Abstract
Molecular pathology diagnostics to subclassify diseases based on pathogenesis are increasingly common in clinical translational medicine. Molecular pathological epidemiology (MPE) is an integrative transdisciplinary science based on the unique disease principle and the disease continuum theory. While it has been most commonly applied to research on breast, lung, and colorectal cancers, MPE can investigate etiologic heterogeneity in non-neoplastic diseases such as cardiovascular diseases, obesity, diabetes mellitus, drug toxicity, and immunity-related and infectious diseases. This science can enhance causal inference by linking putative etiologic factors to specific molecular biomarkers as outcomes. Technological advances increasingly enable analyses of various -omics, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, metagenomics, microbiome, immunomics, interactomics, etc. Challenges in MPE include sample size limitations (depending on availability of biospecimens or biomedical / radiological imaging), need for rigorous validation of molecular assays and study findings, and paucities of interdisciplinary experts, education programs, international forums, and standardized guidelines. To address these challenges, there are ongoing efforts such as multidisciplinary consortium pooling projects, the International Molecular Pathological Epidemiology (MPE) Meeting Series, and the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE)-MPE guideline project. Efforts should be made to build biorepository and biobank networks, and worldwide population-based MPE databases. These activities match with the purposes of the Big Data to Knowledge (BD2K), Genetic Associations and Mechanisms in Oncology (GAME-ON), and Precision Medicine Initiatives of the United States National Institute of Health. Given advances in biotechnology, bioinformatics, and computational / systems biology, there are wide open opportunities in MPE to contribute to public health.
Keywords: epidemiologic methods, informatics, integrative epidemiology, molecular pathologic epidemiology, translational epidemiology, unique tumor principle
Introduction: disease nosology
For many centuries, patients have been grouped into disease classifications based on the similarity and dissimilarity of their health problems. The primary purpose of assigning a specific disease name to a group of patients is to better predict the natural history (disease course) and the patient’s response to a particular treatment or intervention. Disease classification systems have typically been based on symptomatology, affected organ system, and/or impaired physiologic function (often in combination). Examples of function-based or functional system-based disease classification include: neoplastic disease, auto-immune disease, metabolic disease, etc. Occasionally, when a definitive etiology is identified in patients with similar symptoms, the etiologic factor is used to name the disease; such examples include influenza, burn, hypothermia, lead poisoning, etc. However, many chronic diseases are multifactorial in origin, and their causes are often not well understood. Establishing the etiology of such complex diseases requires extensive research. Thus, since the 20th century, epidemiology of chronic complex diseases has developed to investigate this issue. The typical study design in traditional epidemiology aims to examine the relationship between an exposure and a disease which is named or classified based on symptomatology or organ/function system.
As disease classification systems are incorporating newer knowledge of molecular pathogenesis of various diseases, we expect that individualized timely management and treatment of each patient can be realized. That is exactly the goal of precision medicine. Hence, when precision medicine becomes prevalent, there will be detailed data on pathologies and phenotypes among patients with the same disease. Increasing amounts of such data will be available to medical and epidemiologic researchers, and provide ample opportunities. In this article, our discussion focuses on following points: (1) a recent overall trend towards molecular classification of disease in all areas of science including epidemiology and population health sciences; (2) benefits derived from consideration of disease heterogeneity; and (3) future potential of molecular pathological epidemiology (MPE) as ubiquitous population health science. Our intention is not to give detailed review in specific disease areas, but to give an overview on usefulness of the MPE paradigm in a wide variety of disease areas.
Epidemiology as core and universal health science
Epidemiology is the science of study of occurrence, distribution, causes and effects of diseases and health-related conditions. As epidemiology gained influence as the study of virtually all diseases in the mid to late 20th century, it became a core and universal component in public health. Epidemiology has been integrated into various disciplines of population health science, including “clinical epidemiology”, “nutritional epidemiology”, “social epidemiology”, and “environmental epidemiology”. When rigorously conducted, clinical and epidemiologic research can produce new knowledge, and advance medicine and public health. Because the scale and size of epidemiologic studies became larger in the mid to late 20th century, information on diseases was usually extracted from study participants, public health records, and medical records, often without detailed assessments of molecular pathology for practical reasons. In addition, there was no available molecular pathological assay for many diseases. As a result, common disease classification in clinical settings has been used in most epidemiologic studies, although consideration of pathogenesis (i.e., biological mechanisms of disease development) has always been a part of epidemiology as subject matter knowledge.
An important epidemiologic premise is that individuals with the same disease entity have similar causes of the disease, and show similar natural history of the disease and similar responses to treatment or intervention. Under this important assumption (what can be called as “homogeneity premise” or “generalizability premise”), observations in a population study which contains a large number of individuals with a certain disease can be generalized to patients with the same disease in a different set of people in the same background population or in people in a different background population (with possible effect modifications). In this context, traditional epidemiology has contributed to not only the successful identification of major etiologic factors even without molecular sub-classification but also the development of prevention strategies, such as smoking cessation programs.
