Abstract
The superior colliculus (SC) is a midbrain center involved in controlling head and eye movements in response to inputs from multiple sensory modalities. Visual inputs arise from both the retina and visual cortex and converge onto the superficial layer of the SC (sSC). Neurons in sSC send information to deeper layers of SC and to thalamic nuclei that modulate visually guided behaviors. Presently, our understanding of sSC neurons is impeded by a lack of molecular markers that define specific cell types. To better understand the identity and organization of sSC neurons, we took a systematic approach to investigate gene expression within four molecular families: transcription factors, cell adhesion molecules, neuropeptides and calcium binding proteins. Our analysis revealed 12 molecules with distinct expression patterns in mouse sSC: cadherin 7, contactin 3, netrin G2, cadherin 6, protocadherin 20, retinoid-related orphan receptor β, brain-specific homeobox/POU domain protein 3b, Ets variant gene 1, substance P, somatostatin, vasoactive intestinal polypeptide and parvalbumin. Double labeling experiments, by either in situ hybridization or immunostaining, demonstrated that the 12 molecular markers collectively define 10 different sSC neuronal types. The characteristic positions of these cell types divide sSC into 4 distinct layers. The 12 markers identified here will serve as valuable tools to examine molecular mechanisms that regulate development of sSC neuron types. These markers could also be used to examine the connections between specific cell types that form retinocollicular, corticocollicular or colliculothalamic pathways.
Keywords: retinal ganglion cells, transcription factors, cell adhesion molecules, neuropeptides, calcium binding proteins, RRID: nif-0000-00509, RRID: nif-0000-30467, RRID: AB_2167523, RRID: AB_91338, RRID: AB_10000240, RRID: AB_732196, RRID: AB_2298772, RRID: AB_10000344, RRID: AB_1288870, RRID: AB_518614, RRID: AB_2079751, RRID: AB_305869
Introduction
The superior colliculus (SC) is a midbrain center that controls orientating behaviors in response to sensory stimulation (Wurtz and Goldberg, 1971, 1972; Sparks, 1991; Isa and Hall, 2009; Mysore and Knudsen, 2011; Krauzlis et al., 2013). For example, the SC regulates gaze shifts that direct head and eye movements toward behaviorally relevant visual stimuli such as moving objects (Munoz et al., 1991; Guitton, 1992). Close topographic correlation among visual, auditory and somatosensory inputs to the SC coordinates behavioral responses toward spatial cues arising from multiple sensory modalities, and visual inputs influence the spatial alignment of these signals (Dräger and Hubel, 1975; Meredith and Stein, 1983; King et al., 1988; Triplett et al., 2009; Stein et al., 2014). Morphological studies revealed that the SC is composed of a highly organized, six-layer structure (May, 2006). The superficial layer of the SC (sSC) receives visual inputs from the retina and cortex, and contains neurons that project to deeper layers of the SC and to several areas including dorsal and ventral lateral geniculate nucleus, lateral posterior nucleus, parabigeminal nucleus, lateral pontine nucleus and pretectum. Deeper SC layers are interconnected with cortical and subcortical areas involved in auditory, somatosensory and motor function.
Organization of the sSC has been characterized along multiple dimensions. First, the sSC is organized retinotopically and also segregated into eye-specific regions (Constantine-Paton et al., 1990; McLaughlin and O’Leary, 2005; Feldheim and O’Leary, 2010). The development of both retinotopic and eye-specific organizations is regulated by a combination of molecular and activity-dependent mechanisms. Second, limited data describe a vertical columnar organization of sSC based on structural studies of retinal ganglion cell (RGC) axonal arbors or functional studies of sSC neurons (Huberman et al., 2008; Hong et al., 2011; Feinberg and Meister, 2015). Little is known about the developmental origin of this putative columnar organization. Third, a horizontal laminar organization of sSC has been proposed based on axonal projections of RGC types as well as cyto-and myeloarchitecture (Sachs and Schneider, 1984; Hofbauer and Dräger, 1985; Edwards et al., 1986a, 1986b; May, 2006). Such criteria divide sSC into 4 layers in the mouse: the stratum zonale (SZ), a thin layer located just below the pial surface; the stratum griseum superficialis (SGS) where most RGC axons arborize, and which can itself be further divided into upper (uSGS) and lower (lSGS) sublayers; the stratum opticum (SO), where RGC axons enter the SC at a deep level relative to the pial surface. However, the distinction between uSGS and lSGS depends on subjective evaluation in the absence of objective reference markers. Fourth, electrical recording and morphological studies characterize at least 6 different sSC neuronal types (Langer and Lund, 1974; Fukuda et al., 1978; Mooney et al., 1985; Wang et al., 2010; Gale and Murphy, 2014). However, a systematic catalog of cell types has been impeded by a lack of corresponding molecular markers that can distinguish the same types. By contrast, studies of retinal neurons demonstrate the utility of combining molecular and physiological features to facilitate cell type definitions (Hattar et al., 2002; Kim et al., 2008; Huberman et al., 2008; Huberman et al., 2009; Kim et al., 2010; Zhang et al., 2012; Park et al., 2015).
Historically, molecular studies of the mammalian SC and avian or amphibian tectum have focused on the development of retinotopic map formation or eye-specific segregation (Constantine-Paton et al., 1990; McLaughlin and O’Leary, 2005; Feldheim and O’Leary, 2010). However, individual surveys of a small number of molecules produced only a limited cell type classification (e.g., Laemle and Feldman, 1985; Okamoto et al., 1990; Behan et al., 1992; Mize et al., 1992; Ogawa-Meguro et al., 1992; Arai et al., 1993; Behan et al., 1993; Cork et al., 1998; Cork et al., 2000; Soares-Mota et al., 2001). Understanding the diversity of sSC neurons requires a systematic analysis of multiple molecular families. Identifying cell type-specific markers for sSC neurons could help us understand molecular mechanisms that regulate development of sSC neuronal types, as well as specific neuronal connections between sSC and retina, cortex or thalamus. The sSC neuron markers could also provide genetic access to specific cell types, enabling systematic functional analysis.
Here we screened molecular markers that exhibit layer-restricted expression within the mouse sSC and apparently label distinct sSC neuronal cell types. We surveyed diverse gene families and focused on four functional classes based on their presumed importance in neuronal development: 1) transcription factors, 2) cell adhesion molecules, 3) neuropeptides and 4) calcium binding proteins. Initial screening identified 12 molecules with differential expression. Double labeling, either by in situ hybridization or immunostaining, showed that our markers divide sSC into 4 different layers and define at least 10 distinct sSC neuronal types. Given that our markers labeled a small percentage of sSC neurons, we expect that the total number of sSC neuronal types is much greater than 10. These results suggest an unexpected degree of cell type diversity within the sSC.
Materials and Methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee at Yale University and were in compliance with federal guidelines. Samples from 3–4 mice, of either gender, were examined for each condition. Initial screening of molecular expression was performed on a CD1 background (Charles River) at P10-P13 and P18-P20. Subsequent experiments on cell type classification were mainly conducted on a C57/BL6 background (Jackson Laboratory) at P10-P13. No obvious differences were noted between strains. We used two mice, generated at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), in which a reporter was knocked into a retinoid-related orphan receptor β (Rorβ) gene: Rorβ1g/+ and Rorβ2Z/+. In the Rorβ1g/+ mouse, the reading frame sequence of green fluorescent protein (GFP) replaced the Rorβ1 specific exon; whereas in the Rorβ2Z/+ mouse, the reading frame sequence of β-galactosidase replaced the Rorβ2 specific exon (Liu et al., 2013; Fu et al., 2014). In both cases, we used heterozygotes in our analysis, so that Rorβ1 and Rorβ2 function is presumed to be normal. Indeed, no abnormalities were observed in these heterozygotes.
In situ hybridization
In situ hybridization was performed to detect expression of candidate molecules in the sSC at P10-P13 and P18-P20 following the method of Yamagata et al. (2002). Briefly, each DNA template amplified from P1 brain cDNA was subcloned into pGEM Teasy (Promega) or pCR8/GW/TOPO TA vector (Invitrogen). We used the entire coding region of cadherin 7 (NCBI reference sequence: NM_172853), contactin 3 (NM_008779), netrin G2 (NM_133500), cadherin 6 (NM_007666), protocadherin 20 (NM_178685), Rorβ (NM_146095), brain-specific homeobox/POU domain protein 3b (NM_138944), Ets variant gene 1 (NM_007960), substance P (NM_009311), somatostatin (NM_009215) and vasoactive intestinal polypeptide (NM_011702). Riboprobes (anti-sense) were synthesized using either digoxigenin-or fluorescein-labeled UTP. For single labeling with one in situ probe, signals were detected by anti-digoxigenin antibody conjugated to alkaline phosphatase. For double labeling with two in situ probes, peroxidase conjugated antibodies (either anti-digoxigenin-or anti-fluorescein-antibodies) were used and signals were detected by the tyramide signal amplification system (TSA-Plus system, Perkin-Elmer Life Sciences). For double labeling combined with GFP amplification, signals were first visualized by anti-digoxigenin antibody and then developed with anti-GFP antibody. Two different GFP antibodies (chicken and rabbit anti-GFP) were combined to increase detection sensitivity.
For in situ hybridization of Rorβ mRNA, we followed instructions described within the Allen Brain Atlas (RRID: nif-0000–00509) and used Rorβ2 as an in situ probe. We amplified a complete reading frame of Rorβ2 in our screening. The overall sequences of Rorβ1 and Rorβ2 are identical except for short, unique 5′ exons (Liu et al., 2013), such that our in situ probe detects mRNA for both Rorβ1 and Rorβ2 isoforms. The biochemical properties of Rorβ1 and Rorβ2 are similar in transcriptional activation (Fu et al., 2014). Using knockin reporter mice, strong signals were detected in the sSC in the Rorβ1g/+ strain but not in the Rorβ2z/+ strain. Hence, in situ hybridization measures of Rorβ expression represent predominantly the Rorβ1 isoform. For double labeling experiments, the Rorβ in situ probe did not work well, so we switched to analyze Rorβ1 expression in the Rorβ1g/+ mouse: GFP was visualized by immunostaining as a measure of Rorβ1 expression. Below, measures of Rorβ expression based on in situ hybridization refer collectively to Rorβ1 and Rorβ2.
Immunohistochemistry
Mice were anesthetized by intraperitoneal injection of ketamine/xylazine and perfused transcardially with 4% paraformaldehyde (PFA)/PBS. Following perfusion, brains were dissected, post-fixed overnight at 4°C, incubated sequentially first with 15% sucrose/PBS and then 30% sucrose/PBS overnight at 4°C, and parasagittally sectioned with a cryostat (18–20 μm). For vibratome sections (50 μm), tissues were kept in 4% PFA/PBS overnight at 4°C and washed with PBS before sectioning. For immunostaining, brain sections were washed twice with PBS for 5 min each time at room temperature, blocked with 3% donkey serum/0.3–0.5% Triton X-100/PBS for 30 min at room temperature, and incubated with the primary antibodies for 2–3 days at 4°C. Sections were incubated with the secondary antibodies for 2 hr at room temperature. Primary antibodies are listed in Table 1. Secondary antibodies were conjugated to AlexaFluor-488, Cy3 or Cy5 (Jackson Immuno Research Laboratories) and diluted at 1:500.
Table 1.
