Abstract
The modulation of AMPA receptor (AMPAR) content at synapses is thought to be an underlying molecular mechanism of memory and learning. AMPAR content at synapses is highly plastic and is regulated by numerous AMPAR accessory transmembrane proteins such as TARPs, cornichons, and CKAMPs. SynDIG (Synapse Differentiation Induced Gene) defines a family of four genes (SynDIG1-4) expressed in distinct and overlapping patterns in the brain. SynDIG1 was previously identified as a novel transmembrane AMPAR-associated protein that regulates synaptic strength. The related protein SynDIG4 [also known as Prrt1 (Proline rich transmembrane protein 1)] has recently been identified as a component of AMPAR complexes. In this study, we show that SynDIG1 and SynDIG4 have distinct yet overlapping patterns of expression in the central nervous system, with SynDIG4 having especially prominent expression in the hippocampus, and particularly within CA1. In contrast to SynDIG1 and other traditional AMPAR auxiliary subunits, SynDIG4 is de-enriched at the post synaptic density and colocalizes with extrasynaptic GluA1 puncta in primary dissociated neuron culture. These results indicate that although SynDIG4 shares sequence similarity with SynDIG1 it might act through a unique mechanism as an auxiliary factor for extrasynaptic GluA1 containing AMPARs.
Keywords: Prrt1, Synapse Differentiation Induced Gene (SynDIG) family, SynDIG4, AMPA receptor, auxiliary factor, rat brain, excitatory synapse, RRID:SCR_013715, RRID:ScrRes_000161, RRID:nif-0000-30467, RRID:RGD_70508, RRID:AB_2491106, RRID:AB_390919, RRID:AB_10999753, RRID:AB_309885, RRID:AB_2232661, RRID:AB_2113602, RRID:AB_2307331, RRID:AB_887824, RRID:AB_2301751, RRID:AB_2277296
Introduction
The two primary types of ionotropic glutamate receptors at excitatory synapses in the central nervous system are α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs), and N-methyl-D-aspartate receptors (NMDARs). During development, NMDARs are initially recruited to the site of contact between axons and dendrites to establish nascent synapses followed by the subsequent recruitment of AMPARs to generate mature synapses [reviewed in (McAllister, 2007; Hall and Ghosh, 2008; Bury and Sabo, 2010)]. The number of AMPARs at synapses, as well as their subunit composition, is highly dynamic and their regulation at synapses is implicated in several types of plasticity such as long term potentiation (LTP) [reviewed in (Huganir and Nicoll, 2013)] and synaptic scaling [reviewed in (Turrigiano, 2012; Lee et al., 2014)]. AMPAR expression and function is regulated through diverse mechanisms, including dynamic changes in phosphorylation state, and interaction with a large and varied group of interacting proteins acting as AMPAR auxiliary factors. Stargazin was the first auxiliary factor identified in the transmembrane AMPAR regulating protein (TARP) family, and was shown to alter functional properties as well as trafficking to the cell surface of AMPARs (Chen et al., 2000). Since the discovery of TARPs, several other protein families such as Cornichons (CNIHs) and Cysteine-knot AMPAR modulating proteins (CKAMPs) have been identified as auxiliary factors for AMPARs with distinct and overlapping functions (Chen et al., 2000; Schwenk et al., 2009; Diaz, 2010; von Engelhardt et al., 2010; Jackson and Nicoll, 2011; Shanks et al., 2012; Chen et al., 2014; Schwenk et al., 2014)]. For example, both CNIHs and CKAMPs are enriched at the postsynaptic density (PSD) and act to prolong the deactivation of AMPARs. However, CKAMP44 will increase the rate of desensitization of the AMPAR after glutamate stimulation, whereas CNIHs will decrease the rate of desensitization (Shi et al., 2010; von Engelhardt et al., 2010).
SynDIG1 (Synapse Differentiation Induced Gene 1) is a type II transmembrane protein identified in a microarray based approach as a gene involved in neuronal differentiation during pontocerebellar development (Diaz et al., 2002). SynDIG1 interacts with AMPARs in heterologous cells and regulates AMPAR content and clustering at synapses (Kalashnikova et al., 2010). Overexpression of SynDIG1 increased the number of functional AMPAR containing synapses, but not NMDAR containing synapses. Knock down of SynDIG1 showed the opposite effect and decreased the number of functional AMPAR containing synapses by reducing the level of surface labeled AMPAR subunits GluA1 and GluA2 (Kalashnikova et al., 2010). These results suggest a role for SynDIG1 in the regulation of AMPAR surface trafficking. However, a recent study showed that SynDIG1 does not alter AMPAR gating, pharmacology, or surface trafficking when co-transfected in HEK cells (Lovero et al., 2013), indicating that SynDIG1 does not act as a typical AMPAR auxiliary subunit. Interestingly, when Lovero et. al. overexpressed SynDIG1 in slice culture, they saw an increase in the AMPAR and NMDAR mediated synaptic transmission, while knock down of SynDIG1 showed a decrease in both AMPAR and NMDAR mediated transmission, suggesting a role for SynDIG1 in glutamatergic synaptogenesis. However, SynDIG1 distribution is re-localized to spines upon treatment with tetrodotoxin, indicating that it is regulated by activity (Kalashnikova et al., 2010) suggesting that in addition to its role in synapse development, SynDIG1 may also play a role in certain types of plasticity.
Intriguingly, SynDIG4 [also known as Prrt1 (Proline-rich transmembrane protein-1)], a polypeptide whose C-terminal domain exhibits extensive sequence similarity to that of SynDIG1, was identified in three independent proteomic studies as a candidate AMPAR associated protein (von Engelhardt et al., 2010; Schwenk et al., 2012; Shanks et al., 2012; Schwenk et al., 2014) as well as a component of the PSD (Jordan et al., 2004), suggesting that SynDIG4 might also regulate AMPAR synaptic targeting. To date, the sub-cellular expression and distribution of SynDIG4 in brain neurons has not been characterized. In this study, we define SynDIG4 spatial and temporal patterns of expression in the rat central nervous system compared to SynDIG1. In contrast to SynDIG1 and other traditional AMPAR auxiliary subunits, SynDIG4 is de-enriched at the PSD and colocalizes with extrasynaptic GluA1 puncta in primary neuron culture. These results indicate that although SynDIG4 shares sequence similarity with SynDIG1, it likely acts through a unique mechanism as an auxiliary factor for extrasynaptic GluA1 containing AMPARs.
Materials and Methods
Animals
Sprague Dawley timed pregnant rats (RRID:RGD_70508) were purchased from Harlan. Rats were house and maintained in the animal facility at UC Davis. The use and maintenance of animals were carried out according to the guidelines set forth by UC Davis, the NIH, and AALAC.