Another aspect of major contributions of epidemiologic studies to broader biomedical and health sciences is provision of study subjects and materials. Epidemiologic research typically requires intensive efforts to carefully select subjects from a larger population, compile information on environment, lifestyle, behavior, health and diseases, collect biospecimens, and build a large database infrastructure. Such epidemiologic resources can become an enormous asset for transdisciplinary research, and provide a large sample of disease patients (with molecular pathological annotations), which is more representative of the general population than a typical hospital-based convenience sample. It is increasingly feasible to apply advanced omics technologies (such as expression arrays, and next generation sequencing) to those large population-based disease datasets.
Accordingly, along with advances in computing, bioinformatics, genomic medicine and biomedical sciences, modern epidemiology has broadened to integrate these disciplines. This advancement has been opening enormous opportunities, including the evolution of molecular pathological epidemiology.
Recent evolution of molecular pathological epidemiology (MPE)
Molecular classifications are emerging as the new standard in medicine given inherent heterogeneity of disease, especially neoplasms.1 Because epidemiologic analysis is based on the premise that individuals with the same diagnosis have similar causes and disease evolution, it is essential that epidemiologic research rely upon modern molecular classification of disease. Thus, it is increasingly necessary to consider disease heterogeneity and molecular classification more explicitly in epidemiology, with an integrative view of homogeneity and heterogeneity of pathogenic processes.
Analyses of tumor molecular pathology in epidemiologic research began in late 1980s and 1990s, and gained increasing popularity (although still relatively uncommon) throughout the early 2000s, under the umbrella term of “molecular epidemiology” (see references in eAppendix 1). However, a vast majority of molecular epidemiology studies have used molecular analyses of exposures (including germline genetics) and relied on disease data without detailed molecular pathological assessment. For example, genome-wide association studies have commonly used traditional disease classification as the case definition. This situation led to an underestimation of unique features of molecular pathology analysis in epidemiology, and limited the development of concepts and methods.
To address the need to investigate the inherent heterogeneity of pathogenic processes even for a single disease entity, molecular pathologic epidemiology (MPE) has emerged as an integration of molecular pathology and epidemiology.2–4 This concept and paradigm have been the subject of discussion at established international meetings such as Society for Epidemiologic Research (SER),5 American Association for Cancer Research (AACR),6 and American Society of Preventive Oncology (ASPO),7 and their utility have been widely acknowledged.8–66 Recently, MPE has been integrated into lifecourse epidemiology models.67 To expand opportunities and address challenges, the International Molecular Pathological Epidemiology (MPE) Meeting Series started in 2013, and its second meeting was held in Boston in December 4 to 5, 2014.68 MPE research has caveats and challenges (discussed in the later section) as well as unique strengths.3,69 It can (1) demonstrate a relationship between an exposure and specific molecular alterations; (2) refine the effect estimate of the association between an exposure and a specific disease subtype; (3) support causality; (4) uncover risk factors for a specific disease subtype (which may be masked without disease subtyping); (5) identify disease subtypes associated with benefits from lifestyle or pharmacological intervention; and (6) discover and validate molecular biomarkers for risk assessment, early detection, diagnosis, and decision-making on interventions.3,69 A major value of MPE resides in better definition of phenotype to improve our understanding of disease etiologies from host susceptibility and exposures. Because it can link a putative risk factor to specific molecular signatures of the disease, MPE can provide biological evidence for causality.3,69 For example, MPE research has linked cigarette smoking to epigenetic changes such as DNA methylation, and linked MGMT promoter polymorphism (rs16906252) to MGMT promoter methylation and epigenetic silencing (see references in eAppendix 1).70 Additionally, in contrast to traditional epidemiology, which estimates the association with a given disease (i.e., a collection of subtypes), MPE research can reveal a more precise and stronger association for a specific subtype, to further support a causal relationship (discussed in detail in the next paragraph).3,69 This is because, in general, the larger the effect size of association, the more difficult it is for the association to be explained by uncontrolled confounding. Hence, MPE can work synergistically with the field of causal inference in epidemiology.
