Abstract
Type 2 diabetes (T2DM) is a common complex metabolic disorder that has a strong genetic predisposition. During the past decade, progress in genetic association studies has enabled the identification of at least 75 independent genetic loci for T2DM, thus allowing a better understanding of the genetic architecture of T2DM. International collaborations and large-scale meta-analyses of genome-wide association studies have made these achievements possible. However, whether the identified common variants are causal is largely unknown. In addition, the detailed mechanism of how these genetic variants exert their effect on the pathogenesis of T2DM requires further investigation. Currently, there are ongoing large-scale sequencing studies to identify rare, functional variants for T2DM. Environmental factors also have a crucial role in the development of T2DM. These could modulate gene expression via epigenetic mechanisms, including DNA methylation, histone modification and microRNA regulation. There is evidence that epigenetic changes are important in the development of T2DM. Recent studies have identified several DNA methylation markers of T2DM from peripheral blood and pancreatic islets. In this review, we will briefly summarize the recent progress in the genetic and epigenetic research on T2DM and discuss how environmental factors, genetics and epigenetics can interact in the pathogenesis of T2DM.
Introduction
According to the International Diabetes Federation's Diabetes Atlas sixth edition, it was estimated that 382 million people had diabetes in the year 2013 worldwide, and this figure is expected to increase to 592 million in 2035.1 In 2014, 4.9 million deaths were attributable to diabetes. It is also the leading cause of blindness, chronic kidney disease and amputation. Substantial effort to understand the pathophysiology of type 2 diabetes (T2DM) and to develop novel preventive and therapeutic measures for T2DM has been made in recent years.
One of these approaches relies on genetic research. This strategy is based on the findings that T2DM has a genetic predisposition. It has been reported that subjects having T2DM-affected siblings are at a two- to threefold increased risk of developing T2DM compared with the general population.2 Having one parent with diabetes increases the risk of T2DM by 30–40%, and having both parents with diabetes increases the risk by 70%.3 Furthermore, there are specific forms of monogenic diabetes that suggest a genetic etiology in diabetes. It should be noted that understanding the human genetic variation and its implications in disease is one of the core components of the precision medicine initiative.4 However, the explosive increase in the prevalence of T2DM during the past several decades cannot be explained by genetic factors alone, as it is unlikely that our genomes have changed during this relatively short time period.
Since the first genome-wide association study (GWAS) on T2DM in 2007, there has been considerable progress in genetic research on T2DM.5 There are at least 75 independent genetic loci associated with T2DM (Table 1). Technical advances in genotyping method, massive parallel sequencing and the development of global references for human genetic variations such as the 1000 Genomes Project have laid the foundation for these achievements.6, 7 In addition, there has been open collaboration between researchers in this field. Many of the major advances would not have been possible without these collaborative efforts.
Table 1. Common genetic variant association loci for T2DM.
| SNP | Chr | Pos | Genes | Context | Risk allele | Risk allele frequency | P-value | OR | First author | Pubmed ID |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17106184 | 1 | 50,444,313 | FAF1 | Intron | G | 0.92 | 4E-09 | 1.10 | Mahajan A | 24509480 |