Antibodies
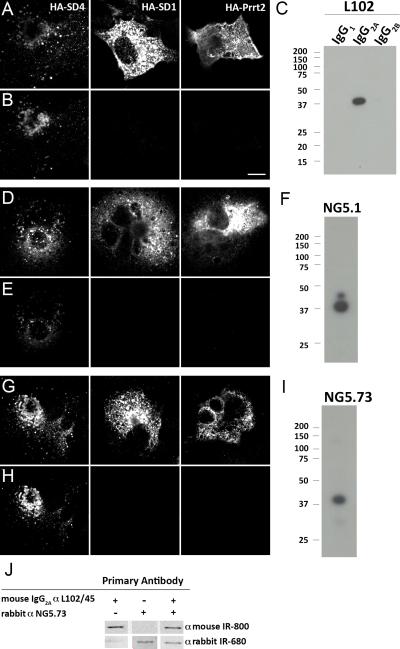
Primary antibodies and dyes used are described in Table 1. In this manuscript, we use three different antibodies targeted to SynDIG4 (Fig. 1). One antibody, L102/45 (NeuroMab; Cat# 73-409; RRID: AB_2491106), is a mouse IgG2A monoclonal antibody that we developed by targeting the first 66 N-terminal amino acids of SynDIG4, and was generated using standard monoclonal antibody approaches (Trimmer et al., 1985; Bekele-Arcuri et al., 1996), with the exception that electrofusion was used to generate the hybridomas, and soluble GST was added during primary monoclonal antibody screening to eliminate signal from anti-GST monoclonal antibodies. Rat anti-HA antibodies (Roche, Cat# 1867431, RRID: AB_390919) were used to identify positively transfected cells (Fig. 1A, D, G). L102/45 shows positive immunoreactivity against recombinant HA-tagged SynDIG4 (HA-SynDIG4) transfected into COS-7 cells (referred to as COS cells going forward) and co-localizes with HA signal, but does not show immunoreactivity against two related proteins, HA-SynDIG1 and HA-Prrt2 (Fig. 1B). A single band is detected on an L102/45 immunoblot at approximately 38 kDa in rat brain membrane homogenate when probed with an anti-IgG2A secondary antibody; this relative electrophoretic mobility is slightly higher than the predicted molecular weight of SynDIG4 of 31.4 kD (Fig. 1C). The other two antibodies, SD4/NG5.1 and SD4/NG5.73, are polyclonal rabbit antibodies received as a gift from Dr. August Smit (Chen et al., 2014). The antigens for NG5.1 and NG5.73 are amino acids 73-86, and amino acids 1-14 of the rat protein sequence, respectively. Both rabbit antibodies show positive and specific immunoreactivity for HA-SynDIG4 (Figure 1D-E, G-H), and predominant bands at 38 kDa on immunoblots of rat brain homogenate (Figure 1F, I). Because of the overlapping antigen regions for L102/45 and NG5.73, we assayed whether the two antibodies compete with each other for the same binding site. Rat brain membrane homogenates were probed with either L102/45, NG5.73, or both antibodies (Fig. 1J). The signal intensities for each of the antibodies in the blot co-probed with both antibodies is not substantially different than the blots probed with single antibodies, indicating that the antibodies do not compete for binding and likely have distinct, non-overlapping epitopes, strengthening the reliability of similar results obtained with both antibodies (Rhodes and Trimmer, 2006).
Table 1.
Primary antibodies Used in This Study
| Target | Host | Isotype | Type (clone) | Immunogen | Source | Cat. No. | RRID | Form | IB dilution | ICC dilution | IHC dilution |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SynDIG4 (L102/45) | mouse | IgG2A | mono (L102/45) |
Amino acids 1-66 rat MSSEKSGLPDSVPHTSPPPYNAPQPPAEPPIPPPQT APSSHHHHHHHYHQSGTATLPRLGAGGLAS |
Trimmer Lab (available at NeuroMab) |
73-409 | AB_2491106 | TC supe | Neat | 1:2 | 1:2 |
| SynDIG4 (NG5.1) | rabbit | poly | RGPSSSATLPRPPH | Smit Lab (GenScript) |
custom | 1:1k | 1:100 | ||||
| SynDIG4 (NG5.73) | rabbit | poly | MSSEKSGLPDSVPH | Smit Lab (GenScript) |
custom | 1:1k | 1:100 | ||||
| HA | rat | mono (3F10) |
Amino acids 76-111 of X47 hemaglutinin 1 YPYDVPDYA |
Roche | 11867431001 | AB_390919 | purified | 1:1k | 1:100 | ||
| β- tubulin | mouse | IgG1 | mono | Amino acids 412-430 bovine SNMNDLVSEYQQYQDATA |
Millipore | 05-661 | AB_309885 | protein G purified |
1:5k | ||
| SynDIG1 | mouse | IgG2A | mono (L42/17) |
Amino acids 1-183 of mouse MDGIIEQKSVLVHSKISDAGKRNGLINTRNFMAE SRDGLVSVYPAPQYQSHRLVASAAPGSLEGGRS EPVQQLLDPNTLQQSVESHYRPNIILYSDGVLRS WGDGVATDCCETTFIEDRSPTKDSLEYPDGKFID LSGDDIKIHTLSYDVEEEEELQELESDYSSDTESE DNFLMMPPRDHLG |
Trimmer Lab (available at NeuroMab) |
75-251 | AB_10999753 | Purified IgG | 1:1k | 1:100 | |
| GluA1 | rabbit | poly | human cytoplasmic domain | Millipore | AB1504 | AB_2113602 | 1:3k | 1:100 | 1:200 | ||
| GluA2 | mouse | IgG1 | mono (L21/32) |
Amino acids 834-883 rat FCYKSRAEAKRMKVAKNAQNINPSSSQNSQNFA TYKEGYNVYGIESVKI |
NeuroMab | 75-002 | AB_2232661 | Purified IgG | 1:3k | 1:100 | |
| PSD-95 | mouse | IgG2A | mono (K28/43) |
Amino acids 77-299 human PSD-95 FSIAGGTDNPHIGDDPSIFITKIIPGGAAAQDGRLR VNDSILFVNEVDVREVTHSAAVEALKEAGSIVRL YVMRRKPPAEKVMEIKLIKGPKGLGFSIAGGVG NQHIPGDNSIYVTKIIEGGAAHKDGRLQIGDKILA VNSVGLEDVMHEDAVAALKNTYDVVYLKVAK PSNAYLSDSYAPPDITTSYSQHLDNEISHSSYLGT DYPTAMTPTSPRRYSPVA |
NeuroMab | 75-028 | AB_2307331 | Purified IgG | 1:20k | ||
| Synaptophysin 1 | mouse | IgG1 | mono (7.2) | cytoplasmic tail | Synaptic Systems |
101011 | AB_887824 | Purified IgG | 1:10k | ||
| vGlut1 | guinea pig |
poly | C-terminus of rat vGlut1 | Millipore | AB5905 | AB_2301751 | serum | 1:500 | 1:500 | ||
| PSD-93 | mouse | IgG1 | mono (N18/30) |
full length rat | NeuroMab | 75-057 | AB_2277296 | Purified IgG | 1:100 | ||
| DAPI | 1:2k |
Figure 1. Demonstration of SynDIG4 antibody specificity.
A, B, D, E, G, H: COS cells expressing recombinant HA-SynDIG4 (SD4) or the related proteins HA-SynDIG1 (SD1) and HA-Prrt2 were immunolabled with antibodies against HA (A, D, G) or against SynDIG4 using L102/45 (B), NG5.1 (E), NG5.73 (H). All three antibodies recognized HA-SD4, but not HA-SD1 or HA-Prrt2. Scale bar = 10 μm.
C, F, I: Immunoblots of rat brain membrane homogenates probed with antibodies against SynDIG4: L102/45 (C), NG5.1 (F), or NG5.73 (I). All three antibodies show a predominant band at Mr = 38 kDa, slightly larger than the predicted molecular weight of SynDIG4 of 31.4 kDa.
J: Immunoblots of rat brain membrane homogenates probed with either L102/45, NG5.73, or both. Signal intensity in the blots co-probed with both antibodies is equivalent to the blots probed with single antibodies, indicating that the antibodies do not compete for the same binding site.