As an illuminating example, a relationship between cigarette smoking and colorectal cancer risk is worth discussion. There has been a debate on whether smoking is a cause of colorectal cancer, and epidemiologic studies have shown that an effect estimate for their association is typically not strong (with a relative risk of approximately 1.2).70 MPE can give clues to this vexing public health problem. Under this MPE paradigm, one can posit that a risk factor (such as smoking) facilitates a disease process (such as carcinogenesis) through a specific molecular mechanism (related to the risk factor), which can lead to the development of a specific molecular subtype of the disease. In fact, MPE studies have shown that smoking is a risk factor for a colorectal cancer subtype described as microsatellite instability (MSI) or CpG island methylator phenotype-high,71 with a relative risk of approximately 2 (see references in eAppendix 1).70 These data with the stronger effect estimate support that smoking is indeed causally associated with colorectal cancer, in particular, MSI-high and CpG island methylator phenotype-high subtypes. A similar phenomenon might underline the rather weak inverse association of omega-3 polyunsaturated fatty acid intake with colorectal cancer risk, as evidence suggests this association might be specific to MSI-high cancer subtype.72
MPE research has high relevance in disease prevention, because such studies have shown that different risk factors have shown to influence risks of different subtypes of one disease.73 For example, smoking and obesity have been suggested as risk factors of colorectal cancer when viewed as one disease in the traditional epidemiology paradigm. However, accumulating evidence from MPE studies indicates that smoking is a risk factor for MSI-high subtype of colorectal cancer, while obesity is a risk factor for non-MSI-high subtype of colorectal cancer (see references in eAppendix 1).3 Thus, under the MPE paradigm, smoking and obesity are risk factors for different disease subtypes, and hence, optimal measures for prevention and early detection are likely different between non-obese heavy smokers and obese never smokers. In fact, the MPE research approach has shown that, compared to non-MSI-high colorectal cancer, MSI-high colorectal cancer (for which smoking is a risk factor) appears to be less effectively prevented by colonoscopy,74 and hence, colonoscopy may not be as effective in smokers as in non-smokers. Thus, MPE research can help us understand etiologic heterogeneity, and importance of tailored preventive strategy, depending on different risk factors and profiles of individuals.
MPE has been commonly applied to cancer research because molecular pathologic analyses are most common at present in neoplastic diseases.75 Cancer indisputably represents a group of heterogeneous diseases.76 Genomic and epigenomic analyses provide ample evidence for enormous diversity in tumor molecular features, not only between individuals, but also between two or more tumors within one individual,77 and even between tumor cells within one tumor.1 In addition, evidence also indicates that the exposome (a totality of exposures, including dietary, lifestyle, microbial, and chemical exposures) and host factors such as immunity influence tumor evolution, adding to further diversity of tumor phenotypes.1 The cancer genome and epigenome as well as the relationships to the exposome and interactome (a totality of molecular interactions) differ from tumor to tumor (even with the same tumor name); hence, the “unique tumor principle” holds true.1 This principle asserts that each tumor is unique in terms of exact molecular mechanisms, as well as tumor evolution and behavior including response to treatment.1 Thus, integrated analyses of exposures, tumor molecular features and host immunity are important.4 Integration of cancer immunity and MPE (immuno-MPE) can give new insights on effects of exposures on tumor-host immune interaction.78
The MPE approach can be integrated into a genome-wide association study design or used to follow up this type of research. For example, a candidate polymorphism identified in a genome-wide association study can be studied for its specificity to disease molecular signatures. This genome-wide association study-MPE approach3 can yield a more specific effect estimate for a given subtype, and examine possible biological implications of candidate cancer susceptibility polymorphisms (references in eAppendix 1). Hence, the genome-wide association study-MPE approach can provide evidence for causality. In addition, integrative research of this type may enable us to uncover additional risk variants (specific to only a disease subtype), which have been missed in studies without disease subtyping.
Unique hypothesis evaluation in molecular pathological epidemiology (MPE)
In order to demonstrate specificity of the association between an exposure of interest and a disease molecular subtype, we need to conduct complex hypothesis evaluation. Figure 1 depicts the simplest scenario where a disease of interest consists of binary subtypes A and B. Hypothesis evaluation #1 or #2 is on the relationship between an exposure of interest and subtype A or B, respectively. Hypothesis evaluation #1 and #2 are similar to hypothesis evaluation in traditional epidemiology, except for the outcome being either subtype A or B, instead of overall disease (subtypes A and B combined). In the depicted scenario, one expects that the relative risk effect size for the hypothesis #1 association is greater than 1, while the relative risk effect size for the hypothesis #2 association is close to 1; hence the findings support specificity of the exposure-subtype A relationship. In addition, molecular pathological epidemiology (MPE) focuses on its unique hypothesis evaluation #3, which is on a difference (heterogeneity) between the relationship of the exposure with subtype A and that with subtype B. Hence, this hypothesis evaluation #3 is referred to as “heterogeneity hypothesis evaluation”. One can evaluate the ratio of relative risk which compares relative risk for the exposure - subtype A association to that for the exposure - subtype B association. If there is etiologic heterogeneity, one expects that the ratio of relative risk is greater or smaller than 1.
Figure 1.