| rs10923931 | 1 | 119,975,336 | NOTCH2, ADAM30 | Intron | T | 0.11 | 4E-08 | 1.13 | Zeggini E | 18372903 |
| rs340874 | 1 | 213,985,913 | PROX1 | Intron | C | 0.52 | 7E-10 | 1.07 | Dupuis J | 20081858 |
| rs780094 | 2 | 27,518,370 | GCKR | Intron | T | 0.39 | 1E-09 | 1.06 | Dupuis J | 20081858 |
| rs7578597 | 2 | 43,505,684 | THADA | Missense | T | 0.90 | 1E-09 | 1.15 | Zeggini E | 18372903 |
| rs243021 | 2 | 60,357,684 | BCL11A | Intergenic | A | 0.50 | 3E-15 | 1.08 | Voight BF | 20581827 |
| rs7560163 | 2 | 150,781,422 | RBM43, RND3 | Intergenic | C | 0.86 | 7E-09 | 1.33 | Palmer ND | 22238593 |
| rs7593730 | 2 | 160,314,943 | ITGB6, RBMS1 | Intron | C | 0.78 | 4E-08 | 1.11 | Qi L | 20418489 |
| rs3923113 | 2 | 164,645,339 | GRB14 | Intergenic | A | 0.74 | 1E-08 | 1.09 | Kooner JS | 21874001 |
| rs7578326 | 2 | 226,155,937 | IRS1 | Intron | A | 0.71 | 5E-20 | 1.11 | Voight BF | 20581827 |
| rs1801282 | 3 | 12,351,626 | PPARG | Missense | C | 0.88 | 6E-10 | 1.16 | Mahajan A | 24509480 |
| rs7612463 | 3 | 23,294,959 | UBE2E2 | Intron | C | 0.87 | 7E-09 | 1.10 | Mahajan A | 24509480 |
| rs831571 | 3 | 64,062,621 | PSMD6 | Intergenic | C | 0.61 | 8E-11 | 1.09 | Cho YS | 22158537 |
| rs4607103 | 3 | 64,726,228 | ADAMTS9 | Intron | C | 0.76 | 1E-08 | 1.09 | Zeggini E | 18372903 |
| rs11717195 | 3 | 123,363,551 | ADCY5 | Intron | T | 0.78 | 2E-08 | 1.09 | Mahajan A | 24509480 |
| rs4402960 | 3 | 185,793,899 | IGF2BP2 | Intron | T | 0.29 | 2E-09 | 1.17 | Saxena R | 17463246 |
| rs16861329 | 3 | 186,948,673 | ST6GAL1 | Intron | G | 0.75 | 3E-08 | 1.09 | Kooner JS | 21874001 |
| rs6808574 | 3 | 188,022,735 | LPP | Intergenic | C | 0.79 | 6E-09 | 1.07 | Mahajan A | 24509480 |
| rs6815464 | 4 | 1,316,113 | MAEA | Intron | C | 0.58 | 2E-20 | 1.13 | Cho YS | 22158537 |
| rs1801214 | 4 | 6,301,295 | WFS1 | Missense | T | 0.73 | 3E-08 | 1.13 | Voight BF | 20581827 |
| rs6813195 | 4 | 152,599,323 | TMEM154 | Intergenic | C | 0.59 | 4E-14 | 1.08 | Mahajan A | 24509480 |
| rs702634 | 5 | 53,975,590 | ARL15 | Intron | A | 0.76 | 7E-09 | 1.06 | Mahajan A | 24509480 |
| rs4457053 | 5 | 77,129,124 | ZBED3 | Intron | G | 0.20 | 3E-12 | 1.08 | Voight BF | 20581827 |
| rs9502570 | 6 | 7,258,384 | RREB1, SSR1 | Intergenic | A | 0.45 | 1E-09 | 1.06 | Mahajan A | 24509480 |
| rs7754840 | 6 | 20,661,019 | CDKAL1 | Intron | C | 0.36 | 4E-11 | 1.12 | Scott LJ | 17463248 |
| rs3132524 | 6 | 31,168,937 | TCF19, POU5F1 | Intron | G | 0.38 | 4E-09 | 1.07 | Mahajan A | 24509480 |
| rs9470794 | 6 | 38,139,068 | ZFAND3 | Intron | C | 0.27 | 2E-10 | 1.12 | Cho YS | 22158537 |
| rs1535500 | 6 | 39,316,274 | KCNK16 | Missense | T | 0.42 | 2E-08 | 1.08 | Cho YS | 22158537 |
| rs1048886 | 6 | 70,579,486 | C6orf57 | Missense | G | 0.18 | 3E-08 | 1.54 | Sim X | 21490949 |
| Rs2191349 | 7 | 15,024,684 | DGKB, TMEM195 | Intergenic | T | 0.52 | 1E-08 | 1.06 | Dupuis J | 20081858 |
| rs864745 | 7 | 28,140,937 | JAZF1 | Intron | T | 0.50 | 5E-14 | 1.10 | Zeggini E | 18372903 |
| rs4607517 | 7 | 44,196,069 | GCK | Intergenic | A | 0.16 | 5E-08 | 1.07 | Dupuis J | 20081858 |
| rs6467136 | 7 | 127,524,904 | PAX4, GCC1 | Intergenic | G | 0.79 | 5E-11 | 1.11 | Cho YS | 22158537 |
| rs791595 | 7 | 128,222,749 | MIR129, LEP | Intron | A | 0.08 | 3E-13 | 1.17 | Hara K | 23945395 |
| rs972283 | 7 | 130,782,095 | KLF14 | Intergenic | G | 0.69 | 2E-10 | 1.07 | Voight BF | 20581827 |
| rs515071 | 8 | 41,661,944 | ANK1 | Missense | G | 0.79 | 1E-08 | 1.18 | Imamura M | 22456796 |
| rs896854 | 8 | 94,948,283 | TP53INP1 | Missense | T | 0.24 | 1E-09 | 1.06 | Voight BF | 20581827 |
| rs13266634 | 8 | 117,172,544 | SLC30A8 | Missense | C | 0.69 | 5E-08 | 1.12 | Zeggini E | 17463249 |
| rs7041847 | 9 | 4,287,466 | GLIS3 | Intron | A | 0.41 | 2E-14 | 1.10 | Cho YS | 22158537 |