All other primary antibodies used in this study are described in Table 1. Antibodies have been characterized in the following ways by the manufacturer, unless otherwise noted: Mouse IgG2A anti-SynDIG1 (NeuroMab, Cat# 75-251, RRID: AB_10999753) stains a 36 kDa band in adult rat membrane lysates (manufacturer), and labels HA-SynDIG1 recombinant protein (Kalishnikova et al., 2010). Mouse IgG1 anti-β-tubulin (Millipore, Cat# 05-661, RRID: AB_309885) stains a single 50 kDa band from A431 cell lysates. Mouse IgG1 anti-GluA2 (NeuroMab, Cat# 75-002, RRID: AB_2232661) recognizes a single band at 90 kDa by immunoblot of mouse, rat, and human brain membrane preparations that was absent in GluA2 knock-out hippocampal lysates. Additionally, the GluA2 antibody shows characteristic hippocampal and post-synaptic staining. Rabbit anti-GluA1 (Millipore, Cat# AB1504, RRID: AB_2113602) antibody recognizes a single band at 100 kDa from rat brain tissue lysates. Mouse IgG2A anti-PSD-95 (NeuroMab, Cat# 75-028, RRID: AB_2307331) is shown to have multiple bands by immunoblot on rat brain membrane lysates ranging from 95-110 kDa due to different phosphorylation states. These bands are absent in PSD-95 knock out lysates. Mouse IgG1 anti-Synaptophysin (Synaptic Systems, Cat# 101011, RRID: AB_887824) recognizes a single band at approximately 38 kDa from crude synaptosomal fraction of rat brain. Guinea pig anti-vGlut1 (Millipore, Cat# AB5905, RRID: AB_2301751) was shown to have positive immunohistochemistry staining in rat brain that was absent when the antibody was preadsorbed with the antigen. Mouse IgG1 anti-PSD-93 (NeuroMab, Cat# 75-057, RRID: AB_2277296) shows a specific band at 110 kDa in mouse, rat, and human brain membrane preparations, that is absent in the PSD-93 knock out membrane preparation.
For all immunocytochemistry and immunohistochemistry experiments, subclass specific fluorophore conjugated secondary antibodies from Molecular Probes and Jackson Immunoresearch were used.
Immunoblots
For all biochemistry experiments, animals were anesthetized with CO2 inhalation administered via compressed gas. Brain tissue was then isolated from rats and homogenized in 0.32 M sucrose, 5 mM Hepes using a dounce homogenizer. Supernatants were collected after low speed centrifugation at 1400 xg, quantified by BCA, and separated on 10% SDS-PAGE. For quantitative immunoblotting, IRDye conjugated secondary antibodies (LI-COR Biosciences) were used and fluorescence signal was detected using a Li-COR imaging system. Fluorescence was quantified using Image Studio Lite version 3.1 (http://www.licor.com, RRID: SCR_013715). HRP-conjugated secondary antibodies (Invitrogen) were used for qualitative immunoblotting.
PSD fractionation
All steps were performed at 4°C. For each preparation, one six month old rat brain was mechanically homogenized in 0.32 M sucrose using a dounce homogenizer, followed by low speed centrifugation at 1400 xg. The supernatant [total homogenate (TH)] was centrifuged at high speed (10 min, 47800 xg). After centrifugation a fraction of the supernatant was kept as the cytoplasmic fraction (C). The pellet was resuspended in 0.32 M sucrose [membrane enriched fraction (M)], then layered over a step sucrose gradient (0.85/1/1.25 M) and ultracentrifuged for 2 hours (hrs) at 246000 xg. The fraction between the 1/1.25 M steps was removed as the synaptosomal fraction (S). This fraction was then incubated with 0.5% Triton X-100 for 15 minutes (min), and ultracentrifuged for 30 min at 324600 xg. A fraction of the supernatant was reserved as the Triton X-100 soluble (TS) fraction. The pellet was resuspended in 0.32 M sucrose and layered over a 1/1.5/2 M sucrose gradient. The fraction between the 1.5/2 M gradients was removed and incubated with an equal volume of 0.5% Triton X-100 for 15 min, followed by ultracentrifugation at 325000 xg for 1 hr. The pellet was resuspended in 40 mM Tris-HCl pH 8.0 and sonicated to aid in resuspension. This fraction is the postsynaptic density (PSD) enriched fraction.
Immunohistochemistry
Rats were deeply anesthetized with 60 mg/kg of sodium pentobarbital and transcardially perfused with phosphate buffered saline (PBS), pH 7.4, followed by 4% formaldehyde, prepared fresh from paraformaldehyde, in 0.1 M sodium phosphate buffer (PB), pH 7.4. Brains were removed and cryoprotected with 10% sucrose for 18 hrs, followed by 48 hrs in 30% sucrose. Following cryoprotection, rat brains were frozen and cut into 30 μm sections using a sliding microtome with a freezing stage. Sections were collected in 0.1 M PB.
Immunohistochemistry was performed on 30 μm thick, free floating, sagittal rat brain sections. Sections were blocked in 10% goat serum (Equitech), 0.3% Triton X-100, 0.1 M PB for 1 hr at room temperature (RT). Sections were incubated with primary antibodies in blocking solution for 3 hrs at RT, and washed 3 × 10 min in 0.1 M PB. Sections were then incubated in secondary antibody for 2 hrs at RT, followed by 2 washes in 0.1 M PB, and one final wash in 0.05 M PB. Sections were then mounted on gelatin coated slides, stained with 0.05% Sudan Black in 70% ethanol, and sealed under coverglass with ProLong Gold.
Low resolution images were collected using a Zeiss Axio Observer Z1 fluorescence microscope using a 10x/0.5 NA objective, and stitched together using Zeiss Zen software. High resolution images were taken with the same microscope using a 100x/1.3 NA oil immersion objective in combination with an Apotome for structured illumination. Signals were adjusted equally by using linear adjustments of levels in Photoshop CS6 (http://www.adobe.com; RRID: ScrRes_000161). All sections within a panel were treated identically.
Immunocytochemistry
COS cells were seeded onto poly-L-lysine coated coverslips and allowed to grow overnight. COS media [DMEM (Life Technologies), 10% FBS (Fisher), 100 U/ mL penicillin, 100 μg/ mL streptomycin (Life Technologies)] was replaced with OptiMEM (Life Technologies), and cells were transfected with 2 μg of total DNA per well using Lipofectamine 2000 (Life Technologies). Cells were incubated at 37°C for 3 hrs before being changed back to COS media and allowed to recover overnight. COS cells were fixed in 4% formaldehyde, prepared fresh from paraformaldehyde, in 1X PBS for 10 min at RT, washed in 1X PBS, permeabilized for 15 min in 0.1% Triton X-100, and blocked for 30 min in 5% nonfat milk powder in 1X PBS. Coverslips were incubated in primary antibody overnight at 4°C, washed in 1X PBS, incubated in secondary antibody at RT for 1 hr, and washed again in 1X PBS. Coverslips were mounted onto glass slides using Flouromount-G (Southern Biotech). For surface labeling, transfected COS cells were placed on ice for 10 minutes, washed once with ice cold 1X PBS, and incubated with primary antibodies for 10 minutes. Cells were washed in ice cold 1X PBS three times, then fixed and stained as indicated above. Images were acquired using a Zeiss LSM700 scanning confocal microscope with identical settings for laser power, photomultiplier gain, and digital offset within each staining set. Signals were adjusted equally for all images by using linear adjustments of levels in Photoshop. All panels within a figure were treated identically.
