±1066 mm3) in 2 of 4 (50%) mice (Figure 3b). These results indicate that miR-96 significantly promotes tumorigenicity of HepG2 cells in a nude mouse xenograft model.
Figure 3.
Overexpression of miR-96 contributes to the progression of hepatocellular carcinoma (HCC). (a) Negative control (NC)- or miR-96-transfected HepG2 cells (1 × 106) were injected subcutaneously into each posterior flank of the mice (n=4). Tumor sizes were measured by a caliper. The mean±s.d. is shown, and data were evaluated using Student's t-test. (b) Images of representative features of tumors 88 days after inoculation.
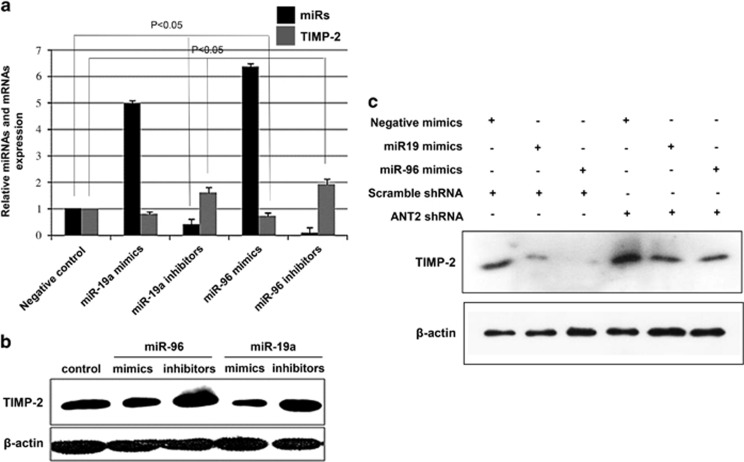
TIMP-2 is a target of miR-19a and miR-96
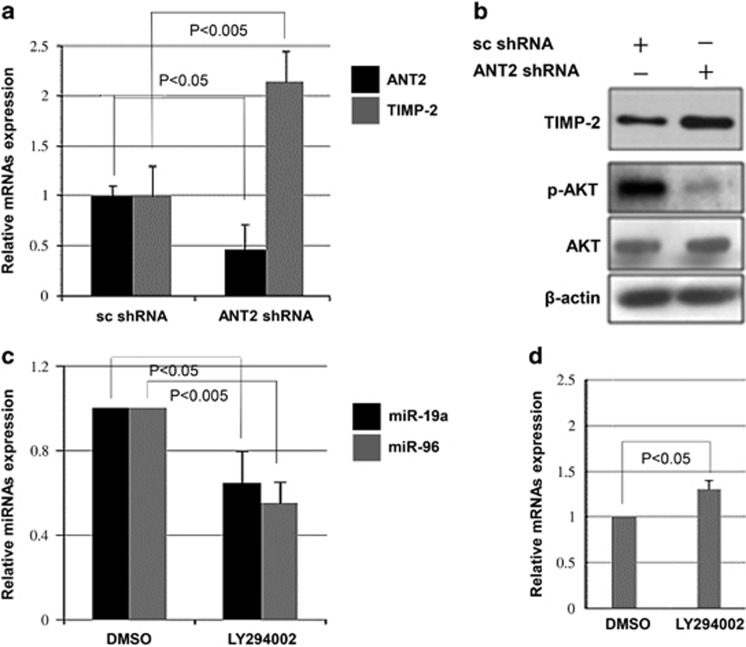
It is generally accepted that miRNAs exert their function by regulating the expression of their downstream target gene(s). Using target prediction programs and an online miRNA database (www.microRNA.org; www.targetscan.org; and pictar.mdc-berlin.de), it was revealed that the 3′-untranslated region of TIMP-2 mRNA has a complementary site for the seed region of miR-19a and miR-96. Previous studies have shown that the TIMP-2 gene, which encodes an endogenous inhibitor of MMP involved in cell invasion and tumorigenesis, is downregulated or silenced in a variety of human cancer cells.25 To verify whether TIMP-2 is a common target of miR-19a and miR-96, Hep3B cells were transiently transfected with miR-19a and miR-96 mimics or inhibitors (Figure 4a). Mimics of miR-19a and miR-96 inhibited the expression levels of TIMP-2, but miR-19a and miR-96 inhibitors induced expression of TIMP-2, suggesting that miR-19a and miR-96 specifically affected TIMP-2 posttranscriptionally. Using western blot analysis, downregulation of TIMP-2 was detected after transfection with miR-19a and miR-96 mimics, whereas TIMP-2 upregulation was observed following transfection with miR-19a and miR-96 inhibitors (Figure 4b). Moreover, to determine whether knockdown of ANT2 deregulates miRNAs, we used ANT2 shRNA to suppress miR-19a and miR-96 in the presence of miR-19a and miR-96 mimics to show that TIMP-2 is indeed a critical target of these miRNAs. The results demonstrated that the treatment with miR-19a and miR-96 mimics without ANT2 shRNA transfection suppressed TIMP-2, but TIMP-2 levels were rescued by ANT2 inhibition (Figure 4c). Taken together, our results indicate that TIMP-2 is a target for both miR-19a and miR-96.