| rs17584499 | 9 | 8,879,118 | PTPRD | Intron | T | 0.06 | 9E-10 | 1.57 | Tsai FJ | 20174558 |
| rs10811661 | 9 | 22,134,095 | CDKN2A, CDKN2B | Intergenic | T | 0.83 | 5E-08 | 1.20 | Saxena R | 17463246 |
| rs13292136 | 9 | 79,337,213 | CHCHD9 | Intergenic | C | 0.90 | 3E-08 | 1.11 | Voight BF | 20581827 |
| rs11787792 | 9 | 136,357,696 | GPSM1 | Intron | A | 0.87 | 2E-10 | 1.15 | Hara K | 23945395 |
| rs12779790 | 10 | 12,286,011 | CDC123, CAMK1D | Intergenic | G | 0.18 | 1E-10 | 1.11 | Zeggini E | 18372903 |
| rs1802295 | 10 | 69,171,718 | VPS26A | UTR-3 | A | 0.26 | 4E-08 | 1.08 | Kooner JS | 21874001 |
| rs12571751 | 10 | 79,182,874 | ZMIZ1 | Intron | A | 0.51 | 2E-10 | 1.09 | Mahajan A | 24509480 |
| rs1111875 | 10 | 92,703,125 | HHEX | Intergenic | C | 0.52 | 6E-10 | 1.13 | Scott LJ | 17463248 |
| rs7903146 | 10 | 112,998,590 | TCF7L2 | Intron | T | 0.30 | 2E-34 | 1.65 | Sladek R | 17293876 |
| rs10886471 | 10 | 119,389,891 | GRK5 | Intron | C | 0.78 | 7E-09 | 1.12 | Li H | 22961080 |
| rs3842770 | 11 | 2,157,440 | INS-IGF2 | Intron | A | 0.23 | 3E-08 | 1.14 | Ng MC | 25102180 |
| rs2237892 | 11 | 2,818,521 | KCNQ1 | Intron | C | 0.61 | 2E-42 | 1.40 | Yasuda K | 18711367 |
| rs5215 | 11 | 17,387,083 | KCNJ11 | Missense | C | 0.27 | 5E-11 | 1.14 | Zeggini E | 17463249 |
| rs1552224 | 11 | 72,722,053 | CENTD2 | Intron | A | 0.45 | 1E-22 | 1.14 | Voight BF | 20581827 |
| rs1387153 | 11 | 92,940,662 | MTNR1B | Intergenic | T | 0.35 | 8E-15 | 1.09 | Voight BF | 20581827 |
| rs1531343 | 12 | 65,781,114 | HMGA2 | Intron | C | 0.21 | 4E-09 | 1.10 | Voight BF | 20581827 |
| rs7961581 | 12 | 71,269,322 | LGR5, TSPAN8 | Intergenic | C | 0.27 | 1E-09 | 1.09 | Zeggini E | 18372903 |
| rs7957197 | 12 | 121,022,883 | HNF1A | Intron | T | 0.89 | 2E-08 | 1.07 | Voight BF | 20581827 |
| rs1727313 | 12 | 123,156,306 | MPHOSPH9 | NcRNA | C | 0.50 | 1E-08 | 1.06 | Mahajan A | 24509480 |
| rs9552911 | 13 | 23,290,518 | SGCG, SACS | Intron | G | 0.93 | 2E-08 | 1.49 | Saxena R | 23300278 |
| rs1359790 | 13 | 80,143,021 | SPRY2 | Intergenic | G | 0.71 | 6E-09 | 1.15 | Shu XO | 20862305 |
| rs7403531 | 15 | 38,530,704 | RASGRP1 | Intron | T | 0.35 | 4E-09 | 1.10 | Li H | 22961080 |
| rs7172432 | 15 | 62,104,190 | C2CD4B, C2CD4A | Intergenic | A | 0.58 | 9E-14 | 1.11 | Yamauchi T | 20818381 |
| rs7178572 | 15 | 77,454,848 | HMG20A | Intron | G | 0.70 | 1E-08 | 1.11 | Perry JR | 22693455 |
| rs11634397 | 15 | 80,139,880 | ZFAND6 | Intergenic | G | 0.44 | 2E-09 | 1.06 | Voight BF | 20581827 |
| rs2028299 | 15 | 89,831,025 | AP3S2 | NcRNA | C | 0.31 | 2E-11 | 1.10 | Kooner JS | 21874001 |
| rs8042680 | 15 | 90,978,107 | PRC1 | Intron; intron | A | 0.74 | 2E-10 | 1.07 | Voight BF | 20581827 |
| rs8050136 | 16 | 53,782,363 | FTO | Intron | A | 0.38 | 1E-12 | 1.17 | Scott LJ | 17463248 |
| rs391300 | 17 | 2,312,964 | SRR | Intron | G | 0.62 | 3E-09 | 1.28 | Tsai FJ | 20174558 |
| rs312457 | 17 | 7,037,074 | SLC16A13 | Intron | G | 0.08 | 8E-13 | 1.20 | Hara K | 23945395 |
| rs4430796 | 17 | 37,738,049 | HNF1B | Intron | G | 0.28 | 2E-11 | 1.19 | Li H | 22961080 |
| rs8090011 | 18 | 7,068,463 | LAMA1 | Intron | G | 0.38 | 8E-09 | 1.13 | Perry JR | 22693455 |
| rs12970134 | 18 | 60,217,517 | MC4R | Intergenic | A | 0.27 | 3E-08 | 1.08 | Mahajan A | 24509480 |
| rs3786897 | 19 | 33,402,102 | PEPD | Intron | A | 0.56 | 1E-08 | 1.10 | Cho YS | 22158537 |
| rs4812829 | 20 | 44,360,627 | HNF4A | Intron | A | 0.29 | 3E-10 | 1.09 | Kooner JS | 21874001 |
| rs5945326 | 23 | 153,634,467 | DUSP9 | Intergenic | A | 0.73 | 3E-10 | 1.27 | Voight BF | 20581827 |
Abbreviations: Chr, chromosome number; Genes, nearby gene reported by the original article noted by the PUBMED ID; NcRNA, non-coding RNA; OR, odds ratio; Pos, physical position based on GRCh38/hg38; SNP, variant rsID; T2DM, type 2 diabetes mellitus; UTR, untranslated region.