Primary neuronal culture
Neurons from P0–P2 rat occipital cortex were dissociated in papain (Worthington) and plated at a density of 15,000/cm2. Neurons were grown on poly-L-lysine-coated glass coverslips suspended over a glial cell feeder layer in media supplemented with N2 (Life Technologies).
Neurons were fixed and stained at 10 days in vitro (DIV) as described above in “Immunocytochemistry” except that the blocking solution was 3% bovine serum albumin (Life Technologies). Images were acquired using either a Zeiss LSM700 scanning confocal microscope or an Olympus Fluoview 1000, with a 63x objective lens with identical settings for laser power, photomultiplier gain, and digital offset within each staining set.
Images were imported into ImageJ software (http://imagej.nih.gov/ij/, RRID: nif-0000-30467) for quantitative analysis of synaptic puncta. Thresholds were established using a subset of images from each image set so that all puncta were included, and the average threshold was applied to the entire data set for quantitative analysis. A mask was created for each channel, and colocalization was defined as any partially overlapping punctate structure. The cell body was excluded from analysis.
Signals were adjusted for all images by using linear adjustments of levels in Photoshop (Adobe Systems). All panels within a figure were treated identically.
Results
In this manuscript, we use three different antibodies against SynDIG4 (L102/45, NG5.1, and NG5.73) to determine expression of SynDIG4 in rat brain. Figure 1 demonstrates that these antibodies are specific for SynDIG4, and do not label related proteins SynDIG1 and Prrt2. Characterization of these antibodies are described in detail in the Methods section.
SynDIG4 is a type-II transmembrane protein
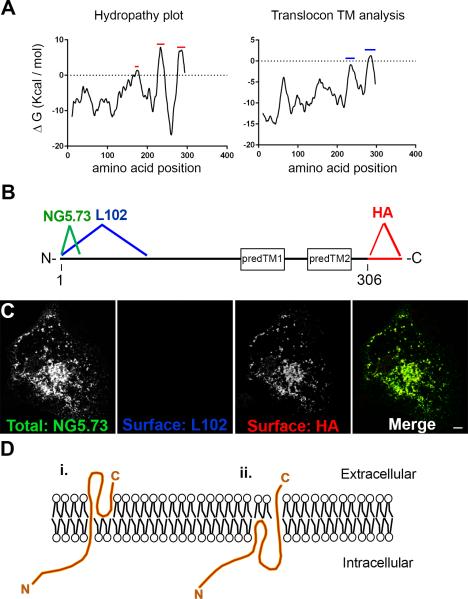
Membrane Protein Explorer (MPEx) was used to generate the hydropathy plot in Figure 2A (http://blanco.biomol.uci.edu/mpex/, RRID: SCR_014077) (Jayasinghe et al., 2001). Using this software, there are three predicted hydrophobic domains, indicated by the red bars. However, using the same software, analysis with translocon transmembrane analysis predict that the last two hydrophobic domains are transmembrane (blue bars) (Snider et al., 2009). To determine whether SynDIG4 is at the cell surface and to determine the protein's topology, SynDIG4 with an HA tag at the C-terminus (SynDIG4-HA) was transfected into COS cells and then live labeled with HA and L102/45 antibodies simultaneously. Figure 2B shows a schematic of the recombinant protein, with the two hydrophobic domains predicted to be transmembrane (predTM1 and predTM2), and the HA tag at the C-terminus of the protein. Epitope regions are indicated for the antibodies used. While the HA antibody will recognize the HA tag at the C-terminus, the L102/45 antibody recognizes the N-terminus of the protein. After fixation and permeabilization, total SynDIG4 protein was labeled using the NG5.73 antibody which does not compete with L102/45 (Figure 1J). Live labeling showed positive staining for the C-terminus of SynDIG4-HA, but signal for the N-terminus is absent (Fig. 2C). These data indicate that SynDIG4 is a type II transmembrane protein, with an extracellular C-terminus and intracellular N-terminus. This topology implies that one of the predicted transmembrane regions does not in fact pass through the membrane completely. We were unable to identify whether the loop region between the two hydrophobic domains was intracellular or extracellular, because tagging this region negatively affected protein expression in heterologous cells (not shown). Therefore, there are two possible models for SynDIG4 topology, with the loop region either extracellular (Fig. 2D.i) or intracellular (Fig. 2D.ii). As SynDIG1 topology is similar to the model shown in Fig. 2D.i, with the first hydrophobic domain passing through the membrane, and the loop region extracellular (Kalashnikova et al., 2010), it is likely that the related protein SynDIG4 is organized similarly.
Figure 2. SynDIG4 topology.
A: Hydropathy plot and translocon TM analysis show three predicted hydrophobic domains (red bars) but only two predicted transmembrane domains (blue bars).
B: Schematic of SynDIG4-HA protein and two predicted transmembrane domains. The antigen sites for NG5.73 and L102 are located near the N-terminus, and the HA tag is on the C-terminus. Regions predTM1 and predTM2 represent the hydrophobic domains that are predicted to be transmembrane.
C: Representative image of COS cells transfected with SynDIG4-HA and surface labeled with L102 (blue) and HA (red) antibodies. NG5.73 (green) was used to stain for total SynDIG4 after fixation and permeabilization. Scale bar = 10 μm.
D: Possible models for SynDIG4 topology, with the C-terminus extracellular, and the N-terminus intracellular. The loop region between the two predicted transmembrane domains could either be extracellular (i) or intracellular. Models are not to scale.
SynDIG4 is a brain specific protein that peaks in expression during synaptogenesis
To assess tissue specific expression of SynDIG4, various tissues were extracted from a P14 rat and probed with two independent SynDIG4 antibodies (Fig. 3). Both SD4/L102 and SD4/NG5.1 antibodies identified one band at ~38 kDa in brain tissue only. This result indicates that SynDIG4, like SynDIG1(Kalashnikova et al., 2010), is a brain specific protein.
Figure 3. SynDIG4 is a brain specific protein.
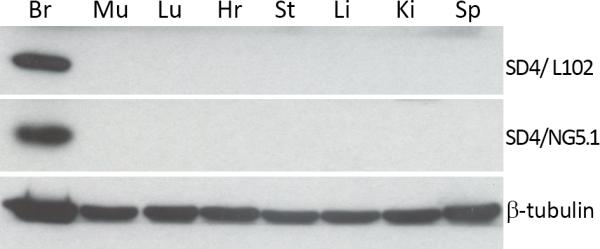
P14 rat tissue homogenates were probed for SynDIG4 with two independent antibodies (L102/45 and SD4/NG5.1). Abbreviations: Br, brain; Mu, muscle; Lu, lung; Hr, heart; St, stomach; Lv, liver; Kd, kidney; Sp, spleen.