Figure 4.
Tissue inhibitor of metalloproteinase-2 (TIMP-2) is a target of miR-19a and miR-96. (a) Hep3B cells were transfected with miR-19a mimics, miR-96 mimics, miR-19a inhibitors and miR-96 inhibitors. Expression of miR-19a, miR-96 and TIMP-2 was examined by quantitative real-time PCR (RT-qPCR) analysis. (b) After 24 h of transfection with miR-19a mimics, miR-96 mimics, miR-19a inhibitors and miR-96 inhibitors, cell extracts were prepared for western blotting. A representative blot from three independent experiments is shown. (c) Hep3B cells were transfected with miR-19a mimics, miR-96 mimics and adenine nucleotide translocase 2 (ANT2) short hairpin RNA (shRNA). Expression of TIMP-2 was examined using western blotting.
miR-19a and miR-96 are regulated by ANT2 through the PI3K/Akt signaling pathway in HCC
We have demonstrated here that ANT2-regulated miR-19a and miR-96 reduce TIMP-2 expression in HCC. To further confirm that ANT2 is involved in the regulation of TIMP-2, the effect of ANT2 knockdown was examined. As expected, the knockdown of ANT2 upregulated the mRNA expression of TIMP-2 in Hep3B cells (Figure 5a). In addition, ANT2 depletion upregulated the protein level of TIMP-2 (Figure 5b). It has been shown that ANT2 shRNA suppresses the PI3K/Akt signaling pathway in breast cancer cells.26 Thus, we wanted to determine whether the PI3K/Akt pathway is inhibited by ANT2 shRNA in HCC cells. The data showed that phosphorylated Akt levels were decreased by knockdown of ANT2 in Hep3B cells (Figure 5b). To assess whether the PI3K/Akt pathway is involved in miR-19a and miR-96 expression, a PI3K inhibitor, LY294002, was used. Treatment with LY294002 successfully inhibited phosphorylated Akt (Supplementary Data 1a) and correspondingly downregulated the expression of miR-19a and miR-96 (Figure 5c) and upregulated TIMP-2 (Figure 5d), suggesting that miR-19a and miR-96 are regulated by the PI3K/Akt signaling pathway in HCC cells. Because LY294002 is a nonspecific inhibitor, we further verified the expression of TIMP-2 after Akt silencing using a dominant-negative PI3K mutant to specifically inhibit the PI3K/Akt pathway. Our data showed that transfection with dominant-negative PI3K significantly decreased the level of pAkt and upregulated TIMP-2, suggesting that Akt activity is inversely correlated with TIMP-2 levels and that the selective targeting of the PI3K/Akt signaling pathway may block potentially invasive cancer cells (Supplementary Data 1b).
Figure 5.
miR-19a and miR-96 are regulated by the phosphatidylinositol-3-kinase (PI3K)/Akt signaling pathway in hepatocellular carcinoma (HCC). (a) After 24 h of transfection with scramble and adenine nucleotide translocase 2 (ANT2) short hairpin RNA (shRNA) in Hep3B cells, total RNA was extracted and subjected to quantitative real-time PCR (RT-qPCR). (b) Hep3B cells were transfected with scramble and ANT2 shRNA for 24 h, and western blotting was performed. β-actin was used as a loading control. (c) The levels of miR-19a and miR-96 were analyzed by RT-qPCR after treatment with LY294002, a PI3K inhibitor. (d) After treating Hep3B cells with LY294002, expression of tissue inhibitor of metalloproteinase-2 (TIMP-2) was examined by RT-qPCR analysis. Three independent experiments were performed, and the mean±s.d. is shown.