Modified from GWAS catalog (https://www.ebi.ac.uk/gwas/). The values of risk allele frequency, P-value and OR are from the first published article as noted by the PUBMED ID. Last accessed 5 November 2015. Variants within 500-kb window were regarded to be in the same loci.
Major critics of genetic research on T2DM highlight the fact that the common variants with a relatively low effect size (odds ratio between 1.10 and 1.40) explain only 10–15% of the T2DM heritability.8 In addition, most of the variants are located at intergenic or intronic region, where it is difficult to explain their functional consequences. In the past several years, efforts were made to reveal the functional role of these genetic variants or those of newly implicated genes in T2DM. On the other hand, there are ongoing studies that investigate additional layers of genomic regulation, such as epigenetics. Environmental factors may exert their effect on epigenetic changes and interact with DNA sequence variations. In this review, we will briefly summarize the recent progress in genetic and epigenetic research on T2DM and discuss how environmental factors, genetics and epigenetics can interact in the pathogenesis of T2DM.
Genetic research on T2DM
Initial GWAS on T2DM
The first report of GWAS on T2DM appeared in early 2007 and identified the novel T2DM-associated loci HHEX/IDE and SLC30A8.5 Soon after, three international consortia reported their own GWAS and simultaneously replicated these results.9, 10, 11 Through collaboration, they confirmed the previous T2DM loci of TCF7L2, PPARG, KCNJ11 and FTO, and identified novel loci for an association in the intergenic region of CDKN2A/2B, as well as the intronic region of CDKAL1 and IGF2BP2. These studies showed that most of the common genetic variants had a modest effect size, with an odds ratio between 1.10 and 1.40, and that multiple variants with a modest effect size would contribute to the genetic etiology of T2DM.
Meta-analysis of GWAS
After the initial success of GWAS, it was recognized that a large sample size is required to identify novel genetic loci. Accordingly, in 2008 the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) study meta-analyzed previous GWAS results encompassing more than 10 000 subjects for an initial genome-wide scan and performed independent replication in more than 53 000 subjects.12 They found six novel loci associated with T2DM in/near JAZF1, CDC123-CAMK1D, TSPAN8-LGR5, THADA, ADAMTS9 and NOTCH2. A further increase in sample size was available in the DIAGRAM+ study in 2012, with a sample size over 101 000. This study identified 12 novel loci associated with T2DM.13 These studies were performed in European descendants.
GWAS on non-Europeans
The first GWASs in East Asians were reported in 2008.14, 15 They found that a variant in KCNQ1 was significantly associated with T2DM in Japanese, Koreans and Chinese. This variant also showed nominal significance in Europeans. Interestingly, meta-analysis of GWAS in Europeans identified a second independent variant in KCNQ1, which was not in linkage disequilibrium with the original variant identified in East Asians.13 The meta-analysis of GWAS in East Asians was led by the Asian Genetic Epidemiology Network (AGEN) in 2011.16, 17 They found eight novel loci associated with T2DM in more than 54 000 cases and controls. These include variants in/near GLIS3, PEPD, FITM2-R3HDML-HNF4A, KCNK16, MAEA, GCC1-PAX4, PSMD6 and ZFAND. GWAS was also performed in South Asians and African Americans.18, 19 In Mexicans and other Latin Americans, a novel locus spanning SLC16A11 and SLC16A13 was significantly associated with T2DM.20 This variant was relatively common in native Americans (minor allele frequency (MAF) of ~50%) and East Asians (MAF of ~10%), but was rare in Europeans and Africans. A functional study suggested that the SLC16A11 gene is involved in triglyceride metabolism. Recently, a meta-analysis of GWAS in African Americans identified two novel loci at HLA-B and INS-IGF2.21
Large-scale genotyping using the Metabochip
To further increase the sample size, a cost-effective custom genotyping array was developed. The ‘Metabochip' included nearly 200 000 variants to investigate the genetic association for metabolic, cardiovascular and anthropometric traits.22 For T2DM, the chip contained ~5000 variants to validate previously suggested loci and ~17 000 additional variants for fine mapping of previously confirmed loci. The Metabochip was used for the genotyping of nearly 150 000 Europeans and successfully identified eight novel loci for T2DM.23 They confirmed 63 T2DM loci, and the sibling relative risk attributable to these loci was estimated to be 1.104. These variants explained 5.7% of the variance in T2DM risk on a liability scale. When a polygenic linear mixed model was applied, the overall genome-wide common variants explained 49–64% of the liability variance in Europeans. In addition, pathway analyses and protein–protein interaction analyses revealed that CREB-binding protein-related transcription, adipokine signaling and cell cycle signaling processes are important in the pathogenesis of T2DM. Another study using Metabochip in an isolated founder population in Greenland identified a common (MAF 17%) nonsense p.Arg684Ter variant in TBC1D4 that was associated with an elevated 2-h glucose and insulin concentration after an oral glucose challenge and was also a risk factor for T2DM.24 A functional study using muscle biopsies of participants suggested that this variant is associated with impaired insulin-dependent glucose disposal.