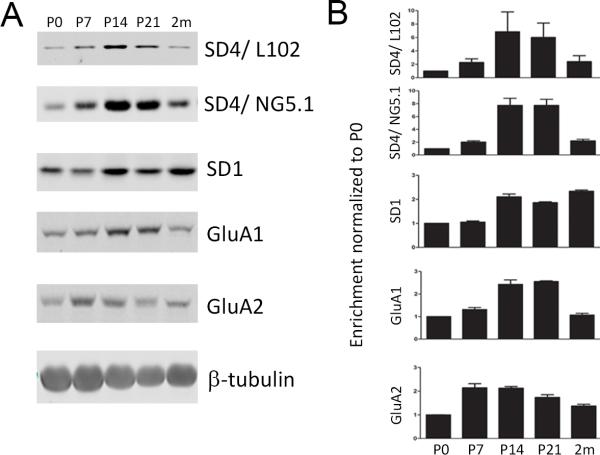
We next examined the respective expression levels of SynDIG1 and SynDIG4 protein during brain development. Total homogenates from the brains of rats at P0, P7, P14, P21, and 2 months of age were probed for SynDIG1 and SynDIG4 (in addition to other synaptic proteins) (Fig. 4A). Signal was normalized to beta-tubulin to control for loading and then normalized again to P0 to determine enrichment in three biological replicates. Both SynDIG4 antibodies show enrichment in signal intensity at P14 and P21, which corresponds to the peak time frame for synapse development. This enrichment is similar to the developmental expression patterns of SynDIG1 and AMPAR subunits GluA1 and GluA2. However, L102/45 and SD4/NG5.1 show 6.9 and 7.7 fold enrichment at P14, respectively, whereas SynDIG1 and GluA1/2 only show 2-3 fold enrichment at P14 (Fig. 4B), indicating the SynDIG4 is highly upregulated during synapse development.
Figure 4. SynDIG4 expression peaks during synaptogenesis.
A: Representative immunoblots of rat brain homogenates prepared from P0, P7, P14, P21, and 2 months are shown.
B: Quantification of (A), normalized to beta-tubulin and compared to P0 for enrichment (n = 3). Multiple blots were run for analysis; each blot had its own beta-tubulin loading control, but for simplicity only one beta-tubulin blot is shown.
SynDIG4 and SynDIG1 have complementary expression patterns
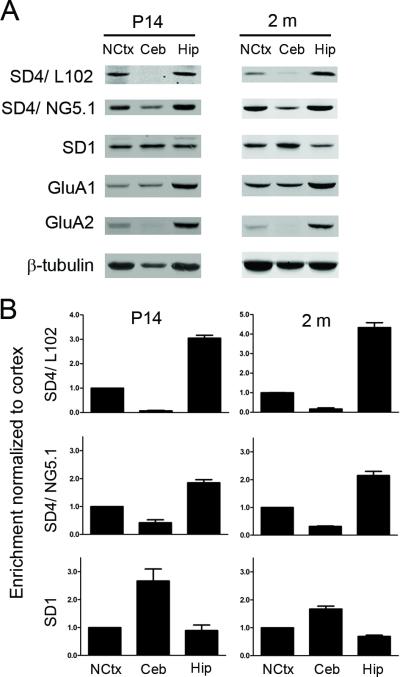
To determine the regional expression of SynDIG4 and SynDIG1 in brain, we probed homogenates prepared from specific brain regions (neocortex, cerebellum, and hippocampus) for SynDIG4 and SynDIG1 expression at two ages P14 and 2 months (Fig. 5A). Signal quantification is normalized to beta-tubulin and then normalized to cortical expression for relative comparison (Fig. 5B). SynDIG4 is expressed in the neocortex and enriched in the hippocampus, while only modestly expressed in the cerebellum when probed with SD4/NG5.1, and almost entirely absent when probed with L102/45. Interestingly, SynDIG1 is enriched in the cerebellum where SynDIG4 expression is lowest. SynDIG1 is also expressed in the neocortex and modestly expressed in the hippocampus. Together, these data suggest that SynDIG1 and SynDIG4 have partially overlapping but distinct relative expression patterns in different brain regions, and these patterns are consistent during two different stages of development.
Figure 5. SynDIG1 and SynDIG4 have complementary patterns of expression in different brain regions.
A: Representative immunoblots of samples prepared from neocortex (NCtx), cerebellar (Ceb), and hippocampal tissue dissected from P14 and 2 month rat brains.
B: Quantification of (A). Signal normalized to beta-tubulin and compared to expression in neocortex (n = 3). Multiple blots were run for analysis; each blot had its own beta-tubulin loading control, but for simplicity only one beta-tubulin blot is shown.
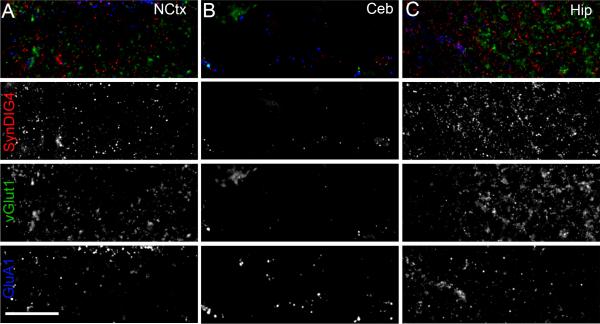
To further analyze SynDIG4 expression in the adult rat brain, immunohistochemistry of free floating sections was performed using the L102/45 antibody. At high resolution, SynDIG4 signal is punctate and most dense in the hippocampus (Fig. 6C), less dense in the neocortex (Fig. 6A), and hardly present in the molecular layer of the cerebellum (Fig. 6B). These results are consistent with the distribution of SynDIG4 expression determined by immunoblotting (Fig. 5).
Figure 6. SyDIG4 puncta are most dense in hippocampal tissue.
High resolution images show increased staining of SynDIG4 puncta in the hippocampus (C) compared to neocortex (NCtx) (A), with little staining in the cerebellum (Ceb) (B). Scale bar = 10 μm.
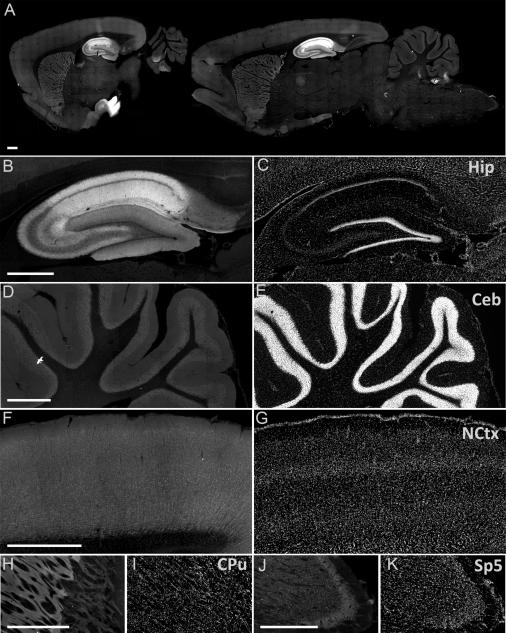
At lower resolution, immunohistochemistry of sagittal sections from adult rat brain shows intense labeling of SynDIG4 in the hippocampus compared to the rest of the brain (Fig. 7A). Higher magnification images show that while SynDIG4 is expressed throughout the hippocampus, it is most highly expressed in the CA1 region and the stratum lacunosum-moleculare of the CA2 region (Fig. 7B). It is also apparent that SynDIG4 staining is relatively absent from the cell bodies and is neuropil in nature, reinforcing that SynDIG4 is a synaptic protein. In the cerebellum, SynDIG4 shows low levels of staining with slightly increased levels in the granular layer compared the molecular layer (Fig. 7D). SynDIG4 staining in the neocortex is higher than cerebellum staining, but does not appear to stain any distinct layers (Fig. 7F). Interestingly, SynDIG4 also shows staining in the caudate putamen (Fig. 7H) and the spinal trigeminal nucleus (Sp5; Fig. 7J). Nuclei are stained with DAPI to aid with orientation (Fig. 7C,E,G,I,K).
Figure 7. SynDIG4 expression is enriched in the hippocampus.
A: Two representative sagittal sections of adult rat brain immunolabeled for SynDIG4 with the L102/45 antibody. Scale bars = 500 μm. Zoomed in panels show SynDIG4 labeling in the hippocampus (Hip, B), cerebellum (Ceb, D), neocortex (NCtx, F), caudate putamen (CPu, H), and the Spinal Trigeminal Nucleus (Sp5, J).