Primary antibodies used in this study.
| Antibody | Immunogen | Source (RRID number) | Dilution factor |
|---|---|---|---|
| Brn3b | Human Brn3b | Santa Cruz Biotechnology, #sc-31989, (RRID: AB_2167523), goat polyclonal | 1:250 |
| ChAT | Human Placental ChAT | Millipore, #AB144P, (RRID:AB_2079751), goat polyclonal | 1:250 |
| GFP | GFP isolated from Aequorea victoria | Millipore, #AB3080P, (RRID: AB_91338), rabbit polyclonal | 1:500 |
| GFP | recombinant GFP | Aves Labs, #GFP-1020, (RRID: AB_10000240), chicken polyclonal | 1:500 |
| ETV1 | synthetic mouse C-terminal peptide | Abcam, #ab36788, (RRID: AB_732196), rabbit polyclonal | 1:2000 |
| MBP | Cow full length protein | Abcam, #ab7349, (RRID: AB_305869), rat monoclonal | 1:500 |
| NeuN | mouse brain nuclei | Millipore, #MAB377, (RRID: AB_2298772), mouse monoclonal | 1:1000 |
| Parvalbumin | rat muscle parvalbumin | Swant, Switzerland, #PV25, (RRID: AB_10000344), rabbit polyclonal | 1:2000 |
| Substance P | substance P-BSA | Fitzgerald Industries International, #10-S15A, (RRID: AB_1288870), rat monoclonal | 1:250 |
| Somatostatin | synthetic somatostatin peptide | BACHEM (Peninsula lab), #T-4103.0050, (RRID: AB_518614), rabbit polyclonal | 1:1000 |
Intraocular eye injection
Mice were anaesthetized by intraperitoneal injection of ketamine/xylazine. A small hole was made in the eye with an insect pin to release intraocular pressure. Cholera toxin b subunit conjugated to either Alexa Fluor-488 or 647 (1–2 μl of 1 mg/ml, Invitrogen) was injected through the same hole using a pressure injector (Harvard Apparatus). After injection, mice were observed to ensure proper recovery. Two to three days after injection, mice were sacrificed by intraperitoneal injection of ketamine/xylazine and perfused transcardially with 4% PFA/PBS.
Image analysis and statistics
Images were acquired using a Zeiss Imager M2 fluorescent microscope and a Zeiss LSM5 Exciter confocal microscope. A z-stack was collected with 1-μm steps and a 20X objective (NA = 0.8). To ensure that analyses were conducted on similar regions, images were taken from a central sSC area that spans ~200 μm from the medial edge to ~200 μm from the lateral edge, and ~520 μm from the rostral edge to ~600 μm from the caudal edge. For cell counting, images (~572 ± 144 μm from the rostral edge) were cropped to either 350 μm × 300 μm or 800 μm × 800 μm (width × height), and image depth was collapsed over 10–11 μm. Data were analyzed using Image J (RRID:nif-0000–30467, National Institutes of Health). Cell numbers were obtained from 7–12 different sections in 3–4 different animals. Data are reported as mean ± SD.
Antibody characterization
Primary antibodies are listed in Table 1.
The goat brain-specific homeobox/POU domain protein 3b (Brn3b) antibody (Santa Cruz Biotechnology, #sc-31989, RRID: AB_2167523) was generated against an internal region of Brn3b of human origin. In western blots, the Brn3b antibody detects the expected size band of 51kDa in mouse eye tissue (manufacturer’s specification). By immunohistochemistry, this antibody labels retinal ganglion cells in mouse retina as expected (Nie et al., 2010; Nadal-Nicolás et al., 2012). We detected similar expression patterns in sSC between in situ hybridization, with a specific probe to Brn3b, and immunostaining with the Brn3b antibody.
The goat choline acetyltransferase (ChAT) antibody (Millipore, #AB144P, RRID: AB_2079751) was generated against human placental ChAT. The staining specificity was demonstrated by the absence of all ChAT staining after preabsorption of an antibody with human placental ChAT (Kha et al., 2000).
The rabbit green fluorescent protein (GFP) antibody (Millipore, #AB3080P, RRID: AB_91338) was generated against purified native GFP from Aequorea victoria. This antibody produces no signals in tissues from wildtype mice (Hong et al., 2011).
The chicken GFP antibody (Aves Labs, #GFP-1020, RRID: AB_10000240) was generated against purified recombinant GFP. Antibodies were analyzed by western blot analysis and immunostaining using transgenic mice expressing the GFP gene product (manufacturer’s specification). No signals were detected in either wildtype mice or in the absence of this antibody (Gautron et al., 2010).
The rabbit Ets variant gene 1 (ETV1) antibody (Abcam, #ab36788, RRID: AB_732196) was generated by a synthetic mouse C-terminal peptide sequence. The specificity of this antibody was demonstrated by western blot in PC3 cells transfected with the siRNA (Vitari et al., 2011). We detected similar expression patterns in sSC between in situ hybridization, with a specific probe to ETV1, and immunostaining with this antibody.
The rat myelin basic protein (MBP) antibody (Abcam, #ab7349, RRID: AB_305869) was generated by a cow full-length protein and recognizes a single band of 19.2 kDa in western blot (manufacturer’s specification). This antibody recognizes MBP present in the myelin sheets of oligodendrocytes and exhibited the same staining pattern in two independent studies (Yang et al., 2013; Sevc et al., 2014).
The mouse neuronal specific nuclear protein (NeuN) antibody (Millipore, #MAB377, RRID: AB_2298772) recognizes a neuron-specific nuclear protein in many vertebrates. In western blot, this antibody recognizes two to three bands in the 46–48 kDa range and possibly another band of approximately 66 kDa (manufacturer’s specification). NeuN staining is detected in neuronal but not in non-neuronal tissues (Mullen et al., 1992; Seelke et al., 2013).
The rabbit parvalbumin (PV) antibody (Swant, #PV25, RRID: AB_10000344) was generated against rat muscle PV and cross-reacts with many other species. This antibody labels a subpopulation of neurons in the normal brain, but not in PV knockout mice (manufacturer’s specification). PV staining patterns were consistent across studies (Bunce et al., 2013; Cai et al., 2013).
The rat substance P (SP) antibody (Fitzgerald Industries International, #10-S15A, RRID: AB_1288870) has no cross-reactivity with Let-or Met-enkephalin, somatostatin or β-endorphin (manufacturer’s specification). The specificity of this antibody (rat) was demonstrated by showing complete overlap with a second antibody to SP (rabbit anti-SP, #20064, Immunostar; Russo et al., 2010). The specificity of the second antibody was confirmed by the absorption test.
The rabbit somatostatin (SST) antibody (BACHEM, #T-4103.0050, RRID: AB_518614) was generated against a synthetic peptide (manufacturer’s specification). The specificity of this antibody was determined by immunohistochemistry (i.e., lack of staining) in SST knockout mice (Saito et al., 2005).
Nomenclature
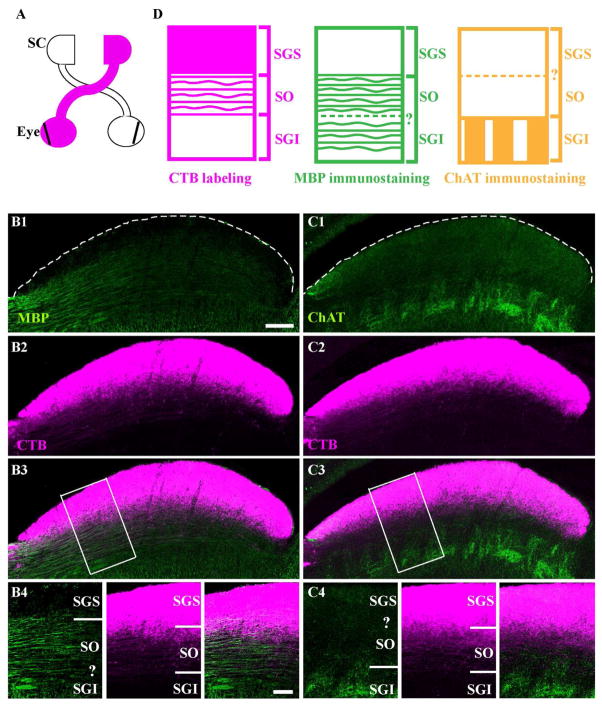
Conventional nomenclature that refers to layer distinction within the sSC is, starting at the pial surface: the stratum zonale (SZ), the stratum griseum superficialis (SGS) and the stratum opticum (SO). In the mouse, the SZ is extremely thin and we did not attempt to distinguish it from the SGS. Hence, our reference to SGS includes the SZ. To visualize SGS and SO layer boundaries, we injected cholera toxin b conjugated to Alexa Fluor dyes (CTB) into an eye (Fig. 1A) and immunostained the contralateral SC with antibodies against myelin-basic protein (MBP, Fig. 1B) and choline actetyltransferase (ChAT) (Fig. 1C). The boundary between SGS and SO was visualized by either an intensity increase of myelinated-fiber staining or by dense labeling of CTB (Fig. 1B). The lower boundary of SO was visualized either by the absence of ChAT-enriched fiber staining or by the deepest retinal fiber labeled by CTB (Fig. 1C). In subsequent experiments, we used CTB labeling to mark the upper and lower boundaries of SO (Fig. 1D).
Figure 1. Visual layer boundaries of mouse superior colliculus (SC).
A. A schematic shows the labeling of the contralateral superficial part of the SC after cholera toxin b subunit (CTB) injection into one eye (magenta). Vibratome sections were subsequently prepared at ~P22 and immunostained. B. A section stained with anti-myelin basic protein (MBP, green) and CTB. The dashed line delineates the pial surface. A boundary between stratum griseum superficiale (SGS) and stratum opticum (SO) is delineated by the boundary between intense labeling of myelinated fibers (B1) and dense labeling of CTB (B2). From the merged image (B3), a higher magnification of the boxed area is shown (B4). The question mark indicates the inability to locate the boundary between SO and stratum griseum intermediale (SGI) based on MBP labeling. C. A section stained with anti-choline acetyltransferase (ChAT, green). The dashed line delineates the pial surface. A boundary between SO and SGI is delineated by the boundary between intense labeling of ChAT-positive fibers (C1) and the deepest retinal fiber labeled by CTB (C2). From the merged image (C3), a higher magnification of the boxed area is shown (C4). The question mark indicates the inability to locate the boundary between SGS and SO based on ChAT labeling. D. Schematic of layer distinctions based on CTB labeling (magenta), MBP staining (green) and ChAT staining (yellow). The dashed lines and question marks indicate the inability to locate certain boundaries. Scale bar: 200 μm (B1-B3 and C1-C3), 100 μm (B4 and C4).
Cellular lamination by soma position in the mouse SC, at the time point when initial screening was conducted (~P10 and ~P18), was less distinguishable by traditional histological methods and became undetectable after in situ hybridization. However, we retrospectively indicated the boundary between putative SGS and putative SO in the initial screening based on co-labeling with CTB and antibodies to the genes selected from our screening. Later, when we conducted double in situ hybridization with molecules selected from the screening, we referred to cellular lamination by a new system for describing lamination composed of four layers, starting at the pial surface. We consider layer 1 as a combination of SZ and upper SGS.
We classified sSC neurons into types according to the cell groups with the most unique patterns of molecular expression. We recognize that these types can be further divided in the future when additional molecular or physiological features can reasonably distinguish the types described here.
Documentation of the screened genes
The genes that were screened in our study, but not pursued below, are listed in supplementary material (Suppl. table 1).
Results
To systematically investigate molecular features of mouse superficial superior colliculus (sSC) neurons, we surveyed diverse gene families. Candidate genes were selected using 3 approaches. First, we examined gene expression in public databases, including the Allen Brain Atlas. Second, we considered genes with layer-specific expression patterns in the cortex, on the assumption that the same or related genes would show layer-specific expression in the sSC. Third, we screened cell surface genes, based on the assumption that such molecules could be involved in intercellular interactions between sSC neurons and retinal ganglion cells (RGCs), cortical neurons or thalamic neurons. Among the entire list of genes, we narrowed down our analysis to 4 molecular families: 1) transcription factors, 2) cell adhesion molecules, 3) neuropeptides and 4) calcium binding proteins.