Trans-ancestry GWAS meta-analysis
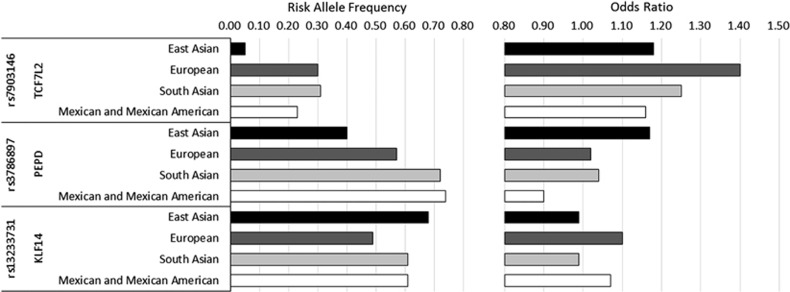
To further identify novel T2DM-associated loci and to fine map the previously reported loci, trans-ancestry GWAS meta-analysis was performed.25 This effort included the meta-analysis of multiple ethnic specific GWAS results obtained from Europeans, East Asians, South Asians and Mexican Americans. The sample size increased to more than 187 000 and successfully identified seven novel T2DM loci. Among the 52 variants that were available in all 4 ancestry groups, 34 showed directional consistency across the 4 groups. However, 3 variants showed a significant difference in the effect size among the 4 ancestry groups (Figure 1). The risk allele frequency of rs7903146 (TCF7L2) was lower in East Asians (5%) compared with Europeans (30%), and the association of this variant with T2DM did not reach genome-wide significance in East Asians. A variant in PEPD, rs3786897, was only significant in East Asians, and a variant in KLF14, rs13233731, was only significant in Europeans. In this study, the authors used the difference in linkage disequilibrium among ethnic groups to refine the association signal in previously confirmed loci. Using this trans-ethnic approach, the 99% credible set of variants and their genomic interval was significantly decreased for loci including JAZF1 and SLC30A8.
Figure 1.
Three variants that showed significant heterogeneity in effect size among the four ancestry groups (modified from Mahajan et al.25).
Sequencing studies
As the common variants only had a modest effect on T2DM, it has been hypothesized that rare- or low-frequency variants with a large functional effect might explain a significant proportion of the T2DM heritability. Technical advances in massive parallel sequencing, variant calling and association testing made it possible to investigate the role of these rare- or low-frequency variants in a large number of samples. As MTNR1B is a relatively small gene with only two exons, it was one of the first genes to be sequenced at a large scale. Experimentally validated loss-of-function variants of MTNR1B were significantly associated with T2DM.26 In a similar manner, rare variants of PPARG, which resulted in decreased adipocyte differentiation, were collectively associated with T2DM.27 Regarding SLC30A8, which encodes an important protein (ZnT8) involved in insulin secretion, rare protein-truncating variants were associated with a decreased risk of T2DM.28 It is currently suggested that SLC30A8 could be a novel therapeutic target for T2DM. In one study, whole-genome sequencing was performed in 2630 Icelanders and was then used as a reference in the imputation of 278 000 additional subjects.29 They found an intronic low-frequency (1.5%) variant in CCND2, which was associated with decreased risk of T2DM. In addition, nonsynonymous variants in PAM (p.Asp563Gly MAF 5% and p.Ser539Trp MAF 0.7%) and a rare (0.2%) frameshift variant in PDX1 (Gly218Alafs*12) were associated with increased risk of T2DM. In the Latino population, whole-exome sequencing was performed in 3756 participants. A rare nonsynonymous variant in HNF1A p.E508K was associated with a fivefold increased risk of T2DM (MAF in T2DM 2.1% vs controls 0.35%).30 Currently, there are ongoing large-scale international collaborative studies to investigate the overall effect of rare, functional variants in the development of T2DM.