C, E, G, I, K: DAPI counterstain of panels B, D, F, H, and J, respectively.
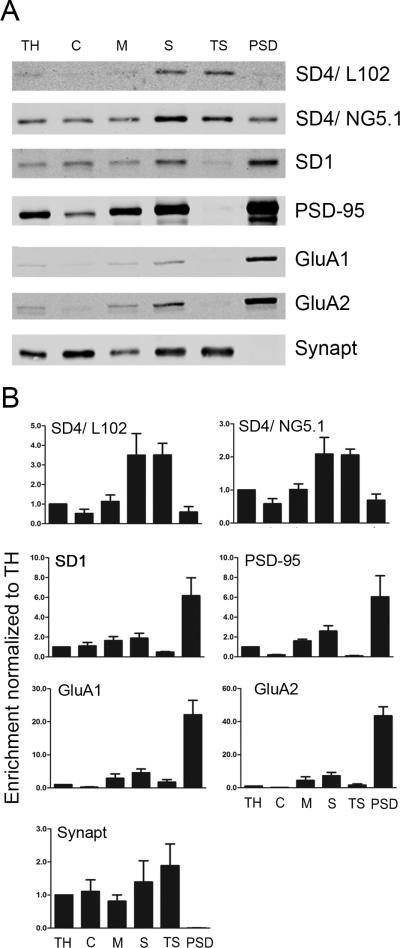
SynDIG4 is de-enriched at the postsynaptic density
To determine the sub-cellular localization of SynDIG4, we used biochemical fractionation to enrich for the PSD in adult rat brain lysates, and assayed for SynDIG1 and SynDIG4 expression with immunoblot (Fig. 8). PSD-95 is used as a positive control as it is an abundant component of the PSD, and is enriched in the PSD fraction, while absent from the detergent soluble fraction. Synaptophysin, a presynaptic protein, is used as a negative control and is absent from the PSD fraction. AMPAR subunits GluA1 and GluA2 also show an expected enrichment in the PSD fraction, along with a modest enrichment of SynDIG1. Intriguingly, while SynDIG4 is enriched in synaptosomes, it is de-enriched at the PSD. This result is unexpected considering that SynDIG4 is a known AMPAR interacting protein, and AMPARs are enriched at the PSD. This result could indicate that either SynDIG4 is pre-synaptic or that SynDIG4 is loosely associated with the postsynaptic structure, rather than tightly embedded in the PSD.
Figure 8. SynDIG4 is de-enriched at the post-synaptic density.
A: Representative blots of adult rat brains (six months old) subjected to sucrose gradient fractionation to enrich for the post-synaptic density (PSD). PSD-95 is used as a positive control to show enrichment at the PSD, and synaptophysin (Synapt) is used as a negative control to show absence in the PSD.
B: Quantification of (A). Signal is compared to total homogenate for enrichment (n = 3). Abbreviations: TH, total homogenate; C, cytosol; M, membrane enriched; S, synaptosomes; TS, triton soluble; PSD, post-synaptic density enriched.
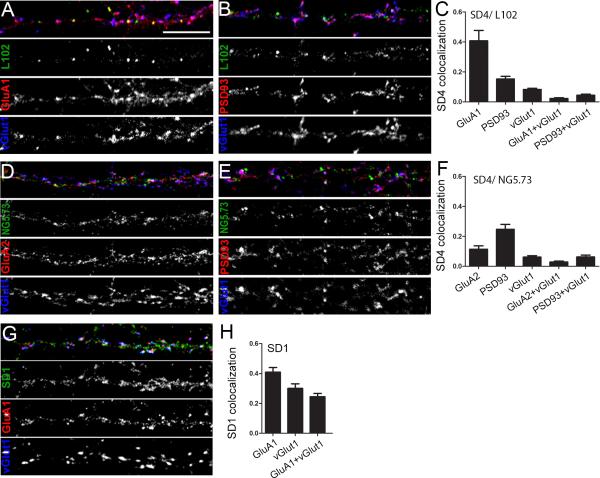
SynDIG4 colocalizes with extrasynaptic AMPARs
To further address the question of SynDIG4 sub-cellular distribution, we used dissociated cortical neurons and two independent antibodies to examine distribution of SynDIG4 with AMPAR subunits and other synaptic markers (Fig. 9). Approximately 40% of L102/45–positive puncta colocalized with the AMPAR subunit GluA1 (Fig. 9A, quantified in Fig. 9C). However, very little SD4/ NG5.1 puncta (11%) colocalizes with GluA2 (Fig. 9D, quantified in Fig. 9F). L102/45 and SD4/NG5.1 also show very little colocalization with vGlut1, 8% and 6%, respectively. Even less colocalization of SynDIG4 is seen at either GluA1/ vGlut1 or GluA2/ vGlut1 synapses, or PSD-93 and vGlut1 defined synapses. These results suggest that SynDIG4 preferentially localizes with a subpopulation of extrasynaptic GluA1 containing AMPARs. Neurons were labeled after permeabilization so it is possible that these GluA1 and SynDIG4 clusters are intracellular although surface labeling of recombinant SynDIG4-HA shows that at least a portion of the protein reaches the surface in heterologous cells (Fig. 2). This observation is a unique aspect of SynDIG4, as other AMPAR auxiliary factors such as Stargazin/ γ-2 are enriched at the PSD and localize to synapses (Chen et al., 2000).
Figure 9. SynDIG4 is enriched at extrasynaptic AMPARs.
A, B, D, E, G: Representative dendrite stretches of dissociated primary cortical neurons (10 DIV) stained with antibodies against SynDIG4 (SD4) or SynDIG1 (SD1) and other synaptic markers. A, B: SD4/ L102/45, D, E: SD4/NG5-73, and G: SD1. GluA1, GluA2, and PSD93 were used as post-synaptic markers, with vGlut1 as a presynaptic marker. Synapses were defined as overlapping pre- and post-synaptic markers.
C, F, H: Quantification of (A-B, D-E, and G), respectively, represented as the fraction of SynDIG1 or SynDIG4 colocalized with the indicated synaptic marker (n = 10 for each staining set). Scale bar = 10 μm.
Discussion
SynDIG4 shares amino acid sequence similarity to SynDIG1, a protein that regulates excitatory synapse development (Kalashnikova et al., 2010), and both have been identified as candidate AMPAR auxiliary factors. Therefore, a main goal of this study was a detailed comparison of developmental expression and sub-cellular localization in rat brain of SynDIG4 and SynDIG1. SynDIG1 is a type II transmembrane protein containing two hydrophobic segments, one of which spans the membrane. While no obvious protein interacting domains are present in SynDIG1, the C-terminal membrane associated region is critical for its interaction with AMPARs (Kalashnikova et al., 2010). Interestingly, SynDIG4 and SynDIG1 share the most sequence similarity throughout the C-terminus; therefore, it is attractive to hypothesize that the C-terminus of SynDIG4 may also be important for its interaction with AMPARs. It should be pointed out, however, that SynDIG4 is highly divergent from SynDIG1 in the SynDIG family, sharing only 35% sequence similarity with SynDIG1. SynDIG4 has a unique proline rich N-terminus shared by the related protein Prrt2 that exhibits only very modest sequence similarity with SynDIG1, but shares 67% sequence similarity to SynDIG4. Proline rich motifs are known to be weak protein-protein interaction domains in multiple signaling molecules, most notably interaction with Src homology (SH) 3 domains which are commonly found in cytoskeleton proteins. EVH1 domains, found in the post-synaptic scaffolding protein Homer, also interact with proline rich motifs [reviewed in (Kay et al., 2000; Freund et al., 2008)]. Live labeling of SynDIG4 suggests that the N-terminus is intracellular, meaning the large proline rich stretch could be an important region for protein-protein interactions.