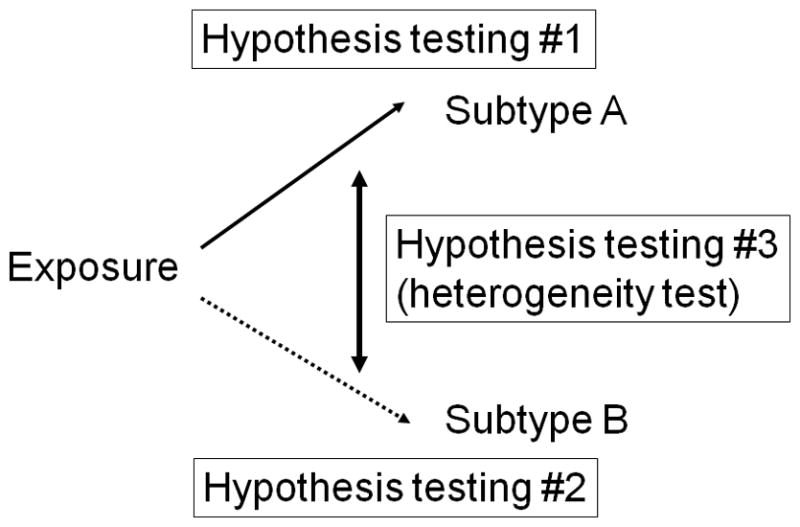
Hypothesis evaluation in molecular pathological epidemiology (MPE). Here, the simplest disease subtyping system with binary subtypes A and B is shown. Note that disease subtyping systems are often more complex than simple dichotomy. Hypothesis evaluation #1 or #2 is on the relationship between an exposure of interest and subtype A or B, respectively. Hypothesis evaluation #3 is unique to MPE, and concerns on a difference (heterogeneity) between the relationship of the exposure with subtype A and that with subtype B. Hence, this hypothesis evaluation #3 is referred to as “heterogeneity hypothesis evaluation” or “heterogeneity evaluation”. See the text for further explanation.
It should be noted that the scenario in Figure 1 addresses the simplest, binary subtypes, and that disease subtyping systems are often more complex than simple dichotomy. For example, disease subtypes may form an ordinal spectrum. Figure 2 depicts a scenario with subtypes classified by an ordinal categorical biomarker, which can theoretically be extended to continuous markers. Many biological functions and tumor biomarkers (including expressions of genes and proteins) are continuous in nature. Furthermore, disease subtypes may be ordinal, non-ordinal, continuous, or defined by more than a single biomarker, and data from various omics analyses (of genome, methylome, epigenome, transcriptome, proteome, metabolome, metagenome, interactome, etc.) can make disease subtyping system even more complex. Epidemiologic analyses need to be performed in various study designs including matched and unmatched case-control studies, case-case studies, prospective cohort studies, and case-cohort studies. It is necessary to develop integrated “epidemiologic design - statistical method” in MPE as new disease subclassification schemes emerge. Thus, new epidemiologic analysis paradigms with statistical methods are constantly needed in MPE. By means of new statistical methods, MPE can synergize advancements of both molecular pathology and epidemiology.
Figure 2.
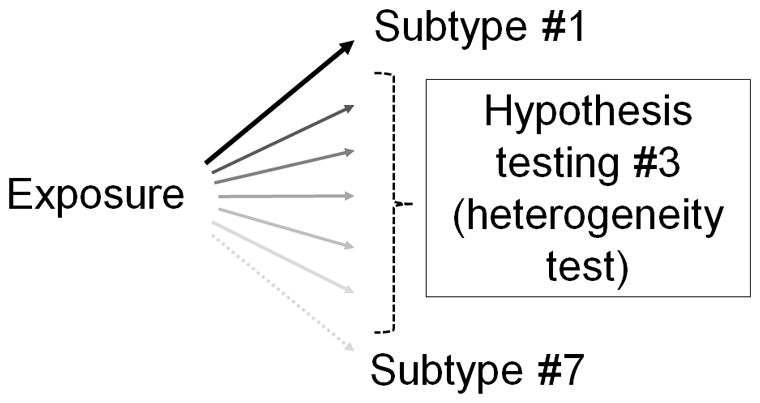
Complex hypothesis evaluation in molecular pathological epidemiology (MPE). Here, shown as an example, the disease subtyping system is ordinal, and more complex than simple dichotomy. Hypothesis evaluation #3 concerns on a difference (heterogeneity) between the relationships of the exposure with ordinal subtypes according to the level of the subtyping biomarker in MPE research. Hence, this heterogeneity evaluation needs to address the ordinal nature of subtyping system. See the text for further explanation.
These unique features of MPE research can amply attest an urgent need for the development of new research frameworks and analytical methodologies as well as research guidelines (which is discussed in the next section).
Challenges in molecular pathological epidemiology
Challenges in molecular pathological epidemiology (MPE) have been discussed in detail elsewhere,3,69 including paucity of interdisciplinary experts, paucity of interdisciplinary education and training programs, paucity of scientific forums focused on MPE, and lack of international guidelines in MPE research.3,69 These challenges are related to each other. Specific caveats of MPE research include selection bias, study sample size and measurement errors, in terms of disease molecular analysis in epidemiologic studies.3,69 In addition, MPE creates opportunities for subgroup analyses, which represents one of its key strengths, but can lead to a higher chance to yield spurious findings.