Glycemic trait analysis
In parallel to efforts aimed at identifying variants of T2DM, several studies investigated the genetic variants associated with quantitative glycemic traits such as fasting glucose, fasting insulin, 2-h glucose after oral glucose challenge and HbA1c. One of the first GWAS of glycemic traits identified that an intronic variant in G6PC2 was significantly associated with the fasting glucose concentration.31 The gene is specifically expressed in pancreatic islets and has been suggested to have a role in glucose-dependent insulin secretion. However, this variant was not associated with the risk of T2DM. By the collaborative efforts of the Meta-Analysis of Glucose and Insulin-related traits Consortium (MAGIC), a variant in MTNR1B was identified to be associated with fasting glucose, but this time the variant was associated with an increased risk of T2DM.32, 33, 34 The MAGIC study also identified an intronic variant of GIPR that was associated with the 2-h glucose concentration.35 In 2010, a large-scale meta-analyses of GWAS in MAGIC identified nine novel loci for fasting glucose (in/near ADCY5, MADD, ADRA2A, CRY2, FADS1, GLIS3, SLC2A2, PROX1 and C2CD4B) and one for fasting insulin (near IGF1).36 It was found that some of the fasting glucose-elevating variants were associated with increased risk of T2DM. However, this overlap was incomplete, suggesting that some variants are affecting the fasting glucose concentration only at the physiologic level. Using Metabochip, the MAGIC study increased their sample size to more than 133 000 and confirmed 53 glycemic trait-associated loci.37 The AGEN consortium identified three variants (in/near PDK1-RAPGEF4, KANK1 and IGF1R) that were associated with fasting glucose in East Asians.38 Finally, there were efforts to identify low- or rare-frequency variants of glycemic traits. An exome array genotyping study in Europeans identified a low-frequency (MAF 1.5%) nonsynonymous variant in GLP1R (p.Ala316Thr), which was associated with fasting glucose and a rare (MAF 0.1%) nonsynonymous variant in URB2 (p.Glu594Val), which was associated with fasting insulin.39 In addition, multiple nonsynonymous variants in G6PC2, which resulted in loss of function, were associated with decreased fasting glucose concentrations. A gene-based variant aggregation test also confirmed that rare nonsynonymous variants in G6PC2 were associated with the fasting glucose concentrations.40
Epigenetic research on T2DM
Introduction to epigenetics
Epigenetics is defined as ‘the study of changes in gene function mitotically and/or meiotically heritable and that do not entail a change in DNA sequence'.41 These changes are influenced by environmental factors and can modulate gene expression independent of DNA sequence. An important characteristic of epigenetic change is that it is reversible and modifiable, making it a possible therapeutic target. Epigenetic regulation is mainly controlled by DNA methylation, histone modification and microRNA. Although epigenetics adds another layer of complexity in understanding the genomic pathogenesis of T2DM, it could provide precious information on how environmental factors contribute to T2DM at the genomic level. In addition, it might explain another significant proportion of the variance in T2DM susceptibility. The field of epigenetic research on T2DM is relatively new due to difficulties in acquiring a large number of homogenous target tissues and the technical limitations in analyzing epigenetic changes at a genome-wide scale. In this review, we will focus on DNA methylation, as it has been the most widely investigated strategy up until now.
Evidence that epigenetic changes contribute to T2DM
There are several lines of evidence suggesting that epigenetic changes have an important role in the pathogenesis of T2DM. First, intrauterine malnutrition has been linked to T2DM. Epidemiologically, offspring born from mothers who experienced famine during pregnancy in the Dutch Hunger Winter or World War II had significantly decreased birth weight and were at high risk of future T2DM.42, 43 Several mechanisms to explain this ‘thrifty phenotype' include altered epigenetic modifications in pancreatic β-cells44 and decreased mitochondrial DNA content.45 These changes could be mediated by low levels of methyl donors and depletion of nucleotide pools. Second, intrauterine exposure to hyperglycemia can lead to diabetes and obesity in offspring. In a study on Pima Indian families, offspring conceived after the mother was diagnosed with T2DM were more likely to have a higher body mass index and a greater risk of T2DM compared with siblings born before their mother developed T2DM.46 Third, environmental factors can affect β-cell function by mediating epigenetic changes. In a recent study, palmitate exposure to human pancreatic islets induced global and specific DNA methylation alterations that resulted in coordinate changes in mRNA expression and decreased insulin secretion.47
Technical advances in epigenetic research
Recent technical advances have accelerated the field of epigenetic research on T2DM. Several DNA methylation arrays are commercially available to investigate epigenome-wide associations. Illumina's Infinium HumanMethylation450 BeadChip interrogates more than 485 000 methylation sites and covers 96% of CpG islands and additional island shores and their flanking regions.48 Another method to investigate genome-wide methylation changes is to perform bisulfite conversion and apply next generation sequencing. An unprecedented resource of information is provided by the Encyclopedia of DNA Elements (ENCODE) project and Roadmap Epigenomics project.49, 50 In the ENCODE project, functional DNA elements were delineated using sequencing studies in a number of cell lines. In the Roadmap Epigenomics project, human tissues and cells were investigated and reference epigenomes for 127 tissues and cell types were generated and are publically available.
Recent progress in epigenetic research on T2DM
One of the first epigenome-wide association studies to identify T2DM-related DNA methylation variations in peripheral blood was reported in 2011.51 DNA from a total of 1169 T2DM cases and controls was assembled in four groups of DNA pools and was investigated with microarray-based methylation assay. They found that known GWAS loci were enriched with differentially methylated sites. In addition, hypomethylation of a CpG site in the FTO gene was significantly associated with the risk of T2DM. Recently, three studies independently reported that differential methylation at a CpG site in TXNIP, cg19693031, was significantly associated with T2DM.52, 53, 54 TXNIP has been shown to be involved in human skeletal muscle glucose uptake and glucotoxicity-induced β-cell apoptosis.55, 56 In the nested case–control study report by Chambers et al., peripheral blood DNA was investigated with the HumanMethylation450 BeadChip in 2664 Indian Asians and further replication was performed in 1141 Europeans. In addition to TXNIP, the authors found that CpG sites in ABCG1, PHOSPHO1, SOCS3 and SREBF1 were significantly associated with future development of T2DM.