Initial screening was conducted at P10-P13 (~P10) and P18-P20 (~P18). We expected that overall development within the sSC would be nearly complete by ~P10 because: 1) Neurogenesis is complete during the embryonic stage and the radial thickness of the sSC reaches ~90% of its mature level by P10 (Edwards et al., 1986b); 2) Layer-restricted axonal arborization of RGCs is complete around eye opening (P12-P14 in mouse; Kim et al., 2010), and sSC neurons acquire adult-like morphology by P15 (Warton and Jones, 1985; Sachs et al., 1986). Expression patterns were examined by a combination of in situ hybridization (ISH) and immunohistochemistry. We focused our analysis on a central sSC area to limit positional variation between experiments (see Methods). We also qualitatively analyzed expression through the entire sSC and found no obvious variation in most cases, with exceptions noted below. The initial screening defined different layers of sSC using traditional nomenclature. Subsequent studies incorporate a new system for defining layers, based on molecular expression patterns (see Methods).
Molecular markers label subsets of sSC neurons
Transcription factors
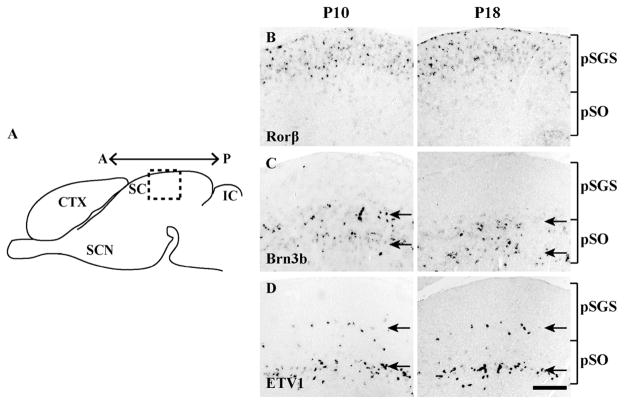
Transcription factors could control the cell fate of specific sSC neuronal types. We screened expression of 10 candidate genes and found that three showed layer-specific expression patterns. Retinoid-related orphan receptor β (Rorβ) was restricted to the superficial layer, whereas expression of Brn3b and ETV1 were absent from this layer (Fig. 2B–D). Brn3b and ETV1 were each expressed within two bands (Fig. 2C–D, respectively). In both cases, these bands were more obvious in the lateral sSC (Fig. 2C–D) relative to the medial sSC (data not shown). The upper band of ETV1+ cells was very sparse, whereas the lower band of cells was dense.
Figure 2. Expression patterns of three transcription factors.
A. A schematic sagittal section of the mouse brain. The boxed area indicates the location where experiments were conducted (~570 ± 140 μm from anterior edge) on the superficial part of the SC (sSC). Also shown are the inferior colliculus (IC), cortex (CTX) and suprachiasmatic nucleus (SCN). A← →P, anterior to posterior axis. B–D. In situ hybridization with each probe was conducted at P10-P13 (left) and P18-P20 (right). The relative position of putative stratum griseum superficiale (pSGS) and putative stratum opticum (pSO) are indicated by brackets. B. Expression of Rorβ is confined to a superficial layer of sSC. C. Expression of Brn3b is confined to a deep layer. Within the region of expression, Brn3b formed two distinct bands in the lateral sSC (shown, arrows); these bands became less distinct in the medial sSC (data not shown). D. Expression of ETV1 is confined to two distinct bands (arrows): a sparse upper band and a denser lower band. In all cases, expression patterns were similar at ~P10 and ~P18. Scale bar: 200 μm.
Cell adhesions molecules
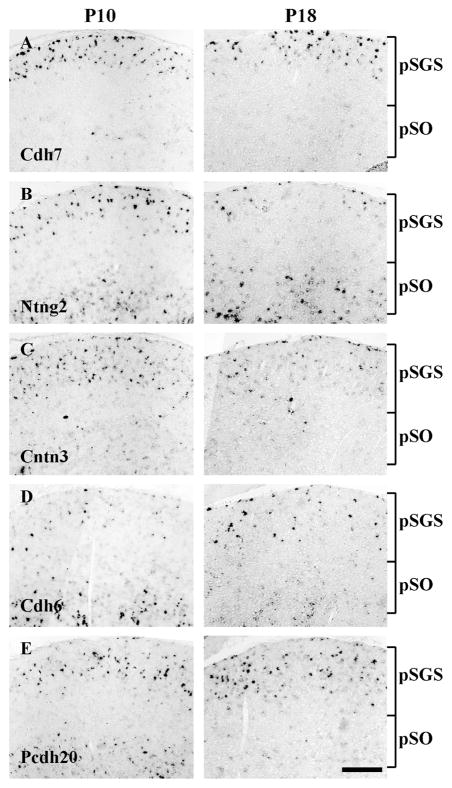
Specific connections between neuronal types can be mediated by neural recognition molecules expressed by presynaptic and postsynaptic neurons, including Sidekicks, Dscams, and Semaphorins (Yamagata et al., 2002; Yamagata and Sanes, 2008; Krishnaswamy et al., 2015; Sun et al., 2015). We screened expression of ~142 genes described earlier (Gu et al., 2015). Among them, we found 5 molecules that showed distinct expression patterns in the sSC. Cadherin 7 (Cdh7) expression was confined to the superficial layer of sSC (Fig. 3A). Netrin G2 (Ntng2) and protocadherin 20 (Pcdh20) expression were each confined to both superficial and deep layers, with a gap in between (Fig. 3B and 3E, respectively). Pcdh20 expression in the deep layer declined by ~P18. Contactin 3 (Cntn3) expression was found in both superficial and deep layers, with more dominant expression in the superficial layer (Fig. 3C). Finally, cadherin 6 (Cdh6) expression was sparse in the superficial layer and more abundant in the deep layer (Fig. 3D). Similar to Pcdh20, expression of Cdh6 in the deep layer declined by ~P18.
Figure 3. Expression patterns of five cell adhesion molecules.
A–E. In situ hybridization with each probe was conducted in the sSC at ~P10 (left) and ~P18 (right). The positions of pSGS and pSO are indicated by brackets. A. Expression of cadherin 7 (Cdh7) was predominantly confined to a superficial layer of sSC. B. Expression of netrin G2 (Ntng2) was found in both superficial and deep layers, with a gap in between. C. Expression of contactin 3 (Cntn3) was most abundant in a superficial layer. D. Expression of cadherin 6 (Cdh6) was detectable in both superficial and deep layers at ~P10. The expression in the deep layer decreased at ~P18. E. Expression of Pcdh20 was found in both superficial and deep layers, with a gap in between at ~P10. The expression in the deep layer decreased at ~P18. Scale bar: 200 μm.
Neuropeptides/Calcium binding proteins
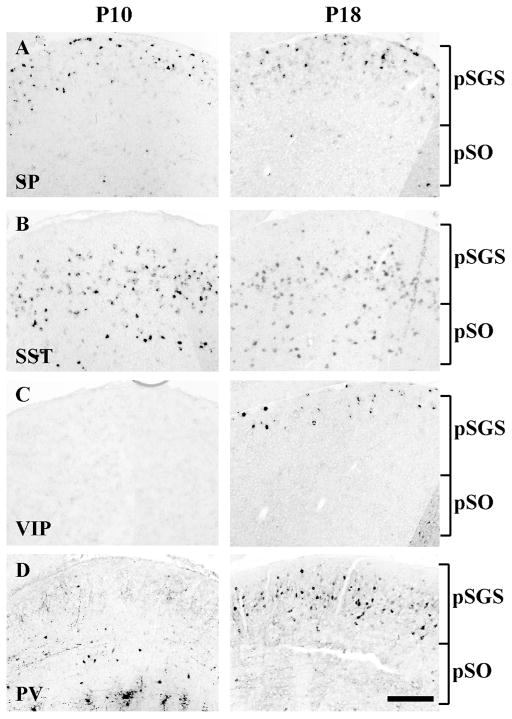
Individual surveys of sSC markers include various neuropeptides and calcium binding proteins identified by antibodies (e.g., Laemle and Feldman, 1985; Okamoto et al., 1990; Behan et al., 1992; Ogawa-Meguro et al., 1992; Arai et al., 1993; Behan et al., 1993; Cork et al., 1998; Cork et al., 2000; Soares-Mota et al., 2001). Given that neuropeptides and calcium binding proteins play important roles in diverse biological phenomena, we reexamined 9 markers to comprehensively determine their expression patterns. We identified 4 molecules that showed selective expression. Substance P (SP) and vasoactive intestinal polypeptide (VIP) expression were each primarily restricted to the superficial layer of sSC (Fig. 4A and C). We occasionally detected SP expression in the deep layer. Somatostatin (SST) expression was found in middle and deeper layers (Fig. 4B), and parvalbumin (PV) expression was found in a superficial layer, but missing near the pial surface (Fig. 4D). Expression of VIP was not present at ~P10 but became prominent by ~P18. Expression of PV was also more detectable at ~P18 relative to ~P10, consistent with cortical studies that described the corresponding neuronal types as late-born populations (Wonders and Anderson, 2006; Miyoshi et al., 2007).
Figure 4. Expression patterns of four neuropeptides/calcium binding proteins.
A–C. In situ hybridization with each probe was conducted in the sSC at ~P10 (left) and ~P18 (right). The positions of pSGS and pSO are indicated by brackets. A. Expression of substance P (SP) was predominantly found in a superficial layer of sSC. B. Expression of somatostatin (SST) was found in middle and deep layers. C. Expression of vasoactive intestinal polypeptide (VIP) was barely detectable at ~P10 but was found in a superficial layer at ~P18. D. Expression of parvalbumin (PV), detected by immunostaining, was sparse at ~P10 but became abundant in a superficial layer, well below the pial surface, at ~P18. Scale bar: 200 μm.
Molecular markers subdivide the sSC into 4 different layers
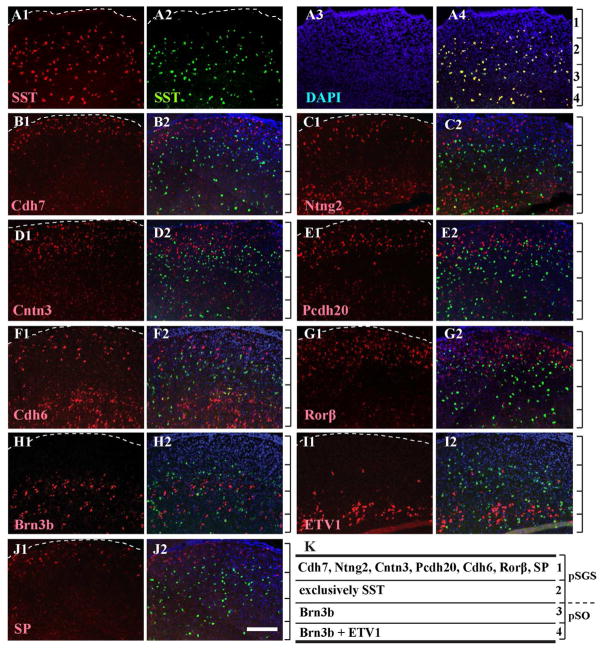
To compare the relative positions of sSC cells that express each molecule of interest, we performed double fluorescent in situ hybridization (dFISH). In each case, we looked for a probe that worked well as either the first (labeled with digoxigenin) or the second probe (labeled with fluorescein: see Methods). Two probes to SST overlapped almost completely with one another: ~95.1% of digoxigenin-labeled cells were fluorescein positive, and ~91.5% of fluorescein-labeled cells were digoxigenin positive (Fig. 5A). Accordingly, we used SST as a reference marker in subsequent dFISH analyses. The five adhesion molecules (Cdh7, Ntng2, Cntn3, Pcdh20, and Cdh6) were all mainly localized to the superficial layer of sSC, above the layer of SST expression (Fig. 5B–F). Rorβ and SP also localized to the superficial layer of sSC, above the layer of SST expression (Fig. 5G and 5J). Pcdh20, Cntn3, Cdh6 and Rorβ had a slightly broader area of expression than Cdh7, Ntng2 and SP. Expression of Ntng2, Cntn3, Cdh6, Pcdh20 and SP also localized below the layer of SST expression. The relative expression of each molecule beneath the SST layer was not evaluated further because it extended below the boundary of the sSC.