To address these challenges, there are several ongoing efforts. For example, to overcome the issues in limited sample sizes and generalizability, pooling and meta-analysis approaches can be utilized.14 To discuss the development and standardization of analysis methodologies as well as interdisciplinary education, a new research forum named the International Molecular Pathological Epidemiology (MPE) Meeting Series was established in 2013,7 and the third international meeting will be held in Boston on May 12 and 13, 2016.
MPE deals with the disease outcome as a collection of heterogeneous subtypes, which are hypothesized to be differentially associated with an exposure of interest (Figures 1 and 2). Thus, analytical methodologies to address this etiological heterogeneity need to be fully developed. However, because MPE-type research had typically been considered to belong to molecular epidemiology, an extension of the STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) guideline, which is termed the STROBE-ME (molecular epidemiology) (references in eAppendix 1), does not adequately address the issues in MPE research. Nonetheless, pioneering efforts to develop new statistical methodologies for analyses of disease heterogeneity, essentially reinforcing the MPE paradigm, have been underway (see references in eAppendix 1).64,79–81 In addition, the “STROBE-MPE” guideline project has been proposed to address issues specific to MPE and standardize analytical methodologies in this area of research.68,69,82
Molecular pathological epidemiology (MPE) for non-neoplastic diseases
When considering various human diseases, neoplastic disease is unique in terms of biology as well as medical practice. Neoplasm is characterized by uncontrolled proliferation of cells. Molecular testing on clonally expanded neoplastic cells with somatic molecular alterations can be relatively easily performed utilizing currently available technologies. In contrast, for many non-neoplastic diseases, molecular alterations are elusive and difficult to measure.83
In disease subclassification, there is always a fine balance in relying on similarities (justifying the same subtype) vs. dissimilarities (justifying different subtypes). Although molecular analysis data are generally lacking for non-neoplastic diseases, molecular heterogeneity must play as important a role in non-neoplastic diseases as it does for neoplastic diseases. This is because heterogeneity of human diseases stems from heterogeneity of all of exposures, genetic factors, molecular alterations of diseased cells, and their interactions (the interactome). Complexity of the exposome and alterations of the interactome influence disease processes. Molecular differences in local tissue environment and microenvironment including microbiota add further complexity.84–86 Therefore, any disease process in one individual is unique, and cannot be exactly the same as that in another individual; this theory has been termed the “unique disease principle”.70,87 This principle asserts that each individual has a unique disease process in terms of exact molecular mechanisms, as well as disease evolution and behavior. The unique disease principle applies to all human illnesses, including not only chronic diseases but also acute diseases. In addition, more broadly, an adverse health condition such as drug toxicity is heterogenous, and can be an outcome variable as typical diseases, though it is not typical to call such a condition as “disease” by itself. Similar to neoplastic diseases, molecular subtyping in non-neoplastic diseases can be done based on pathogenic mechanisms, and incorporated in epidemiologic research under the molecular pathological epidemiology (MPE) paradigm. This idea has been increasingly supported in non-neoplastic diseases.88,89
Despite the uniqueness of each disease process, similarities in pathogenic mechanisms (hence, similarities in disease molecular signatures) can be utilized to guide patient management and treatment,90 and to investigate etiologies,3 based on the fundamental epidemiologic premise (i.e., the “homogeneity” to “generalizability” link).
For any disease or adverse health condition, sub-classification based on detailed pathogenic mechanisms (or interactions between host and diseased cells such as tumor immunity78) is possible. Thus, the MPE principle and methodologies can be applied to gain further insights into heterogeneous etiologies of not only neoplastic diseases but also non-neoplastic diseases. While the MPE approach has been most commonly applied to research on cancers such as colorectal, lung and breast carcinomas, MPE research in non-neoplastic diseases can identify novel factors that might influence initiation and progression of a specific disease subtype, and thus help us to develop more effective prevention and treatment strategies.
Molecular pathological epidemiology (MPE) to assess severity and stage of disease
As an example of analysis of disease heterogeneity, here we discuss severity and stage of disease. A continuum of severity (i.e., mild to severe disease) and stage (i.e., early to advanced disease) is almost universally present in human illnesses, and can be measured by subjective perception of a patient (pain, discomfort, functional disability, duration of illness, etc.) or by objective radiological, physiological, pathological and/or other laboratory tests. In many diseases including cancers, patient survival or long-term clinical outcome is often used as a surrogate of disease severity metric in population-based studies; we will discuss caveats of this approach in the next section. Severity of disease most likely (but imperfectly) reflects the magnitude of molecular and pathological changes. In clinical practice, patients with any disease are commonly subclassified based on the degree of disease severity and/or stage, to predict behavior of the disease, and its response to a particular treatment or intervention.