Discussion
Overexpression of ANT2 in the inner mitochondrial membrane of cancer cells has been associated with glycolysis and proliferation.27 Numerous studies have shown that cancers can be characterized by major metabolic changes that generate energy through predominantly glycolytic mechanisms, in contrast to the normal dual oxidative and glycolytic processes, even in the presence of oxygen, a phenomenon known as the Warburg effect.13 Because cancer cells prefer utilizing the glycolytic pathway, the role of ANT2 could be crucial in cancer cells as ANT2 imports glycolytically generated adenosine triphosphate into the mitochondria, contributing to maintaining the mitochondrial membrane potential and preventing apoptosis. Because the expression of ANT2 is closely linked to the mitochondrial bioenergetics of tumors, we have previously shown that ANT2 knockdown impairs the glycolytic metabolism in cancer cells and results in decreased cell viability and induced apoptotic death.14
In this study, our results demonstrate that knockdown of ANT2 can specifically impair HCC progression through the regulation of miR-19a and miR-96. Increased expression of miR-19a and miR-96 has been observed in various types of cancer,28, 29, 30, 31, 32 and our study of miR-19a and miR-96 in hepatocellular tumors emphasizes their fundamental oncogenic behaviors. As the role of miR-96 in HBV-associated HCC has already been reported,24 we investigated its role in HBV-negative HCC that remains largely unknown. In our study, we used HepG2 as a representative HBV-negative HCC cell line to confirm the oncogenic role of miR-96 in HCC progression. The results showed that miR-96 promoted tumor growth of HepG2 cells in vitro and in vivo, implying that miR-96 functions as an oncomiR regardless of HBV status.
PI3K/Akt activation is involved in the regulation of the MMP family that is associated with increased cancer invasion, metastasis and angiogenesis.33 This suggests that the selective targeting of the PI3K/Akt signaling pathway may block potentially invasive cancer cells. Indeed, our previous study in breast cancer cells demonstrated that the PI3K/Akt signaling pathway was inactivated by ANT2 shRNA, resulting in downregulation of MT1-MMP, MMP2 and MMP9 activity.26 In this study, we showed that knockdown of ANT2 reduced phosphorylated Akt levels and upregulated the mRNA expression and protein levels of TIMP-2 in HCC, suggesting that targeting ANT2 in HCC cells may be a successful anticancer strategy. However, the exact role of ANT2 in the PI3K/Akt signaling pathway remains to be elucidated. A recent study demonstrated that inhibition of aerobic glycolysis perturbs the Akt/mammalian target of rapamycin/hypoxia-inducible factor-1α axis.34 Because glycolytic metabolism and the PI3K/Akt signaling pathway are highly intertwined, it is likely that interruption of glycolysis in cancers via ANT2 suppression will result in downregulation of the PI3K/Akt signaling pathway.
In summary, the knockdown of ANT2 led to downregulation of miR-19a and miR-96 and ultimately resulted in the suppression of HCC. These results suggest a possible beneficial application for the treatment of HCC.