Figure 5. Relative distribution of nine markers compared to expression pattern of SST.
A–J. Double fluorescent in situ hybridization (dFISH) was conducted in sSC at ~P10 using SST as a common second probe (labeled with fluorescein, green) and counterstained with DAPI (blue). The first probes (labeled with digoxigenin) are shown in red. The dashed line delineates the pial surface. Relative positions of four layers, defined by the collective expression patterns (see part K), are indicated by the brackets to the right of the combined images. A. Two in situ probes to SST (A1 and A2) overlapped well, which validates the use of SST in subsequent panels (merged, A4; DAPI alone, A3). B. Expression of Cdh7 (B1, red) is found above the region of SST expression (B2, merged). C–G. Expression of Ntng2 (C1), Cntn3 (D1), Pcdh20 (E1), Cdh6 (F1) and Rorβ (G1) are also primarily located above the region of SST expression (C2, D2, E2, F2 and G2; merged). H–I. Expression of Brn3b and ETV1 (H1 and I1) are primarily located within and below the region of SST expression (H2 and I2, merged). The upper band of ETV1 expression was not used as a reference for the layer distinctions because of the sparse labeling. J. Expression of SP (J1) is predominantly located above the region of SST expression (J2, merged). K. Schematic showing layer-enriched expression of each molecule. Brackets show layers 1–4, as distinguished by molecular markers; a dashed line shows the boundary between pSGS and pSO. Scale bar: 200 μm.
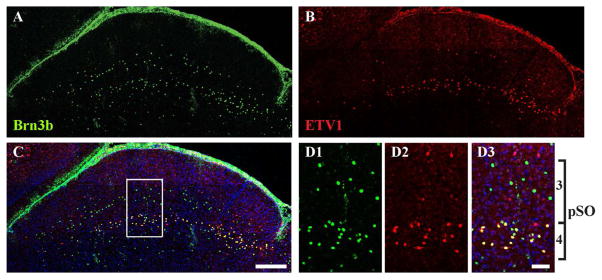
Brn3b and ETV1 expressions were barely noticeable above the layer of SST expression (Fig. 5H–I). In the layer of SST expression, there was no clear overlap (within cells) between SST and Brn3b expression or between SST and ETV1 expression. Upper bands of Brn3b and ETV1 expression were each localized within the SST layer. The lower band of ETV1 expression was particularly well defined. To compare Brn3b and ETV1 expression, we conducted double immunostaining and confirmed that Brn3b and ETV1 each form two bands of expression within sSC (Fig. 6). Interestingly, some neurons within the lower band co-expressed Brn3b and ETV1 (Fig. 6D). Consequently, we quantified the degree of overlap below.
Figure 6. Direct comparison of expression patterns for Brn3b and ETV1.
Sections of sSC at ~P10 were stained with antibodies to both Brn3b (green, A) and ETV1 (red, B) and counterstained with DAPI (blue). The merged image is shown in C. The boxed area in C is shown at higher magnification in D1 (Brn3b), D2 (ETV1) and D3 (merged). Both bands of Brn3b+ and ETV1+ cells were visible. Individual cells were labeled by either one (red or green) or both antibodies (yellow). Layers 3 and 4 together comprise pSO (Figure 5K). Scale bar: (A–C) 200 μm, (D) 50 μm.
A selective grouping of sSC neurons, based on location of our molecular markers allowed us to divide sSC into 4 layers (Fig. 5K). Layer 1 is adjacent to the pia and defined by expression of Cdh7, Ntng2, Cntn3, Pcdh20, Cdh6, Rorβ and SP. Layer 2 is defined by almost exclusive SST expression and extends to the upper band of Brn3b expression. Layer 3 is defined by the region bracketed by the two bands of Brn3b expression, and Layer 4 is defined by the lower bands of Brn3b and ETV1 expression. The upper band of ETV1 expression was not considered as a reference for layer distinction because of its sparse labeling. We did not include PV or VIP expression in our analysis because dFISH methods worked poorly at P18, which is when PV and VIP expression became strong enough to analyze.
Comparison of two different layer-classification methods in sSC
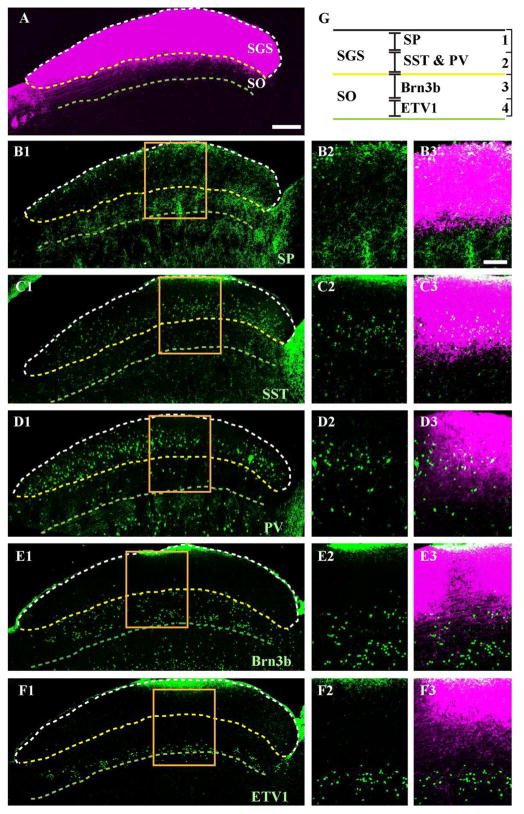
A traditional classification using axonal arborization of RGCs divides sSC into 4 layers (Sachs and Schneider, 1984; Hofbauer and Dräger, 1985; Edwards et al., 1986a; May, 2006): (1) the stratum zonale (SZ), a thin layer located just below the pial surface where a small number of RGC axons project; the stratum griseum superficialis (SGS), where most RGC axons arborize into either (2) an upper (uSGS) or (3) lower (lSGS) sublayer; and (4) the stratum opticum (SO), where RGC axons enter the SC. As mentioned above (see Methods), we did not attempt to distinguish SZ from SGS and refer to them collectively as SGS. To compare the classification based on RGC axonal arborization to our current classification based on soma position of sSC neurons, we sought commercially available antibodies for our sSC neuron markers. Suitable antibodies were available for SP, SST, PV, Brn3b and ETV1. We examined the positions of these antibody labels relative to the pattern of RGC axonal projections labeled by CTB injected into the eye (Fig. 7A). Our analysis revealed that: 1) Layer 1 (defined by SP expression) corresponds to uSGS (Fig. 7B); 2) Layer 2 (defined by SST or PV expression) corresponds to lSGS (Fig. 7C and D); 3) Layer 3 (defined by the region within the bands of Brn3b expression) sits just below the boundary dividing SGS and SO (Fig. 7E); and 4) Layer 4 (defined by the lower bands of Brn3b and ETV1 expression) corresponds to the lower boundary of SO (Fig. 7F). The lower boundary of SO was defined by the deepest retinal fibers labeled by CTB.
Figure 7. Relationship between sSC layers defined by either sSC cell markers or retinal ganglion cell axons.
Vibratome sections at ~P18 were immunostained with specified antibodies (green) after injection of CTB (magenta) into the contralateral eye. A. A sagittal section of sSC labeled with CTB. The pial surface is delineated by a dashed white line, the lower border of stratum griseum superficiale (SGS) by a dashed yellow line and the lower border of stratum opticum (SO) by a dashed green line. B1. A section labeled with anti-SP (green). The retinorecipient layers of sSC are indicated by dashed lines based on CTB labeling, as in A. The higher magnification of the boxed area is shown in B2 (SP) and B3 (merged; SP and CTB labeling). C–F. Same format as B. but with anti-SST, anti-PV, anti-Brn3b and anti-ETV1. G. The schematic shows relative positions of sSC layers defined by either sSC cell markers (layers 1–4) or RGC axons (SGS, SO). Scale bar: (A, B1, C1, D1, E1 and F1) 200 μm, (B2-B3, C2-C3, D2-D3, E2-E3 and F2-F3) 100 μm.
Classification of sSC neuronal types by systematic analysis of molecular expression
Electrical recording and random dye injection define 6–7 different types of sSC neurons, although it is not expected that this is a comprehensive classification (Langer and Lund, 1974; Fukuda et al., 1978; Mooney et al., 1985; Wang et al., 2010; Gale and Murphy, 2014). In this study, we investigated whether molecular features could also distinguish cell types. We focused our analysis at P10-P13, when double labeling methods (either dFISH or combined ISH and immunostaining) worked well. We assume that our results can be extrapolated to the adult sSC for two reasons. First, neurogenesis is complete during the embryonic stage and SC radial growth reaches ~90% of its adult value by P10 (Edwards et al., 1986b). Second, we found that development of NeuN+ cells (i.e., neurons) reached a level that was indistinguishable between the two time points of interest: P10, 3.5 × 105 ± 8.1 × 103 mm−3; P18, 3.4 × 105 ± 1.5 × 104 mm−3.
Layer 1
Most markers we identified were located within layer 1 where the majority of RGC axons terminate. We first sought probes among our candidates that could serve as a reference marker for sSC cell type classification. Many genes were detectable by ISH but suitable antibodies for the corresponding protein were not available. We found two genes that could be used for double labeling experiments, Rorβ and Ntng2. Double labeling for Rorβ was enabled by a knockin reporter line described below, and Ntng2 could be used in dFISH experiments as the second probe.
Rorβ
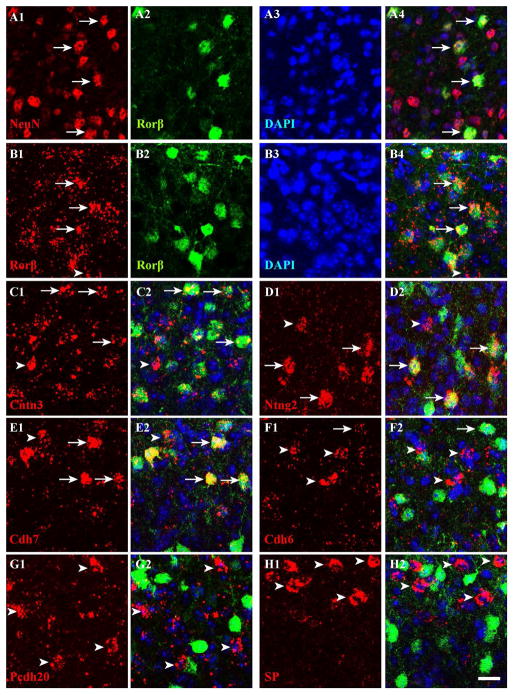
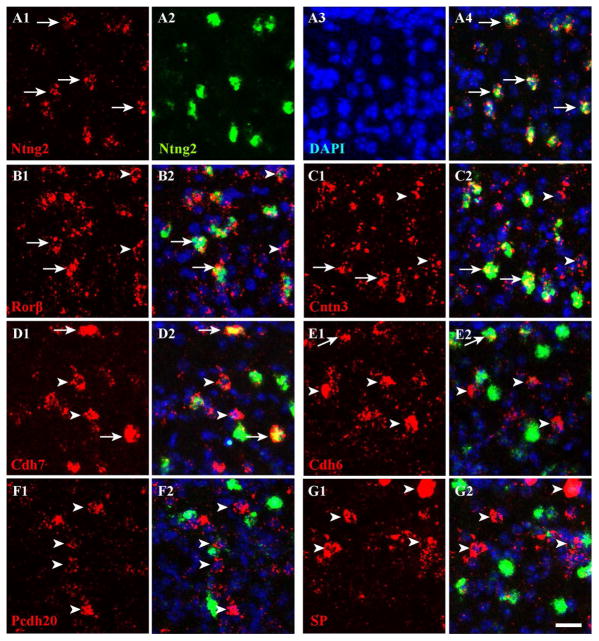
To visualize Rorβ expression, the in situ probe did not work as the second probe, and so we instead used transgenic mice in which the Rorβ1-specific exon is replaced with GFP (Rorβ1g/+) (see Methods). We amplified GFP signals with antibodies in the Rorβ1g/+ mouse, and confirmed that Rorβ1 is highly expressed in sSC (Fig. 8B). Double immunostaining with GFP and NeuN antibodies revealed that most GFP+ cells (~93%) were NeuN+, whereas only ~20% of NeuN+ cells were GFP+ (Fig. 8A). Rorβ apparently marks a small subpopulation of sSC neurons. Most Rorβ in situ signals (~81%) overlapped with GFP signals in the Rorβ1g/+ mouse and vice versa (~84%), suggesting that expression of the transgene in Rorβ1g/+ mice mimics endogenous Rorβ expression (Fig. 8B). Double labeling by ISH and immunostaining with anti-GFP antibodies revealed that most Ntng2+, Cntn3+ and Cdh7+ were Rorβ+ (~82%, ~76% and ~70% respectively; Fig. 8C–E), suggesting that Ntng2, Cntn3 and Cdh7 are highly expressed by Rorβ+ neurons. Among Rorβ+ neurons, ~64% of cells were Cntn3+, ~52% were Ntng2, and ~53% were Cdh7+. A small number of Cdh6+, Pcdh20+ and SP+ cells were Rorβ+ (~14%, ~2.3% and 3.4%, respectively; Fig. 8F–H). Among Rorβ+ neurons, ~7.9% were Cdh6+, ~1.4% were Pcdh20+, and ~2.2% were SP+. These data indicate that Cdh6, Pcdh20 and SP expressions primarily label cell populations distinct from Rorβ+ neurons in sSC. A summary of numerical values, with SD for each measurement, is presented in Table 2.