Potential bias due to use of clinical outcome as surrogate of severity
Lethality (or mortality) is often used in epidemiology and clinical research as a surrogate of severity, aggressiveness, or detrimental effect of disease. Lethality status is known only after follow-up of a diseased patient, but not at the time of diagnosis (unless the disease is diagnosed after the patient’s death). Due to heterogeneity in biologic characteristics of a disease in each individual, some seemingly malignant diseases do not lead to death of patients. This phenomenon of heterogeneity of clinical outcomes is common across various diseases, but particularly evident in neoplasms. Therefore, investigators are interested in classifying neoplasms into lethal and nonlethal subtypes, and identifying risk factors for the lethal, aggressive subtypes.
One notable caveat is that an exposure (which can be measured after diagnosis of cancer) can modify tumor behavior, and at the same time, can be associated with the same exposure variable measured before or at the time of cancer diagnosis. Thus, the exposure can appear to be causally associated with aggressive (or indolent) subtype of cancer, but this association can be biased by the effect of the same exposure (after cancer diagnosis) on tumor behavior. It is difficult to predict how and when this bias can substantially impact results. When severity of disease is used as an endpoint heterogeneity variable, severity of disease ideally should not depend on disease course (such as lethality) after disease diagnosis. In addition to disease course, other surrogates of disease severity such as disease stage are free from this type of bias derived from follow-up data, and can be used to assess whether an exposure is associated with a disease differentially according to its severity. At the very least, investigators must be aware of potential bias caused by assessing disease severity using disease follow-up data.
Disease continuum
An important aspect of diseases is that, for centuries, disease designation has been set for convenience, considering symptomatology and affected organ system or physiologic function. As discussed above, each traditional disease entity consists of a group of heterogeneous pathologic processes. Moreover, under the MPE paradigm, different disease entities may have pathologically overlapping features, and related etiologies. For example, uncontrolled clonal expansion of a specific type of immune cells can manifest not only as a neoplastic disease, but also as an autoimmune disease. In addition, the immune system has been known to play a major role in most human diseases; not only infectious, autoimmune, and inflammatory diseases, but also neoplastic diseases. Neoplastic diseases often cause para-neoplastic syndromes with symptoms which may be often observed in non-neoplastic disease conditions. Morbid obesity and diabetes mellitus represent disease entities and are associated with other diseases such as cardiovascular diseases, and neoplastic diseases, and there are also shared risk factors between diabetes mellitus (or obesity) and neoplastic diseases. Importantly, the “disease continuum” concept encompasses the complex interrelationship with shared risk factors between seemingly diverse diseases, e.g., neoplastic diseases and diabetes mellitus. Microorganisms and other infectious agents (including human microbiota) are associated not only with infectious diseases, but also with many other diseases including inflammatory, autoimmune, and neoplastic diseases. Eventually, pathophysiologies of multiple diseases in each individual of a study population need to be considered as the phenome (a totality of phenotypes). In the future, new epidemiologic and statistical methodologies need to be developed, in order to decipher multidimensional phenotypic parameters and biomarker measurements.
Intermediary phenotype
Molecular disease subtyping is commonly performed when an individual has symptoms of a well-developed disease (for many diseases), but the promise of early detection of disease implies the need for measurement of markers of disease before symptoms develop. Thus, it is increasingly common to analyze molecular signatures before a disease is fully developed, by means of imaging techniques or tests on available biospecimens such as biopsy tissue, blood, urine, sputum, or other body fluids. We use the term “intermediary phenotype” to indicate a phenotype between non-disease state and fully developed disease, as illustrated by the example of screening biomarkers above, although not all persons with this phenotypes will go on to develop disease. A similar term “intermediate phenotype” has also been used, but it can be confused with a middle-level phenotype (of a fully developed disease) between phenotypes at extreme ends.
Molecular pathological epidemiology (MPE) can utilize such intermediary variables related to a certain disease.70 For example, premalignant lesions such as colorectal adenomas or Barrett’s esophagus can provide excellent opportunities to examine the role of intermediary phenotypes in population-based studies.91 In normal tissue or peripheral blood, epigenetic, metabolomic or other molecular changes can be used as intermediary variables in studying disease development.70 We should be aware that, besides a hypothesis that intermediary changes are caused by an exposure, there exists an alternative hypothesis that the exposure may drive pathogenesis in a certain state of normal tissue (with the intermediary changes). Intermediary variables may also be useful in the identification of etiologic field effect.92 This model can explain an elevated cancer risk in contralateral breast of patients with breast cancer.92 Etiologic field effect refers to a field of susceptibility of tumor development and progression through influences of etiologic factors and interactions of those factors with host and tumor cells, within local microenvironment.92 It should be noted that intermediary variables may not completely predict occurrence or severity of disease in the future. Thus, the association between the intermediary variables and disease (or disease subtypes) should be examined and established.