Because epigenetic regulation is tissue specific, it is crucial to investigate a homogenous tissue or cell type from a T2DM target organ. In this regard, pancreatic islets are one of the most thoroughly investigated cell clusters. In 2010, Stitzel et al. investigated the genome-wide analysis of epigenetic markers of DNase I hypersensitivity sites, histone modifications and CCCTC factor binding in primary human pancreatic islets.57 They found over 34 000 distal regulatory elements, among which 47% were specific to islets. In addition, known T2DM genetic loci were enriched in these regulatory elements. Similarly, it has been reported that known T2DM loci were enriched in pancreatic islet enhancer clusters.58 One of the first studies to compare genome-wide methylation in human pancreatic islets between 15 T2DM cases and 34 non-diabetic controls was reported in 2014.59 A total of 1649 CpG sites and 853 genes, including known T2DM loci of TCF7L2, FTO and KCNQ1, were identified to be differentially methylated in T2DM using the HumanMethylation450 BeadChip. In addition, 102 genes showed both differential methylation and gene expression in T2DM islets. Some of these genes affected pancreatic β-cell and α-cell functions. Regarding human skeletal muscle, it has been shown that T2DM subjects had cytosine hypermethylation of PGC-1α and decreased mitochondrial content in vastus lateralis muscle.60 In addition, acute exercise induced promoter hypomethylation and a coordinate increase in the expression of PGC-1α, PDK4 and PPAR-δ in skeletal muscle.61 In human adipose tissue, 6 months of exercise training resulted in differential DNA methylation and mRNA expression in 197 genes, including RALBP1, HDAC4 and NCOR2.62 A total of 39 genes in previously confirmed obesity or T2DM loci had CpG sites that were differentially methylated after exercise training. Furthermore, knockdown of the candidate Hcad4 and Ncor2 in adipocytes resulted in increased lipogenesis in vitro. These findings imply that there are tissue-specific epigenetic changes in T2DM that are at least in part co-regulated by DNA sequence variations.
Interaction between environment, genetics and epigenetics in T2DM
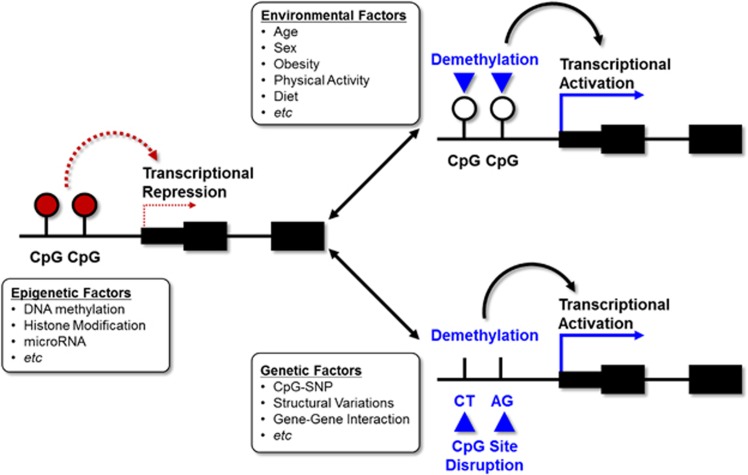
The pathogenesis of T2DM is thought to involve a complex interaction between genetic and environmental factors. Although there could be various mechanisms for a gene–environment interaction, we will specifically focus on epigenetic modulations. Environmental factors can exert their effect by modulating epigenetic changes. Epigenetic change can also be determined by genetic factors. There is substantial evidence for these interactions. In an interesting report by Kim et al., the authors explicitly showed how environmental factors such as obesity could change the promoter methylation state of adiponectin, a well-known adipokine that regulates insulin sensitivity.63 In the obese condition, DNA methyltransferase 1 is activated and hypermethylates a specific region of the adiponectin gene, leading to decreased expression. It should be noted that confirmed genetic loci (rs17300539 and rs266729) for the plasma adiponectin concentration reside at the CpG site of the adiponectin promoter and can either introduce or remove the CpG site according to the genotype (termed CpG-single nucleotide polymorphism (SNP)). Regarding resistin, another well-known adipokine, promoter DNA variants were associated with decreased methylation of cg02346997, which is also located at the resistin promoter.64 The methylation state of cg02346997 was associated with a significantly decreased plasma resistin concentration. These findings highlight the complex interaction between the environment, genetics and epigenetics. The model for these interactions is depicted in Figure 2. Epigenetics can modulate tissue-specific gene expression through DNA methylation, histone modification and microRNA. These epigenetic regulations can be influenced by environmental factors such as age, obesity, physical activity and diet. Furthermore, DNA sequence variations such as CpG-SNP, structural variations and gene–gene interactions can also modulate epigenetic regulation.
Figure 2.
The complex interaction between various environmental factors, genetics and epigenetics.