Acknowledgments
This work was supported by the Global Core Research Center (GCRC) Grant (No. 2012-0001190) from the National Research Foundation (NRF), Ministry of Education, Science and Technology (MEST), Republic of Korea.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch 2008; 452: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med 2009; 60: 167–179. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med 2008; 359: 2720–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Gusv Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res 2008; 14: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of microRNAs in human liver cancers. Semin Oncol 2011; 38: 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 2013; 57: 840–847. [DOI] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou WP, Wang ZX, Ding GS, Dong QZ et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011; 19: 232–243. [DOI] [PubMed] [Google Scholar]
- Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol 2012; 227: 275–285. [DOI] [PubMed] [Google Scholar]
- Schonfeld P, Schild L, Bohnensack R. Expression of the ADP/ATP carrier and expansion of the mitochondrial (ATP + ADP) pool contribute to postnatal maturation of the rat heart. Eur J Biochem 1996; 241: 895–900. [DOI] [PubMed] [Google Scholar]
- Chevrollier A, Loiseau D, Reynier P, Stepien G. Adenine nucleotide translocase 2 is a key mitochondrial protein in cancer metabolism. Biochim Biophys Acta 2011; 1807: 562–567. [DOI] [PubMed] [Google Scholar]
- Jang JY, Choi Y, Jeon YK, Kim CW. Suppression of adenine nucleotide translocase-2 by vector-based siRNA in human breast cancer cells induces apoptosis and inhibits tumor growth in vitro and in vivo. Breast Cancer Res 2008; 10: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Jeon YK, Choi Y, Kim CW. Short-hairpin RNA-induced suppression of adenine nucleotide translocase-2 in breast cancer cells restores their susceptibility to TRAIL-induced apoptosis by activating JNK and modulating TRAIL receptor expression. Mol Cancer 2010; 9: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Jeon YK, Lee CE, Kim CW. ANT2 suppression by shRNA may be able to exert anticancer effects in HCC further by restoring SOCS1 expression. Int J Oncol 2013; 42: 574–582. [DOI] [PubMed] [Google Scholar]
- Jang JY, Lee YS, Jeon YK, Lee K, Jang JJ, Kim CW. ANT2 suppression by shRNA restores miR-636 expression, thereby downregulating Ras and inhibiting tumorigenesis of hepatocellular carcinoma. Exp Mol Med 2013; 45: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer 2003; 3: 489–501. [DOI] [PubMed] [Google Scholar]
- Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell 1997; 91: 439–442. [DOI] [PubMed] [Google Scholar]
- Goldberg GI, Strongin A, Collier IE, Genrich LT, Marmer BL. Interaction of 92-kDa type IV collagenase with the tissue inhibitor of metalloproteinases prevents dimerization, complex formation with interstitial collagenase, and activation of the proenzyme with stromelysin. J Biol Chem 1992; 267: 4583–4591. [PubMed] [Google Scholar]
- Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol 1997; 74: 111–122. [PubMed] [Google Scholar]
- Ispanovic E, Haas TL. JNK and PI3K differentially regulate MMP-2 and MT1-MMP mRNA and protein in response to actin cytoskeleton reorganization in endothelial cells. Am J Physiol Cell Physiol 2006; 291: C579–C588. [DOI] [PubMed] [Google Scholar]
- Liu M, Iavarone A, Freedman LP. Transcriptional activation of the human p21(WAF1/CIP1) gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J Biol Chem 1996; 271: 31723–31728. [DOI] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 2008; 47: 1955–1963. [DOI] [PubMed] [Google Scholar]
- Pulukuri SM, Patibandla S, Patel J, Estes N, Rao JS. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene 2007; 26: 5229–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Jeon YK, Kim CW. Degradation of HER2/neu by ANT2 shRNA suppresses migration and invasiveness of breast cancer cells. BMC Cancer 2010; 10: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrollier A, Loiseau D, Chabi B, Renier G, Douay O, Malthiery Y et al. ANT2 isoform required for cancer cell glycolysis. J Bioenerg Biomembr 2005; 37: 307–316. [DOI] [PubMed] [Google Scholar]
- Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS ONE 2010; 5: e15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 2009; 284: 23204–23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa-miR-96 up-regulates MAP4K1 and IRS1 and may function as a promising diagnostic marker in human bladder urothelial carcinomas. Mol Med Rep 2012; 5: 260–265. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang Z, Yang S, Zhang W, He S, Hu C et al. TNF-alpha is a novel target of miR-19a. Int J Oncol 2011; 38: 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pezzolesi MG, Platzer P, Waite KA, Eng C. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet 2008; 82: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33. [DOI] [PubMed] [Google Scholar]
- Woo YM, Shin Y, Lee EJ, Lee S, Jeong SH, Kong HK et al. Inhibition of aerobic glycolysis represses Akt/mTOR/HIF-1 alpha axis and restores tamoxifen sensitivity in antiestrogen-resistant breast cancer cells. PLoS ONE 2015; 10: e0132285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.