Future potential of molecular pathological epidemiology (MPE)
Molecular pathology tests have become routine clinical practice, and been utilized for multiple purposes, including screening, risk prediction in family members, prognostication, minimal disease monitoring, and decision making of treatment and management option.87 Therefore, data from routine molecular pathology testing have been accumulating in hospitals and health care systems around the world. Just as we have utilized disease data in population-based studies, data on molecular and other forms of pathology tests may be utilized in population-based studies; hence, the molecular pathological epidemiology (MPE) approach can become routine research practice.87
As advancement of biomedical sciences is increasingly rapid, disease characterization has been increasingly sophisticated. Next generation sequencing technologies are becoming routine tools in anatomic and clinical pathology laboratories. Multigene-panel sequencing test is performed in germline DNA or DNA from neoplastic cells. In the future, exome and genome sequencing will be routine clinical practice across the world. Data from these tests will be available, and epidemiologic research should incorporate data on molecular characterization of disease. To achieve this, we must address various regulatory and ethical issues, including how to protect privacy of patients.
Molecular subtyping of diseases can be integrated into pharmacoepidemiology to form the integrative field of pharmaco-MPE. For example, pharmaco-MPE research enabled the identification of aspirin as a possible treatment option for PIK3CA-mutated subtype of colorectal cancer, but not for PIK3CA-wild-type subtype.93,94 Because of toxicity often associated with aspirin, biomarkers that are identified to predict benefits using the MPE approach are useful in selecting an appropriate group of patients.95–98 This pharmaco-MPE approach has also been applied to statin use and colorectal cancer subtypes,99,100 and has the potential to be used with other medications and diseases as well.
In the future, pathology must transform itself into diagnostic science that can examine cells, tissues, and bioactive molecules in vivo. Analysis of the in vivo interactome is a major future goal of pathology. As pathology will change into in vivo diagnostic science, epidemiology and MPE can also transform into an integrative population health science where investigators can follow study subjects real-time with examination of the in vivo interactomes around the whole body.
Conclusions
Molecular pathological epidemiology (MPE) has the potential to expand and transform epidemiology in both neoplastic and non-neoplastic diseases; that is, virtually all disease areas. The fundamentally heterogeneous nature of disease pathogenesis necessitates this paradigm shift in epidemiology and study of health and disease in general. This MPE approach helps us to link risk factors to specific molecular signatures present in disease, refine effect estimates for specific disease subtypes, and enhance causal inference. In the near future, advances in molecular pathology will make disease molecular data more widely available to researchers, and the MPE paradigm will become ubiquitous and play important roles in population health sciences. Because heterogeneity of disease is an undeniable fundamental phenomenon, research on diseases (including non-neoplastic diseases) needs to take into account this essential nature of disease. Therefore, the MPE paradigm should become ubiquitous in all areas in epidemiology and population health sciences. Towards these goals, the International Molecular Pathological Epidemiology (MPE) Meeting Series will continue to be held.68
There are growing numbers of biobank / biorepository networks, and population-based MPE databases (“big data”) around the world. Along with these efforts, we can also facilitate transformation of population health science, integrated with not only basic biology and medicine but also social, behavioral, economic, environmental and ecologic sciences. These activities motivated by MPE efforts match with the purposes of the BD2K (Big Data to Knowledge), GAME-ON (Genetic Associations and Mechanisms in Oncology), and Precision Medicine Initiatives of the U.S.A. National Institute of Health. Given advancements in genomics, epigenomics, proteomics, metabolomics, metagenomics, immunomics, interactomics, and other omics technologies as well as bioinformatics and computational and systems biology, there are wide open opportunities in MPE to study virtually all human diseases and contribute to medicine and public health.
Acknowledgments
Funding: This work was supported by grants from the United States of America (U.S.A.) National Institute of Health (NIH) [R35 CA197735 (to SO), R01 CA151993 (to SO), K07 CA190673 (to RN), R01 CA137178 (to ATC), and K24 DK098311 (to ATC)]. AN was supported by a Fellowship Grant from Japan Society for Promotion of Science. PL is a Scottish Government Clinical Academic Fellow. ATC is Damon Runyon Clinical Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Funding agencies did not have any role in the decision to submit the manuscript for publication, or the writing of the manuscript.
Footnotes
Conflict of Interest: ATC previously served as a consultant for Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, and Pfizer Inc. The work was not funded by Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, or Pfizer Inc. All of the other authors declare no conflict of interest.