In this study, we show that SynDIG4 is a brain specific protein that is developmentally regulated in the rat central nervous system. Previously, Schwenk et al. (2014) used a mass spectrometry-based proteomics approach to examine the molecular abundance of AMPAR subunits and known AMPAR auxiliary factors in different brain regions, as well as throughout different stages of development. They found an enrichment of SynDIG4 protein at P14, with further enrichment at P28. Consistent with these results, we see an enrichment of SynDIG4 at P14 and P21. We did not assay animals at P28, but it is likely that the levels of SynDIG4 continue to increase beyond P21 and possibly up until 2 months in age (P60), where we observe a decrease in SynDIG4 expression. Additionally, we show that SynDIG1 and SynDIG4 have complementary expression patterns in the brain regions examined. SynDIG4 expression is highly enriched in the hippocampus of both P14 and two month old animals, with a marked decrease of expression in the cerebellum. Conversely, SynDIG1 is highly enriched in the cerebellum, with only modest expression in the hippocampus when compared to cortical expression. Decreased expression of SynDIG4 in the cerebellum and enrichment in the hippocampus is consistent with previous proteomics studies (Chen et al., 2014; Schwenk et al., 2014).
Interestingly, Chen et al. (2014) found that while SynDIG4 is robustly co-immunoprecipitated with AMPAR subunits GluA1-3 in the hippocampus, very little association was seen with AMPARs in the cerebellum. In fact, SynDIG4 appeared to preferentially interact with the voltage-gated potassium channel subunits KCNC1 and KCNC3 and the post-synaptic scaffolding protein Homer3 in the cerebellum. KCNC1/3 are primarily localized to the axon [reviewed in (Trimmer, 2015)], where they are involved in action potential firing in the cerebellum (Hurlock et al., 2009). Therefore, it is possible that SynDIG4 could play distinct roles in different brain regions by acting as an auxiliary factor for different ion channel complexes. Furthermore, SynDIG1 may have a more prominent role in regulating AMPARs in the cerebellum compared with SynDIG4. Further experiments are needed to test these possibilities.
A striking result from our study is that SynDIG4, unlike SynDIG1, is de-enriched at the PSD, indicating that SynDIG4's predominant localization is either extrasynaptic or pre-synaptic. Based on the proteomics data from Chen et al. (2014), it is possible that SynDIG4 may have different subcellular localizations in different brain regions. Since our PSD fractionation experiments were performed with whole brain, the observed de-enrichment of SynDIG4 at the PSD may reflect a collection of various subcellular localizations. However, at least in primary cortical neurons, SynDIG4 has a dendritic localization where it predominantly co-localizes with GluA1. In contrast, SynDIG4 shows very little co-localization with other synaptic markers, indicating that SynDIG4 is primarily associated with extrasynaptic AMPARs.
While the majority of AMPAR auxiliary subunits are enriched at synapses, differential subcellular localization is also seen in the TARP family of proteins. For example, Stargazin/γ-2 is enriched at synapses (Chen et al., 2000) while γ-4 is enriched at extrasynaptic membranes, and γ-8 is more evenly distributed in both synaptic and extrasynaptic membranes (Ferrario et al., 2011). We found that in cortical neurons, SynDIG4 is primarily associated with extrasynaptic GluA1 containing AMPARs. GluA1 has been shown to insert into extrasynaptic sites, where it is part of a highly mobile population of AMPARs that can traffic to synapses by lateral diffusion (Passafaro et al., 2001; Heine et al., 2008). Others have shown an impairment of LTP, distance-dependant synaptic scaling, and synapse unsilencing in the GluA1−/− mouse, indicating an important role for GluA1 containing AMPARs in different types of plasticity (Zamanillo et al., 1999; Andrasfalvy et al., 2003; Selcher et al., 2012). Since the majority of extrasynaptic AMPARs are GluA1/2 heteromers (Andrasfalvy et al., 2003; Lu et al., 2009) and SynDIG4 also shows very little co-localization with GluA2, it is an intriguing possibility that SynDIG4 has a preference for a small sub-population of extrasynaptic GluA1 homomers. GluA1 homomers are Ca2+ permeable AMPARs (CP-AMPARs) that are enriched at perisynaptic sites (He et al., 2009; Ferrario et al., 2011). CP-AMPARs are thought to be recruited to the synapse in the early phase of LTP, where they are later replaced with GluA1/2 heteromers to stabilize the change in synaptic strength (Plant et al., 2006; Tanaka and Hirano, 2012). There are also reports that indicate CP-AMPARs are stabilized at perisynaptic sites by phosphorylation of Ser-845 on GluA1, and that upon induction of long-term depression (LTD) this site is de-phosphorylated (Davies et al., 2008; He et al., 2009). However, LTD was not impaired in GluA1−/− mice (Selcher et al., 2012). Interestingly, a recent report demonstrates that LTP does not require GluA1, or any AMPAR subunit for that matter, but rather shows that LTP is dependent on an extrasynaptic pool of glutamate receptors regardless of type (Granger et al., 2013). In fact, when GluA1-3 was replaced with GluK1 and Neto, potentiation still occurred (Adesnik and Nicoll, 2007; Granger et al., 2013). These differences may be reconciled by the fact that GluA1 is present in nearly all extrasynaptic receptors, and in the GluA1−/− mouse, extrasynaptic AMPARs are almost completely absent (Andrasfalvy et al., 2003).
Our data indicate that although SynDIG4 shares sequence similarity with SynDIG1, it has distinct expression in brain, and might act through a unique mechanism as an auxiliary factor for extrasynaptic GluA1 containing AMPARs. Given the importance of extrasynaptic GluA1 in synaptic plasticity and its prominent expression in hippocampus, it would be intriguing to test whether SynDIG4 has a functional role in hippocampal LTP.
Acknowledgments
We thank A. Smit for antibodies and A.K. McAllister for primary cortical neuron cultures. L.M.K. was supported by the National Institutes of Health Training Grant T32 GM007377. These studies were supported by funds to E.D. from the National Institutes of Health (NIH) Director's New Innovator Award Program (DP2 OD006479-01), the National Science Foundation (NSF 1322302), and a Pilot Grant from the UC Davis Academic Senate Research Programs and in part by funds to J.S.T. from NIH grant R01 NS42225.
This work was supported by grants to E.D. from the National Institutes of Health Director's New Innovator Award Program (DP2 OD006479-01) and the National Science Foundation (1322302), and to J.S.T. from the National Institutes of Health (R01 NS042225).
Role of authors: All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: LMK and ED. Acquisition of data: LMK, HIB, MOL, and MP. Analysis and interpretation of data: LMK. Drafting of the manuscript: LMK. Critical revision of the manuscript for important intellectual content: ED and JST. Statistical analysis: LMK. Obtained funding: ED, JST. Administrative, technical, and material support: SWT and JST. Study supervision: ED.
Footnotes
Conflict of interest: The authors declare no competing financial interests exist.