Figure 8. Rorβ marks a subpopulation of sSC neurons.
A. Expression of Rorβ was detected by anti-GFP in the Rorβ1g/+ mouse (see Methods). All images in this figure were acquired in layer 1. Sections of sSC at ~P10 were stained with antibodies to NeuN (red, A1) and GFP (green, A2) and counterstained with DAPI (A3). The merged image (A4) shows that Rorβ+ cells are neurons (i.e., NeuN+; arrows). B–H. Double labeling was conducted at ~P10 in the Rorβ1g/+ mouse by ISH (first probes, digoxigenin, red) and immunostaining (second probe, anti-GFP, green) and counterstained with DAPI (blue). Arrows indicate overlapping expression and arrowheads indicate expression by the first probe only. Arrows are missing in G and H because overlapping expression between the two genes is rare (<5%). B. An in situ probe to Rorβ (B1, red) and anti-GFP signal (B2, green) highly overlapped (B4, merged; B3, DAPI alone). C–E. The majority of cells expressing Cntn3 (C1), Ntng2 (D1) and Cdh7 (E1) were Rorβ+ (merged; C2, D2, E2). F–H. Very few cells expressing Cdh6 (F1), Pcdh20 (G1) and SP (H1) were Rorβ+ (merged; F2, G2, H2). Scale bar: 20 μm.
Table 2.
Relationship among sSC neuronal markers in layer 1.
| Gene (X) | % (Rorβ+ ∩ X+) / Rorβ+ | % (Rorβ+ ∩ X+) / X+ | % (Ntng2+ ∩ X+) / Ntng2+ | % (Ntng2+ ∩ X+) / X+ |
|---|---|---|---|---|
| Rorβ | 83.6 ± 5.3 | 81.1 ± 3.0 | 78.6 ± 4.6* | 50.3 ± 8.1# |
| Ntng2 | 51.5 ± 6.4# | 81.8 ± 4.3* | 82.5 ± 3.4 | 89.3 ± 1.9 |
| Cdh7 | 53.3 ± 3.5 | 70.1 ± 5.4 | 45.8 ± 3.7 | 43.5 ± 3.9 |
| Cntn3 | 64.4 ± 3.0 | 75.8 ± 4.0 | 58.9 ± 9.7 | 50.3 ± 5.5 |
| Cdh6 | 7.9 ± 2.3 | 14.3 ± 3.4 | 9.6 ± 2.1 | 18.0 ± 3.6 |
| Pcdh20 | 1.4 ± 1.4 | 2.3 ± 2.1 | 2.3 ± 1.3 | 2.5 ± 1.6 |
| SP | 2.2 ± 1.9 | 3.4 ± 3.1 | 4.9 ± 2.5 | 7.8 ± 4.3 |
Summary of results from double In situ hybridization using either Rorβ (2nd and 3rd columns) or Ntng2 (4th and 5th columns) as the second probe. The number in each case indicates the average overlap (∩) between the second probe and each first probe (X) or vice versa. Data (mean ± SD) were averaged across 7–12 sections from at least 3 independent experiments. Symbols (*, #) indicate cases where two different second probes produced similar degrees of overlapping expression between Rorβ and Ntng2, suggesting that our double In situ hybridization results are highly reproducible.
Ntng2
To classify further the Rorβ+ population, we used a second probe to Ntng2 in dFISH experiments. We confirmed that two probes to Ntng2 overlapped strongly (~89% of digoxigenin-labeled cells were fluorescein-positive; and ~83% of fluorescein-labeled cells were digoxigenin-positive; Fig. 9A). Subsequent experiments showed that Ntng2 was significantly co-expressed with Rorβ (~79%; Fig. 9B). This result matches the overlap between Ntng2 and Rorβ when Rorβ/GFP was used as the second probe (i.e., detected by GFP signals in Rorβ1g/+ mouse; ~82%, Fig. 8D), suggesting that our double labeling quantification yields reliable results. Approximately half of Ntng2+ cells were Cntn3+ (~59%) or Cdh7+ (~46%; Fig. 9C–D). Ntng2 labels ~50% of Rorβ+, ~44% of Cdh7+, and ~50% of Cntn3+ cells. Two different approaches (using either Ntng2 or Rorβ/GFP as a second probe) confirmed that Ntng2 marks roughly half of Rorβ+ neurons (~50% with Ntng2 versus ~52% with Rorβ/GFP). A small percentage of Pcdh20+ and SP+ cells were Ntng2+ (~2.5% and ~7.8%, respectively; Fig. 9F–G). Pcdh20 and SP also labeled a small percentage of Ntng2+ cells (~2.3% and ~4.9%, respectively). These data were anticipated from previous observations that: 1) the majority of Ntng2 (~80%) cells belong to the Rorβ+ population, and 2) overlap between either Pcdh20 or SP and Rorβ is minimal (~1.4% or ~2.2%, respectively; Table 2). Interestingly, ~18% of Cdh6+ cells were Ntng2+ and ~9.6% of Ntng2+ cells were Cdh6+, suggesting that some cells express both Cdh6+ and Ntng2. A summary of numerical values is presented in Table 2.
Figure 9. Ntng2 marks a subpopulation of Rorβ+ neurons in sSC.
A–G. The dFISH was conducted in sSC at ~P10 using Ntng2 as a second probe (labeled with fluorescein, green) and counterstained with DAPI (blue). The first probes (labeled with digoxigenin) are shown in red. All images in this figure were acquired in layer 1. A. Two in situ probes to Ntng2 (A1 and A2) highly overlapped (merged, A4; DAPI alone, A3). Here and in subsequent panels, arrows indicate examples of cells with overlapping expression, and arrowheads indicate expression by the first probe only. Arrows are missing in F and G because overlapping expression between the two genes is rare (<8%). B–D. Approximately half of Rorβ+ (B1), Cntn3+ (C1) and Cdh7+ (D1) cells were Ntng2+ (merged; B2, C2, D2). E–G. Very few cells expressing Cdh6 (E1), Pcdh20 (F1) and SP (G1) were Ntng2+ (merged; E2, F2, G2). Scale bar: 20 μm.
Layers 2, 3 and 4
Layers 2, 3 and 4 are distinguished by almost exclusive SST expression (layer 2), strong Brn3b expression (layer 3) and lower bands of Brn3b and ETV1 expression (layer 4) (Fig, 5 and 7). All of the labeled cells were neurons, as opposed to glia, because double immunostaining showed that SST+, Brn3b+ and ETV1+ cells were NeuN+ (~97%, ~98% and ~99%, respectively; data not shown). We next tested whether SST, Brn3b and ETV1 label individual cell types with dFISH experiments. SST expression was almost mutually exclusive with the expression of Brn3b or ETV1 (Fig. 5H–I and Table 3). Furthermore, ~62% of ETV1+ neurons were Brn3b+ and ~15% of Brn3b+ neurons were ETV1+. These data distinguish five different types of sSC neurons in layers 2–4: SST+, Brn3b+/ETV1+, Brn3b+/ETV1-, Brn3b−/ETV1+, and cells negative for all three markers (Table 3).
TABLE 3.
Relationship among sSC neuronal markers in layers 2, 3 and 4.
| Gene (X) | % (SST+ ∩ X+) / SST+ | % (Brn3b+ ∩ X+) / Brn3b+ | % (ETV1+ ∩ X+) / ETV1+ |
|---|---|---|---|
|
| |||
| SST | N. D. | 1.3 ± 0.5 | 1.2 ± 1.2 |
| Brn3b | 1.6 ± 0.6 | N. D. | 61.9 ± 6.3 |
| ETV1 | 0.5 ± 0.6 | 14.5 ± 2.7 | N. D. |
Summary of results from double In situ hybridization or immunostaining experiments. The number in each case indicates the average overlap (∩) between the second label and each first label (X). Data (mean ± SD) were averaged across 7–10 sections from at least 3 independent experiments.
Adult expression of selected genes
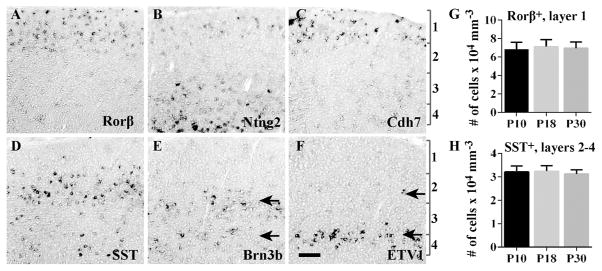
To confirm that our analysis of cell type classification at ~P10 can be extrapolated to the adult sSC, we examined expression of Rorβ, Ntng2, Cdh7, SST, Brn3b and ETV1 at P30. In general, the in situ signals were more difficult to visualize at P30, but a qualitative analysis revealed that all six genes show detectable expression (Fig. 10A–F). We did not analyze co-expression by in situ hybridization at P30 because the double labeling did not work well at this age. For Rorβ and SST, signals were sufficiently strong at P30 that cells could be counted and compared to earlier stages. For both molecules, cell number remains consistent at P10, P18 and P30 (Fig. 10G–H); for Rorβ, 6.9 ×104 ± 7.4 ×103 mm−3 at P10 versus 7.0 × 104 ± 6.6 ×103 mm−3 at P30; for SST, 3.2 ×104 ± 2.6 ×103 mm−3 at P10 versus 3.1 × 104 ± 1.8 ×103 mm−3 at P30. These data suggest that genes selected in our study will be valuable tools for cell type classification in adult sSC.
Figure 10. Adult expression of selected molecules used for cell type classification.
A–F. In situ hybridization was conducted in the sSC at P30 for six probes: Rorβ (A), Ntng2 (B), Cdh7 (C), SST (D), Brn3b (E) and ETV1 (F). Two distinct bands of Brn3b or ETV1 expression are indicated by arrows. The positions of layers 1–4 are indicated by brackets. G–H. The number of Rorβ+ (G) and SST+ cells (H) remains consistent between P10 and P30. Rorβ+ cells were identified by GFP signals in the Rorβ1g/+ mouse and counted within layer 1. SST+ cells were analyzed by in situ signals and counted within layers 2–4. Data (mean ± SD) were averaged across 7–12 sections from at least 3 independent experiments. Scale bar: 100 μm.