Summary and future directions
There has been great success in identifying genetic variants associated with T2DM using large-scale GWAS and their meta-analyses. So far, at least 75 T2DM-associated common genetic variants have been identified. However, detailed mechanisms on how these variants exert their effects on the pathogenesis of T2DM have been largely lacking. Although international collaborative studies on whole-exome and whole-genome sequencing are expected to be reported soon, it remains unknown to what extent the heritability of T2DM could be explained by low- or rare-frequency functional genetic variants. Once we have detailed information on both rare and common genetic variations and their effects on T2DM, it will become necessary to shift our focus to another layer of genomics that could encompass environmental factors, such as epigenetics. As the technology to investigate genome-wide epigenetic changes is expected to further advance, it will be important to collect high-quality tissues or cells that have an important role in the pathogenesis of T2DM. Future research should focus on the interaction between various environmental factors, genetics and epigenetics. Through these efforts, we hope that detailed information on these multilayered genomics could provide a foundation for precision medicine and improve outcomes in the prevention and treatment of T2DM.
Acknowledgments
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grant no. HI14C0060).
The authors declare no conflict of interest.
References
- Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 2014; 103: 137–149. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X, Sundquist K, Sundquist J. Familial risks for type 2 diabetes in Sweden. Diabetes Care 2010; 33: 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes 2000; 49: 2201–2207. [DOI] [PubMed] [Google Scholar]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015; 372: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445: 881–885. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med 2010; 363: 2339–2350. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007; 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007; 316: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University,, Novartis Institutes of BioMedical Research, Saxena R, Voight BF, Lyssenko V et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007; 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010; 42: 579–U155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008; 40: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008; 40: 1092–1097. [DOI] [PubMed] [Google Scholar]
- Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet 2012; 44: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go MJ, Hwang JY, Park TJ, Kim YJ, Oh JH, Kim YJ et al. Genome-wide association study identifies two novel Loci with sex-specific effects for type 2 diabetes mellitus and glycemic traits in a korean population. Diabetes Metab J 2014; 38: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 2011; 43: 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ND, McDonough CW, Hicks PJ, Roh BH, Wing MR, An SS et al. A genome-wide association search for type 2 diabetes genes in African Americans. PLoS ONE 2012; 7: e29202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGMA Type 2 Diabetes ConsortiumSIGMA Type 2 Diabetes ConsortiumWilliams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014; 506: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ND, Ng MC, Hicks PJ, Mudgal P, Langefeld CD, Freedman BI et al. Evaluation of candidate nephropathy susceptibility genes in a genome-wide association study of African American diabetic kidney disease. PLoS ONE 2014; 9: e88273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet 2012; 8: e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012; 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 2014; 512: 190–193. [DOI] [PubMed] [Google Scholar]
- DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014; 46: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond A, Clement N, Fawcett K, Yengo L, Vaillant E, Guillaume JL et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 2012; 44: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci USA 2014; 111: 13127–13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, Thorleifsson G, Beer NL, Jacobs SB, Grarup N, Burtt NP et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet 2014; 46: 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet 2014; 46: 294–298. [DOI] [PubMed] [Google Scholar]
- SIGMA Type 2 Diabetes Consortium, Estrada K, Aukrust IA, Bjorkhaug L, Burtt NP, Mercader JM et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 2014; 311: 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 2008; 320: 1085–1088. [DOI] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009; 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009; 41: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009; 41: 89–94. [DOI] [PubMed] [Google Scholar]
- Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 2010; 42: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010; 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012; 44: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Sim X, Wu Y, Liang J, Tabara Y, Hu C et al. Genome-wide association meta-analysis identifies novel variants associated with fasting plasma glucose in East Asians. Diabetes 2015; 64: 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM et al. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet 2015; 11: e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun 2015; 6: 5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science 2001; 293: 1103–1105. [DOI] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 1992; 35: 595–601. [DOI] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet 1998; 351: 173–177. [DOI] [PubMed] [Google Scholar]
- Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 2008; 118: 2316–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Park KS, Pak YK, Lee HK. The role of mitochondrial DNA in the development of type 2 diabetes caused by fetal malnutrition. J Nutr Biochem 2005; 16: 195–204. [DOI] [PubMed] [Google Scholar]
- Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes 2000; 49: 2208–2211. [DOI] [PubMed] [Google Scholar]
- Hall E, Volkov P, Dayeh T, Bacos K, Ronn T, Nitert MD et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med 2014; 12: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics 2009; 1: 177–200. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet 2012; 21: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 2015; 3: 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schottker B et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1500 older adults. Diabetologia 2015; 59: 130–138. [DOI] [PubMed] [Google Scholar]
- Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW Jr., Goring HH et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet 2015; 24: 5330–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med 2007; 4: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology 2005; 146: 2397–2405. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab 2010; 12: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 2014; 46: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 2014; 10: e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 2009; 10: 189–198. [DOI] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 2012; 15: 405–411. [DOI] [PubMed] [Google Scholar]
- Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 2013; 9: e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AY, Park YJ, Pan X, Shin KC, Kwak SH, Bassas AF et al. Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nat Commun 2015; 6: 7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatochi M, Ichihara S, Yamamoto K, Ohnaka K, Kato Y, Yokota S et al. Epigenome-wide association study suggests that SNPs in the promoter region of RETN influence plasma resistin level via effects on DNA methylation at neighbouring sites. Diabetologia 2015; 58: 2781–2790. [DOI] [PubMed] [Google Scholar]