Literature Cited
- Adesnik H, Nicoll RA. Conservation of glutamate receptor 2-containing AMPA receptors during long-term potentiation. J Neurosci. 2007;27(17):4598–4602. doi: 10.1523/JNEUROSCI.0325-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrasfalvy BK, Smith MA, Borchardt T, Sprengel R, Magee JC. Impaired regulation of synaptic strength in hippocampal neurons from GluR1-deficient mice. J Physiol. 2003;552(Pt 1):35–45. doi: 10.1113/jphysiol.2003.045575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekele-Arcuri Z, Matos MF, Manganas L, Strassle BW, Monaghan MM, Rhodes KJ, Trimmer JS. Generation and characterization of subtype-specific monoclonal antibodies to K+ channel alpha- and beta-subunit polypeptides. Neuropharmacology. 1996;35(7):851–865. doi: 10.1016/0028-3908(96)00128-1. [DOI] [PubMed] [Google Scholar]
- Bury LA, Sabo SL. How it's made: the synapse. Molecular interventions. 2010;10(5):282–292. doi: 10.1124/mi.10.5.5. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408(6815):936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chen N, Pandya NJ, Koopmans F, Castelo-Szekelv V, van der Schors RC, Smit AB, Li KW. Interaction Proteomics Reveals Brain Region-Specific AMPA Receptor Complexes. Journal of proteome research. 2014 doi: 10.1021/pr500697b. [DOI] [PubMed] [Google Scholar]
- Davies KD, Goebel-Goody SM, Coultrap SJ, Browning MD. Long term synaptic depression that is associated with GluR1 dephosphorylation but not alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor internalization. J Biol Chem. 2008;283(48):33138–33146. doi: 10.1074/jbc.M803431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E. Regulation of AMPA receptors by transmembrane accessory proteins. European Journal of Neuroscience. 2010;32(2):261–268. doi: 10.1111/j.1460-9568.2010.07357.x. [DOI] [PubMed] [Google Scholar]
- Diaz E, Ge Y, Yang YH, Loh KC, Serafini TA, Okazaki Y, Hayashizaki Y, Speed TP, Ngai J, Scheiffele P. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron. 2002;36(3):417–434. doi: 10.1016/s0896-6273(02)01016-4. [DOI] [PubMed] [Google Scholar]
- Ferrario CR, Loweth JA, Milovanovic M, Wang X, Wolf ME. Distribution of AMPA receptor subunits and TARPs in synaptic and extrasynaptic membranes of the adult rat nucleus accumbens. Neuroscience letters. 2011;490(3):180–184. doi: 10.1016/j.neulet.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund C, Schmalz HG, Sticht J, Kuhne R. Proline-rich sequence recognition domains (PRD): ligands, function and inhibition. Handbook of experimental pharmacology. 2008;(186):407–429. doi: 10.1007/978-3-540-72843-6_17. [DOI] [PubMed] [Google Scholar]
- Granger AJ, Shi Y, Lu W, Cerpas M, Nicoll RA. LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature. 2013;493(7433):495–500. doi: 10.1038/nature11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BJ, Ghosh A. Regulation of AMPA receptor recruitment at developing synapses. Trends Neurosci. 2008;31(2):82–89. doi: 10.1016/j.tins.2007.11.010. [DOI] [PubMed] [Google Scholar]
- He K, Song L, Cummings LW, Goldman J, Huganir RL, Lee HK. Stabilization of Ca2+-permeable AMPA receptors at perisynaptic sites by GluR1-S845 phosphorylation. Proc Natl Acad Sci U S A. 2009;106(47):20033–20038. doi: 10.1073/pnas.0910338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine M, Groc L, Frischknecht R, Beique JC, Lounis B, Rumbaugh G, Huganir RL, Cognet L, Choquet D. Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science. 2008;320(5873):201–205. doi: 10.1126/science.1152089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80(3):704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlock EC, Bose M, Pierce G, Joho RH. Rescue of motor coordination by Purkinje cell-targeted restoration of Kv3.3 channels in Kcnc3-null mice requires Kcnc1. J Neurosci. 2009;29(50):15735–15744. doi: 10.1523/JNEUROSCI.4048-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The Expanding Social Network of Ionotropic Glutamate Receptors: TARPs and Other Transmembrane Auxiliary Subunits. Neuron. 2011;70(2):178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe S, Hristova K, White SH. Energetics, stability, and prediction of transmembrane helices. J Mol Biol. 2001;312(5):927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3(9):857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Kalashnikova E, Lorca RA, Kaur I, Barisone GA, Li B, Ishimaru T, Trimmer JS, Mohapatra DP, Diaz E. SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron. 2010;65(1):80–93. doi: 10.1016/j.neuron.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14(2):231–241. [PubMed] [Google Scholar]
- Lee KF, Soares C, Beique JC. Tuning into diversity of homeostatic synaptic plasticity. Neuropharmacology. 2014;78:31–37. doi: 10.1016/j.neuropharm.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Lovero KL, Blankenship SM, Shi Y, Nicoll RA. SynDIG1 Promotes Excitatory Synaptogenesis Independent of AMPA Receptor Trafficking and Biophysical Regulation. PLoS One. 2013;8(6):e66171. doi: 10.1371/journal.pone.0066171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62(2):254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passafaro M, Piech V, Sheng M. Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nature neuroscience. 2001;4(9):917–926. doi: 10.1038/nn0901-917. [DOI] [PubMed] [Google Scholar]
- Plant K, Pelkey KA, Bortolotto ZA, Morita D, Terashima A, McBain CJ, Collingridge GL, Isaac JT. Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nature neuroscience. 2006;9(5):602–604. doi: 10.1038/nn1678. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J Neurosci. 2006;26(31):8017–8020. doi: 10.1523/JNEUROSCI.2728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B, Schulte U. Regional Diversity and Developmental Dynamics of the AMPA-Receptor Proteome in the Mammalian Brain. Neuron. 2014;84(1):41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, Klocker N, Schulte U, Fakler B. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74(4):621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klocker N. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323(5919):1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Selcher JC, Xu W, Hanson JE, Malenka RC, Madison DV. Glutamate receptor subunit GluA1 is necessary for long-term potentiation and synapse unsilencing, but not long-term depression in mouse hippocampus. Brain Res. 2012;1435:8–14. doi: 10.1016/j.brainres.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR, 3rd, Nakagawa T. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 2012;1(6):590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A. 2010;107(37):16315–16319. doi: 10.1073/pnas.1011706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider C, Jayasinghe S, Hristova K, White SH. MPEx: a tool for exploring membrane proteins. Protein Sci. 2009;18(12):2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Hirano T. Visualization of subunit-specific delivery of glutamate receptors to postsynaptic membrane during hippocampal long-term potentiation. Cell Rep. 2012;1(4):291–298. doi: 10.1016/j.celrep.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Trimmer JS. Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron. 2015;85(2):238–256. doi: 10.1016/j.neuron.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS, Trowbridge IS, Vacquier VD. Monoclonal antibody to a membrane glycoprotein inhibits the acrosome reaction and associated Ca2+ and H+ fluxes of sea urchin sperm. Cell. 1985;40(3):697–703. doi: 10.1016/0092-8674(85)90218-1. [DOI] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harbor perspectives in biology. 2012;4(1):a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327(5972):1518–1522. doi: 10.1126/science.1184178. [DOI] [PubMed] [Google Scholar]
- Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Kaiser KM, Koster HJ, Borchardt T, Worley P, Lubke J, Frotscher M, Kelly PH, Sommer B, Andersen P, Seeburg PH, Sakmann B. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science. 1999;284(5421):1805–1811. doi: 10.1126/science.284.5421.1805. [DOI] [PubMed] [Google Scholar]