Discussion
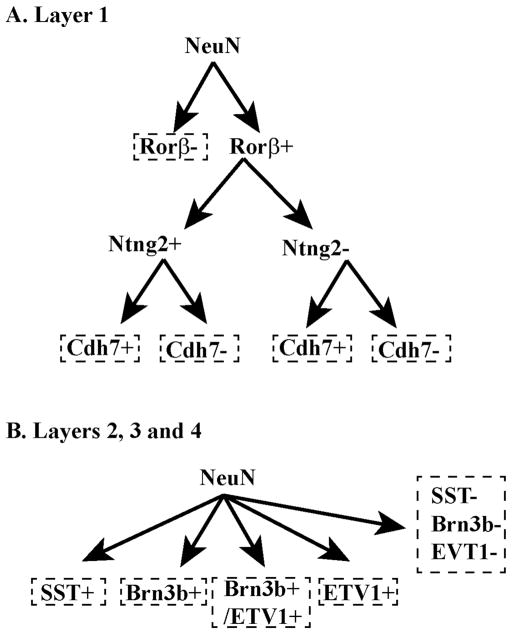
In this study, we took a systematic approach to examine molecular features of mouse sSC neurons and found 12 molecules with distinct expression patterns. We conducted dFISH and immunohistochemistry to directly compare the expression patterns of each molecule. Our results show that 1) sSC can be divided into four distinct layers based on molecular expression (Figs. 5–7) and 2) combinatorial expression of candidate molecules defines at least 10 different types of sSC neuron (Fig. 11).
Figure 11. Summary of molecularly-defined sSC neuronal cell types.
A. Neurons in layer 1 can be divided into two groups: Rorβ+ and Rorβ-. Rorβ+ neurons can be divided again into two groups: Ntng2+ and Ntng2-. Ntng2+ cells can be subdivided into two groups: Cdh7+ and Cdh7-. Ntng2− cells could be also subdivided based on Cdh7 expression, although we could not confirm this classification with our current technique. The combined expression pattern for Ntng2 and Cdh7 divides Rorβ+ neurons into four groups: (1) Ntng2+/Cdh7+, (2) Ntng2+/Cdh7-, (3) Ntng2−/Cdh7+, and (4) Ntng2−/Cdh7-. B. Expression of SST, Brn3b and ETV1 distinguishes subpopulations of neurons in layers 2, 3, and 4. Very few (if any) cells express both SST and Brn3b or ETV1, whereas some cells express both Brn3b and ETV1. Together, neurons in layers 2, 3 and 4 can be subdivided into 5 types: (1) SST+ only, (2) Brn3b+ only, (3) ETV1+ only, (4) both Brn3b+ and ETV1+, and (5) cells negative for all three markers.
Layer-restricted expression of molecular markers
The mouse sSC, unlike the chicken tectum, lacks clear subdivisions when viewed by conventional histological methods (Karten et al., 1982; Yamagata and Sanes, 2006). However, transgenic mice with labeled RGC types revealed that RGC axons and axonal arborizations divide sSC into four apparent layers (Huberman et al., 2008; Huberman et al., 2009; Kim et al., 2010; Hong et al., 2011). This finding prompted us to search for molecular markers that likewise label different sSC neurons in a layer-restricted pattern. Indeed, molecular expression patterns divide the sSC into four layers (Figs. 5–7). Comparison between axonal arborizations of RGCs and sSC neuronal markers revealed that the majority of RGCs initially enter the sSC and run longitudinally within layers 3 and 4 before terminating within layers 1 and 2 (Fig. 7).
Compared to the cortex, relatively little is known about molecular mechanisms that regulate development of laminar boundaries in sSC. An important next step will be to examine the timing of neurogenesis and cell migration of sSC neuron types within each layer identified here. We assume that specific types of RGCs interact with specific types of sSC neuron in distinct layers. Therefore, it is likely that the establishment of layers within sSC affects arborization patterns of RGC axons. The developmental sequence of sSC cell types within specific layers may provide insights into the sequence of cell type-specific connections between RGCs and sSC neurons.
Molecular markers classify a minimum of 10 different types of sSC neurons
We aimed to identify molecular markers that classify different types of sSC neurons. Such markers will be useful for investigating the development of sSC neurons or specific neuronal connections between sSC and retina, cortex or thalamus. In the future, the identified molecular markers can be used for visualizing and characterizing functional properties of individual sSC neurons.
Rorβ distinguishes two types of sSC neuron, Rorβ+ and Rorβ− (Fig. 11A). Expression of Ntng2 further divides Rorβ+ neurons into two types. The dFISH with the Ntng2 probe revealed that Ntng2+ cells can be subdivided based on Cdh7 expression. We speculate that Ntng2− cells could likewise be subdivided based on Cdh7 expression, although we could not confirm this with our current technique. Collectively, this would result in at least 4 different types within the Rorβ+ population: 1) Ntng2+/Cdh7+, 2) Ntng2+/Cdh7-, 3) Ntng2−/Cdh7+, and 4) Ntng2−/Cdh7-. We could not characterize the extent of Cdh7 and Cntn3 co-expression because none of our probes for either molecule worked well as a second probe in dFISH experiments. In general, we chose Cdh7 over Cntn3 expression for cell type classification because Cdh7 in situ signals are relatively more distinct than Cntn3 signals. We expect that Cntn3 expression could eventually demarcate additional types when double labeling experiments become possible. At present, we consider Rorβ− cells as a single group and distinguish a total of five types of sSC neurons in layer 1.
Neurons in layers 2, 3 and 4 consist of five different types (Fig. 11B): 1) SST+, 2) Brn3b+/ETV1+, 3) Brn3b+/ETV1-, 4) Brn3b−/ETV1+, and 5) cells negative for all three markers. We combined upper and lower bands of Brn3b neurons for cell type classification. Based on differential expression of ETV1 among Brn3b neurons, however, we speculate that Brn3b+ cells comprise two different types and their laminar position might also contribute to functional diversity. Analysis and comparison of knockout mice in which Brn3b or ETV1 are deleted would provide insights into developmental contributions of each molecule to sSC neuronal type specification.
Molecular markers from our systematic screening distinguish at least 10 different types of sSC neuron. Given that our markers label a small percentage of sSC neurons (~20%), we expect that the total number of sSC neuronal types is much greater than 10. This predicted level of cell type diversity exceeds previous estimates based on dye filling and physiology (Langer and Lund, 1974; Fukuda et al., 1978; Mooney et al., 1985; Wang et al., 2010; Gale and Murphy, 2014). To correlate our data with cell types identified in the literature, the morphological analysis of sSC neurons that are labeled by our markers should be conducted. Combining multiple approaches to examine molecular, morphological and physiological properties will provide us with more comprehensive tools to classify sSC neuronal types.
Caveats to the cell type analysis
For cell type classification in layer 1, antibodies to the molecules of interest were not available and so we primarily used in situ hybridization. We were conservative when identifying labeled cells using this technique, which might result in incomplete identification of positive cells for a given probe. Indeed, we conducted double labeling experiments with two markers for the same molecule (either Rorβ or Ntng2; Figs. 8 and 9) and found only ~80% overlap (Table 2). We therefore assume that ~80% overlap may be a maximum outcome under our experimental conditions. When the overlap of probes to two different molecules is less than 80%, it implies that there are at least two types of cells. For example, only ~50% of Rorβ+ cells were Ntng2+ (Table 2), suggesting that the Rorβ+ population comprises at least two types, Ntng2+ and Ntng2-(Fig. 11). In the case of double labeling for Rorβ and Ntng2, we obtained a nearly identical degree of overlap with two different approaches, using either Rorβ or Ntng2 as the second probe (~52% vs. ~50%), suggesting that our results are reliable (Table 2). Our analysis likely underestimates the true number of different types of sSC neuron with expression of any of our molecules, due to the limited number of possible combinations we could test with dFISH experiments.
We conducted double labeling experiments for cell classification at P10-P13 tissues and assumed that our results can be extrapolated to the adult sSC for three reasons. First, neurogenesis is complete during the embryonic stage (Edwards et al., 1986b). Furthermore, our analysis showed no change in NeuN+ positive cell number between P10 and P18. Second, overall development of sSC structures is complete by P15 (Warton and Jones, 1985; Sachs et al., 1986). Third, we also qualitatively analyzed expression of genes used for cell type classification (Rorβ, Ntng2, Cdh7, SST, Brn3b and ETV1) and found that expression of each gene remains consistent up to P30. Moreover, quantification of Rorβ+ or SST+ cell number revealed no changes between P10 and P30 (Fig. 10). Therefore, our analysis should apply to adult cell type classification.
We confirmed that Rorβ, SST, Brn3b and ETV1 label sSC neurons, as opposed to glia, by showing co-expression of NeuN (Fig. 8). Most of the markers are also likely expressed in neurons because: 1) A significant portion of Cntn3+, Ntng2+ and Cdh7+ cells are from the Rorβ+ population, suggesting that the majority of these cells are neurons (Fig. 8; Table 2); 2) SP, PV and VIP label neurons in other systems, including cortex and retina (Bagnoli et al., 2003; Markram et al., 2004; Huang et al., 2007; Park et al., 2015); 3) Cdh6 and Pcdh20 likewise are expressed in neurons in other systems, including the retina and the olfactory system (Lee et al., 2008; Kay et al., 2011).
Future applications of sSC markers
The molecular features of mouse sSC neurons had been explored primarily with markers using commercially available antibodies (e.g., PV). Expression patterns of Rorβ, Brn3b and ETV1 were also reported previously (Turner et al., 1994; Yoneshima et al., 2006; Harris et al., 2014). However, earlier studies failed to examine the relative distribution of multiple cell markers. Moreover, systematic investigation of cell adhesion molecule expression had not been conducted. In this context, it is noteworthy that most markers identified in layer 1, where RGC axons terminate, are cell adhesion molecules (Fig. 5). Selective wiring between presynaptic and postsynaptic neurons in many systems depends on homophilic or heterophilic interactions among cell adhesion molecules. Of particular interest in our study are the expression patterns of Cdh7, Cntn3, Pcdh20 and Ntng2. For example, overexpression of Cntn3 in the chicken mediates layer-specific targeting of retinal neurons (Yamagata and Sanes, 2012). Moreover, homophilic interaction of Cdh7 or heterophilic interaction of Ntng2 regulates development of the cerebellar mossy fiber circuit or hippocampal Schaffer collateral pathways, respectively (Nishimura-Akiyoshi et al., 2007; Kuwako et al., 2014; Matsukawa et al., 2014). Identifying the expression patterns of putative binding partners of Cdh7, Cntn3, Pcdh20 and Ntng2 in the retina, cortex, or thalamus will be informative for future studies. Such data will provide insights into the functional contribution of each molecule to wiring in the visual system.
Live imaging of the tectum of lower vertebrates reveals diverse mechanisms that regulate development of retinotectal pathways or function of tectal neurons (Ramdya and Engert, 2008; Baier and Scott, 2009; Xiao et al., 2011; Gabriel et al., 2012; Nikolaou et al., 2012; Ruthazer et al., 2013). Mouse sSC is also anatomically accessible to optical imaging and recent technical advances enable functional measurements of mouse sSC neurons in vivo (Ackman et al., 2012; Feinberg and Meister, 2015; Inayat et al., 2015). Multiple transgenic lines are available that allow type-specific manipulation of sSC neurons: for example, SP-Cre, VIP-Cre, Rorβ-Cre, and ETV1-Cre mice in which Cre expression is controlled under the endogenous expression of each molecule (commercially available from Jackson Laboratory). Experiments using optogenetics and in vivo imaging in these Cre lines will enhance our understanding of functional specification of sSC neuron types.
Specific sSC neuronal markers could facilitate the analysis of sSC output pathways that modulate visually guided behaviors including eye and head movements. Anatomical tracing or electrical recording reveal that sSC neurons project to the deeper layers of SC or to several areas including dorsal lateral geniculate nucleus (dLGN), lateral posterior nucleus (LP) and parabigeminal nucleus (PBGN). (Graham and Berman, 1981; Mooney et al., 1988; Harting et al., 1991; Lane et al., 1997; May, 2006; Lyon et al., 2010; Bickford et al., 2015). The sSC neurons confined to specific layers appear to project selectively to distinct targets. For example, neurons in uSGS project to dLGN and neurons in lSGS and SO project to LP. In some studies, the morphology and function of sSC neurons in distinct layers was identified, but in many studies, classification of sSC output neurons was speculative due to a limited ability to label specific cell types. Recent studies using transgenic mice in which distinct sSC cell types are molecularly tagged overcome such obstacles and demonstrate specific sSC output pathways using optogenetics (Gale and Murphy, 2014; Shang et al., 2015). Our markers could be used in a similar way and provide efficient tools for characterizing sSC output pathways.
Here, we used a candidate approach as a cost-effective way to identify genes that classify specific types of neurons. However, this approach relies on existing information and limits the identification of novel gene families. Moreover, our study focused on classification of the Rorβ+ population that comprises only ~20% of sSC neurons. In the future, a more comprehensive gene expression profile such as a genome-wide association study could generate more markers and identify additional neuronal types in sSC.
Supplementary Material
Acknowledgments
Grant Sponsor: This research was funded by R00 EY019355, R01 EY014454, the Whitehall Foundation, the E. Matilda Ziegler Foundation and an unrestricted grant from Research to Prevent Blindness to Yale University. This study was also supported in part by the intramural research program at NIDDK at the National Institutes of Health.
We thank members of the Kim and Demb labs for constructive discussions. We also thank Joshua Sanes (Harvard University) for his advice on the manuscript.
Footnotes
Conflict of Interest: The authors have no competing interests.
Role of Authors: All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and Design: H.W.B., S.H.K. and I.-J.K. Acquisition of data: H.W.B., S.H.K. H.J.A. I.-J.K. Analysis and interpretation of data: H.W.B., S.H.K., J.B.D. and I.-J.K. Drafting of manuscript: J.B.D. and I.-J.K. Critical revision of the manuscript for important intellectual content: D.F., J.B.D. and I.-J.K Statistical analysis: H.W.B. and I.-J.K. Obtained funding: D.F., J.B.D. and I.-J.K. Technical and material support: H.L. and D.F. Study supervision: I.-J.K.
Literature Cited
- Ackman JB, Burbridge TJ, Crair MC. Retinal waves coordinate patterned activity throughout the developing visual system. Nature. 2012;490(7419):219–225. doi: 10.1038/nature11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M, Arai R, Sasamoto K, Kani K, Maeda T, Deura S, Jacobowitz DM. Appearance of calretinin-immunoreactive neurons in the upper layers of the rat superior colliculus after eye enucleation. Brain Res. 1993;613(2):341–346. doi: 10.1016/0006-8993(93)90924-c. [DOI] [PubMed] [Google Scholar]
- Bagnoli P, Dal Monte M, Casini G. Expression of neuropeptides and their receptors in the developing retina of mammals. Histol Histopathol. 2003;18(4):1219–1242. doi: 10.14670/HH-18.1219. [DOI] [PubMed] [Google Scholar]
- Baier H, Scott EK. Genetic and optical targeting of neural circuits and behavior-zebrafish in the spotlight. Curr Opin Neurobiol. 2009;19(5):553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan M, Jourdain A, Bray GM. Calcium binding protein (calbindin D28k) immunoreactivity in the hamster superior colliculus: ultrastructure and lack of co-localization with GABA. Exp Brain Res. 1992;89(1):115–124. doi: 10.1007/BF00229008. [DOI] [PubMed] [Google Scholar]
- Behan M, Appell PP, Kime N. Postnatal development of substance-P immunoreactivity in the rat superior colliculus. Vis Neurosci. 1993;10(6):1121–1127. doi: 10.1017/s0952523800010221. [DOI] [PubMed] [Google Scholar]
- Bickford ME, Zhou N, Krahe TE, Govindaiah G, Guido W. Retinal and Tectal “Driver-Like” Inputs Converge in the Shell of the Mouse Dorsal Lateral Geniculate Nucleus. J Neurosci. 2015;35(29):10523–10534. doi: 10.1523/JNEUROSCI.3375-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce JG, Zikopoulos B, Feinberg M, Barbas H. Parallel prefrontal pathways reach distinct excitatory and inhibitory systems in memory-related rhinal cortices. J Comp Neurol. 2013;521(18):4260–4283. doi: 10.1002/cne.23413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhang Q, Wang C, Zhang Y, Ma T, Zhou X, Tian M, Rubenstein JL, Yang Z. Nuclear receptor COUP-TFII-expressing neocortical interneurons are derived from the medial and lateral/caudal ganglionic eminence and define specific subsets of mature interneurons. J Comp Neurol. 2013;521(2):479–497. doi: 10.1002/cne.23186. [DOI] [PubMed] [Google Scholar]
- Constantine-Paton M, Cline HT, Debski E. Patterned activity, synaptic convergence, and the NMDA receptor in developing visual pathways. Annu Rev Neurosci. 1990;13:129–154. doi: 10.1146/annurev.ne.13.030190.001021. [DOI] [PubMed] [Google Scholar]
- Cork RJ, Baber SZ, Mize RR. CalbindinD28k-and parvalbumin-immunoreactive neurons form complementary sublaminae in the rat superior colliculus. J Comp Neurol. 1998;394(2):205–217. [PubMed] [Google Scholar]
- Cork RJ, Calhoun T, Perrone M, Mize RR. Postnatal development of nitric oxide synthase expression in the mouse superior colliculus. J Comp Neurol. 2000;427(4):581–592. [PubMed] [Google Scholar]
- Dräger UC, Hubel DH. Physiology of visual cells in mouse superior colliculus and correlation with somatosensory and auditory input. Nature. 1975;253(5488):203–204. doi: 10.1038/253203a0. [DOI] [PubMed] [Google Scholar]
- Edwards MA, Schneider GE, Caviness VS., Jr Development of the crossed retinocollicular projection in the mouse. J Comp Neurol. 1986a;248(3):410–421. doi: 10.1002/cne.902480309. [DOI] [PubMed] [Google Scholar]
- Edwards MA, Caviness VS, Jr, Schneider GE. Development of cell and fiber lamination in the mouse superior colliculus. J Comp Neurol. 1986b;248(3):395–409. doi: 10.1002/cne.902480308. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Meister M. Orientation columns in the mouse superior colliculus. Nature. 2015;519(7542):229–232. doi: 10.1038/nature14103. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, O’Leary DD. Visual map development: bidirectional signaling, bifunctional guidance molecules, and competition. Cold Spring Harb Perspect Biol. 2010;2(11):a001768. doi: 10.1101/cshperspect.a001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Liu H, Ng L, Kim JW, Hao H, Swaroop A, Forrest D. Feedback induction of a photoreceptor-specific isoform of retinoid-related orphan nuclear receptor β by the rod transcription factor NRL. J Biol Chem. 2014;289(47):32469–32480. doi: 10.1074/jbc.M114.605774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Suzuki DA, Iwama K. A four group classification of the rat superior collicular cells responding to optic nerve stimulation. Jpn J Physiol. 1978;28(3):367–384. doi: 10.2170/jjphysiol.28.367. [DOI] [PubMed] [Google Scholar]
- Gabriel JP, Trivedi CA, Maurer CM, Ryu S, Bollmann JH. Layer-specific targeting of direction-selective neurons in the zebrafish optic tectum. Neuron. 2012;76:1147–1160. doi: 10.1016/j.neuron.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Gale SD, Murphy GJ. Distinct representation and distribution of visual information by specific cell types in mouse superficial superior colliculus. J Neurosci. 2014;34(40):13458–13471. doi: 10.1523/JNEUROSCI.2768-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautron L, Lee C, Funahashi H, Friedman J, Lee S, Elmquist J. Melanocortin-4 receptor expression in a vago-vagal circuitry involved in postprandial functions. J Comp Neurol. 2010;518(1):6–24. doi: 10.1002/cne.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J, Berman N. Origins of the projections of the superior colliculus to the dorsal lateral geniculate nucleus and the pulvinar in the rabbit. Neurosci Lett. 1981;26(2):101–106. doi: 10.1016/0304-3940(81)90333-5. [DOI] [PubMed] [Google Scholar]
- Gu Z, Imai F, Kim IJ, Fujita H, Katayama Ki, Mori K, Yoshihara Y, Yoshida Y. Expression of the immunoglobulin superfamily cell adhesion molecules in the developing spinal cord and dorsal root ganglion. PLoS One. 2015;10:e0121550. doi: 10.1371/journal.pone.0121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton D. Control of eye-head coordination during orienting gaze shifts. Trends Neurosci. 1992;15(5):174–179. doi: 10.1016/0166-2236(92)90169-9. [DOI] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, Smith KA, Dang C, Sunkin S, Bernard A, Oh SW, Madisen L, Zeng H. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harting JK, Huerta MF, Hashikawa T, van Lieshout DP. Projection of the mammalian superior colliculus upon the dorsal lateral geniculate nucleus: organization of tectogeniculate pathways in nineteen species. J Comp Neurol. 1991;304(2):275–306. doi: 10.1002/cne.903040210. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295(5557):1065–1670. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A, Dräger UC. Depth segregation of retinal ganglion cells projecting to mouse superior colliculus. J Comp Neurol. 1985;234(4):465–474. doi: 10.1002/cne.902340405. [DOI] [PubMed] [Google Scholar]
- Hong YK, Kim IJ, Sanes JR. Stereotyped axonal arbors of retinal ganglion cell subsets in the mouse superior colliculus. J Comp Neurol. 2011;519(9):1691–1711. doi: 10.1002/cne.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Di Cristo G, Ango F. Development of GABA innervation in the cerebral and cerebellar cortices. Nat Rev Neurosci. 2007;8(9):673–686. doi: 10.1038/nrn2188. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59(3):425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MBBarres BA. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62(3):327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inayat S, Barchini J, Chen H, Feng L, Liu X, Cang J. Neurons in the most superficial lamina of the mouse superior colliculus are highly selective for stimulus direction. J Neurosci. 2015;35(20):7992–8003. doi: 10.1523/JNEUROSCI.0173-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isa T, Hall WC. Exploring the superior colliculus in vitro. J Neurophysiol. 2009;102(5):2581–2593. doi: 10.1152/jn.00498.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten HJ, Reiner A, Brecha N. Laminar organization and origins of neuropeptides in the avian retina and optic tectum. In: Chan-Palay V, Palay SL, editors. Cytochemical Melthods in Neuroanatomy. New York: A. R. Liss; 1982. pp. 189–204. [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31(21):7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kha HT, Finkelstein DI, Pow DA, Lawrence AJ, Horne MK. Study of projections from the entopeduncular nucleus to the thalamus of the rat. J Comp Neurol. 2000;426(3):366–377. [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452(7186):478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AJ, Hutchings ME, Moore DR, Blakemore C. Developmental plasticity in the visual and auditory representations in the mammalian superior colliculus. Nature. 1988;332(6159):73–76. doi: 10.1038/332073a0. [DOI] [PubMed] [Google Scholar]
- Krauzlis RJ, Lovejoy LP, Zénon A. Superior colliculus and visual spatial attention. Annu Rev Neurosci. 2013;36:165–182. doi: 10.1146/annurev-neuro-062012-170249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524(7566):466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwako K, Nishimoto Y, Kawase S, Okano HJ, Okano H. Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep. 2014;9(1):311–323. doi: 10.1016/j.celrep.2014.08.063. [DOI] [PubMed] [Google Scholar]
- Laemle LK, Feldman SC. Somatostatin (SRIF)-like immunoreactivity in subcortical and cortical visual centers of the rat. J Comp Neurol. 1985;233(4):452–462. doi: 10.1002/cne.902330404. [DOI] [PubMed] [Google Scholar]
- Lane RD, Allan DM, Bennett-Clarke CA, Howell DL, Rhoades RW. Projection status of calbindin-and parvalbumin-immunoreactive neurons in the superficial layers of the rat’s superior colliculus. Vis Neurosci. 1997;14(2):277–286. doi: 10.1017/s095252380001141x. [DOI] [PubMed] [Google Scholar]
- Langer TP, Lund RD. The upper layers of the superior colliculus of the rat: a Golgi study. J Comp Neurol. 1974;158(4):418–435. doi: 10.1002/cne.901580404. [DOI] [PubMed] [Google Scholar]
- Lee W, Cheng TW, Gong Q. Olfactory sensory neuron-specific and sexually dimorphic expression of protocadherin 20. J Comp Neurol. 2008;507(1):1076–1086. doi: 10.1002/cne.21569. [DOI] [PubMed] [Google Scholar]
- Liu H, Kim SY, Fu Y, Wu X, Ng L, Swaroop A, Forrest D. An isoform of retinoid-related orphan receptor β directs differentiation of retinal amacrine and horizontal interneurons. Nat Commun. 2013;4:1813. doi: 10.1038/ncomms2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon DC, Nassi JJ, Callaway EM. A disynaptic relay from superior colliculus to dorsal stream visual cortex in macaque monkey. Neuron. 2010;65(2):270–279. doi: 10.1016/j.neuron.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5(10):793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Matsukawa H, Akiyoshi-Nishimura S, Zhang Q, Luján R, Yamaguchi K, Goto H, Yaguchi K, Hashikawa T, Sano C, Shigemoto R, Nakashiba T, Itohara S. Netrin-G/NGL complexes encode functional synaptic diversification. J Neurosci. 2014;34(47):15779–15792. doi: 10.1523/JNEUROSCI.1141-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PJ. The mammalian superior colliculus: laminar structure and connections. Prog Brain Res. 2006;151:321–378. doi: 10.1016/S0079-6123(05)51011-2. [DOI] [PubMed] [Google Scholar]
- McLaughlin T, O’Leary DD. Molecular gradients and development of retinotopic maps. Annu Rev Neurosci. 2005;28:327–355. doi: 10.1146/annurev.neuro.28.061604.135714. [DOI] [PubMed] [Google Scholar]
- Meredith MA, Stein BE. Interactions among converging sensory inputs in the superior colliculus. Science. 1983;221: 389–391. doi: 10.1126/science.6867718. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27(29):7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mize RR, Luo Q, Butler G, Jeon CJ, Nabors B. The calcium binding proteins parvalbumin and calbindin-D 28K form complementary patterns in the cat superior colliculus. J Comp Neurol. 1992;320(2):243–256. doi: 10.1002/cne.903200208. [DOI] [PubMed] [Google Scholar]
- Mooney RD, Klein BG, Rhoades RW. Correlations between the structural and functional characteristics of neurons in the superficial laminae and the hamster’s superior colliculus. J Neurosci. 1985;5(11):2989–3009. doi: 10.1523/JNEUROSCI.05-11-02989.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RD, Nikoletseas MM, Ruiz SA, Rhoades RW. Receptive-field properties and morphological characteristics of the superior collicular neurons that project to the lateral posterior and dorsal lateral geniculate nuclei in the hamster. J Neurophysiol. 1988;59(5):1333–1351. doi: 10.1152/jn.1988.59.5.1333. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck Cr, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116(1):201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Munoz DP, Pélisson D, Guitton D. Movement of neural activity on the superior colliculus motor map during gaze shifts. Science. 1991;251(4999):1358–1360. doi: 10.1126/science.2003221. [DOI] [PubMed] [Google Scholar]
- Mysore SP, Knudsen EI. The role of a midbrain network in competitive stimulus selection. Curr Opin Neurobiol. 2011;21:653–660. doi: 10.1016/j.conb.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolás FM, Jiménez-López M, Salinas-Navarro M, Sobrado-Calvo P, Alburquerque-Béjar JJ, Vidal-Sanz M, Agudo-Barriuso M. Whole number, distribution and co-expression of brn3 transcription factors in retinal ganglion cells of adult albino and pigmented rats. PLoS One. 2012;7(11):e49830. doi: 10.1371/journal.pone.0049830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie D, Di Nardo A, Han JM, Baharanyi H, Kramvis I, Huynh T, Dabora S, Codeluppi S, Pandolfi PP, Pasquale EB, Sahin M. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13(2):163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou N, Lowe AS, Walker AS, Abbas F, Hunter PR, Thompson ID, Meyer MP. Parametric functional maps of visual inputs to the tectum. Neuron. 2012;76(2):317–324. doi: 10.1016/j.neuron.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A. 2007;104(37):14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Meguro R, Itoh K, Mizuno N. Substance P-, vasoactive intestinal polypeptide-, and cholecystokinin-like immunoreactive fiber projections from the superior colliculus to the dorsal lateral geniculate nucleus in the rat. Exp Brain Res. 1992;89(1):59–66. doi: 10.1007/BF00229001. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Okamura H, Takahashi Y, Akagi Y, Yanaihara N, Ibata Y. Contrary effect of eye enucleation on VIP-immunoreactive neurons in the suprachiasmatic nucleus and the superior colliculus of the rat. Neurosci Lett. 1990;112(2–3):137–142. doi: 10.1016/0304-3940(90)90192-c. [DOI] [PubMed] [Google Scholar]
- Park SJ, Borghuis BG, Rahmani P, Zeng Q, Kim IJ, Demb JB. Function and Circuitry of VIP+ Interneurons in the Mouse Retina. J Neurosci. 2015;35(30):10685–10700. doi: 10.1523/JNEUROSCI.0222-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdya P, Engert F. Emergence of binocular functional properties in a monocular neural circuit. Nat Neurosci. 2008;11:1083–1090. doi: 10.1038/nn.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo D, Bombardi C, Grandis A, Furness JB, Spadari A, Bernardini C, Chiocchetti R. Sympathetic innervation of the ileocecal junction in horses. J Comp Neurol. 2010;518(19):4046–4066. doi: 10.1002/cne.22443. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Schohl A, Schwartz N, Tayakoli A, Tremblay M, Cline HT. In vivo time-lapse imaging of neuronal development in Xenopus. Cold Spring Harb Protoc. 2013;9:804–809. doi: 10.1101/pdb.top077156. [DOI] [PubMed] [Google Scholar]
- Sachs GM, Schneider GE. The morphology of optic tract axons arborizing in the superior colliculus of the hamster. J Comp Neurol. 1984;230(2):155–167. doi: 10.1002/cne.902300202. [DOI] [PubMed] [Google Scholar]
- Sachs GM, Jacobson M, Caviness VS., Jr Postnatal changes in arborization patterns of murine retinocollicular axons. J Comp Neurol. 1986;246(3):395–408. doi: 10.1002/cne.902460308. [DOI] [PubMed] [Google Scholar]
- Saito T, Iwata N, Tsubuki S, Takaki Y, Takano J, Huang SM, Suemoto T, Higuchi M, Saido TC. Somatostatin regulates brain amyloid beta peptide Abeta42 through modulation of proteolytic degradation. Nat Med. 2005;11(4):434–439. doi: 10.1038/nm1206. [DOI] [PubMed] [Google Scholar]
- Seelke AM, Dooley JC, Krubitzer LA. Differential changes in the cellular composition the developing marsupial brain. J Comp Neurol. 2013;521(11):2602–2620. doi: 10.1002/cne.23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevc J, Matiašová A, Kútna V, Daxnerová Z. Evidence that the central canal lining of the spinal cord contributes to oligodendrogenesis during postnatal development and adulthood in intact rats. J Comp Neurol. 2014;522(14):3194–3207. doi: 10.1002/cne.23590. [DOI] [PubMed] [Google Scholar]
- Shang C, Liu Z, Chen Z, Shi Y, Wang Q, Liu S, Li D, Cao P. BRAIN CIRCUITS. A parvalbumin-positive excitatory visual pathways to trigger fear responses in mice. Science. 2015;348(6242):1472–1477. doi: 10.1126/science.aaa8694. [DOI] [PubMed] [Google Scholar]
- Soares-Mota M, Henze I, Mendez-Otero R. Nitric oxide synthase-positive neurons in the rat superior colliculus: colocalization of NOS with NMDAR1 glutamate receptor, GABA, and parvalbumin. J Neurosci Res. 2001;64(5):501–507. doi: 10.1002/jnr.1102. [DOI] [PubMed] [Google Scholar]
- Sparks DL. The neural control of orienting eye and head movements. In: Humphrey DR, Freund H-J, editors. Motor Control: Concepts and Issues. Dahlem Konferenzen. Chichester: John Wiley & Sons Ltd; 1991. pp. 263–275. [Google Scholar]
- Stein BE, Stanford TR, Rowland BA. Development of multisensory integration from the perspective of the individual neuron. Nat Rev Neurosci. 2014;15(8):520–535. doi: 10.1038/nrn3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Brady CM, Cahill H, Al-Khindi T, Sakuta H, Dhande OS, Noda M, Huberman AD, Nathans J, Kolodkin AL. Functional assembly of accessory optic system circuitry critical for compensatory eye movements. Neuron. 2015;86(4):971–984. doi: 10.1016/j.neuron.2015.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JW, Owens MT, Yamada J, Lemke G, Cang J, Stryker MP, Feldheim DA. Retinal input instructs alignment of visual topographic maps. Cell. 2009;139(1):175–185. doi: 10.1016/j.cell.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EE, Jenne KJ, Rosenfeld MG. Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron. 1994;12(1):205–218. doi: 10.1016/0896-6273(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, Mohan S, Pandita A, Hongo JA, Arnott D, Wertz IE, Gao WQ, French DM, Dixit VM. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474(7351):403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- Wang L, Sarnaik R, Rangarajan K, Liu X, Cang J. Visual receptive field properties of neurons in the superficial superior colliculus of the mouse. J Neurosci. 2010;30(49):16573–16584. doi: 10.1523/JNEUROSCI.3305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton SS, Jones DG. Postnatal development of the superficial layers in the rat superior colliculus: a study with Golgi-Cox and Klüver-Barrera techniques. Exp Brain Res. 1985;58(3):490–502. doi: 10.1007/BF00235865. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wurtz RH, Goldberg ME. Superior colliculus cell responses related to eye movements in awake monkeys. Science. 1971;171(3966):82–84. doi: 10.1126/science.171.3966.82. [DOI] [PubMed] [Google Scholar]
- Wurtz RH, Goldberg ME. The primate superior colliculus and the shift of visual attention. Invest Ophthalmol. 1972;11(6):441–450. [PubMed] [Google Scholar]
- Xiao T, Staub W, Robles E, Gosse NJ, Cole GJ, Baier H. Assembly of lamina-specific neuronal connections by slit bound to type IV collagen. Cell. 2011;146(1):164–176. doi: 10.1016/j.cell.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110(5):649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Dulac C, Roth KA, Sanes JR. Labeled lines in the retinotectal system: markers for retinorecipient sublaminae and the retinal ganglion cell subsets that innervate them. Mol Cell Neurosci. 2006;33(3):296–310. doi: 10.1016/j.mcn.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451(7177):465–459. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci. 2012;32(41):14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31(5):434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshima H, Yamasaki S, Voelker CC, Molnár Z, Christophe E, Audinat E, Takemoto M, Nishiwaki M, Tsuji S, Fujita I, Yamamoto N. Er81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137(2):401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim IJ, Sanes JR, Meister M. The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proc Natl Acad Sci U S A. 2012;109(36):E2391–E2398. doi: 10.1073/pnas.1211547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.