References
- 1.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12:621–628. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: The evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–367. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology-analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuller LH, Bracken MB, Ogino S, Prentice RL, Tracy RP. The role of epidemiology in the era of molecular epidemiology and genomics: Summary of the 2013 AJE-sponsored Society of Epidemiologic Research Symposium. Am J Epidemiol. 2013;178:1350–1354. doi: 10.1093/aje/kwt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogino S. Molecular pathological epidemiology (MPE): Overview of its paradigm and wide applicability even without tumor tissue [abstract]. Cancer Prev Res (Phila); Proceedings of the Twelfth Annual AACR International Conference on Frontiers in Cancer Prevention Research; 2013 Oct 27–30; National Harbor, MD. 2013. p. CN06-01. [Google Scholar]
- 7.Epplein M, Bostick RM, Mu L, Ogino S, Braithwaite D, Kanetsky PA. Challenges and Opportunities in International Molecular Cancer Prevention Research: An ASPO Molecular Epidemiology and the Environment and International Cancer Prevention Interest Groups Report. Cancer Epidemiol Biomarkers Prev. 2014;23:2613–2617. doi: 10.1158/1055-9965.EPI-14-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathology Research International. 2011;2011:902674. doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes LA, Simons CC, van den Brandt PA, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP) PLoS One. 2011;6:e18571. doi: 10.1371/journal.pone.0018571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs R, Voorneveld P, Kodach L, Hardwick J. Cholesterol metabolism and colorectal cancers. Curr Opin Pharmacol. 2012;12:690–695. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: Progress and problems. Biochim Biophys Acta. 2012;1825:77–85. doi: 10.1016/j.bbcan.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Iwagami S, Baba Y, Watanabe M, et al. Pyrosequencing Assay to Measure LINE-1 Methylation Level in Esophageal Squamous Cell Carcinoma. Ann Surg Oncol. 2012;19:2726–2732. doi: 10.1245/s10434-011-2176-3. [DOI] [PubMed] [Google Scholar]
- 13.Limburg PJ, Limsui D, Vierkant RA, et al. Postmenopausal Hormone Therapy and Colorectal Cancer Risk in Relation to Somatic KRAS Mutation Status among Older Women. Cancer Epidemiol Biomarkers Prev. 2012;21:681–684. doi: 10.1158/1055-9965.EPI-11-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes LA, Williamson EJ, van Engeland M, et al. Body size and risk for colorectal cancers showing BRAF mutation or microsatellite instability: a pooled analysis. Int J Epidemiol. 2012;41:1060–1072. doi: 10.1093/ije/dys055. [DOI] [PubMed] [Google Scholar]
- 15.Ku CS, Cooper DN, Wu M, et al. Gene discovery in familial cancer syndromes by exome sequencing: prospects for the elucidation of familial colorectal cancer type X. Mod Pathol. 2012;25:1055–1068. doi: 10.1038/modpathol.2012.62. [DOI] [PubMed] [Google Scholar]
- 16.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshiol J, Lin SW. Can Tissue-Based Immune Markers be Used for Studying the Natural History of Cancer? Ann Epidemiol. 2012;22:520–530. doi: 10.1016/j.annepidem.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fini L, Grizzi F, Laghi L. In: Adaptive and Innate Immunity, Non Clonal Players in Colorectal Cancer Progression. Ettarh R, editor. InTech; 2012. pp. 323–340. [Google Scholar]
- 19.Gay LJ, Mitrou PN, Keen J, et al. Dietary, lifestyle and clinico-pathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk Study. J Pathol. 2012;228:405–415. doi: 10.1002/path.4085. [DOI] [PubMed] [Google Scholar]
- 20.Galon J, Franck P, Marincola FM, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med. 2012;10:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer-reinterpreting paradigms. Nat Rev Clin Oncol. 2012;9:561–570. doi: 10.1038/nrclinonc.2012.137. [DOI] [PubMed] [Google Scholar]
- 22.Dogan S, Shen R, Ang DC, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-related KRAS-mutant Cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitz MR, Caporaso NE, Sellers TA. Integrative cancer epidemiology--the next generation. Cancer Discov. 2012;2:1087–1090. doi: 10.1158/2159-8290.CD-12-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanmuganathan R, Nazeema Banu B, Amirthalingam L, Muthukumar H, Kaliaperumal R, Shanmugam K. Conventional and Nanotechniques for DNA Methylation Profiling. J Mol Diagn. 2013;15:17–26. doi: 10.1016/j.jmoldx.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Rosty C, Young JP, Walsh MD, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26:825–834. doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- 26.Weijenberg MP, Hughes LA, Bours MJ, Simons CC, van Engeland M, van den Brandt PA. The mTOR Pathway and the Role of Energy Balance Throughout Life in Colorectal Cancer Etiology and Prognosis: Unravelling Mechanisms Through a Multidimensional Molecular Epidemiologic Approach. Curr Nutr Rep. 2013;2:19–26. doi: 10.1007/s13668-012-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchanan DD, Win AK, Walsh MD, et al. Family History of Colorectal Cancer in BRAF p.V600E mutated Colorectal Cancer Cases. Cancer Epidemiol Biomarkers Prev. 2013;22:917–926. doi: 10.1158/1055-9965.EPI-12-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnett-Hartman AN, Newcomb PA, Potter JD, et al. Genomic aberrations occuring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73:2863–2872. doi: 10.1158/0008-5472.CAN-12-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez MC, Santos JC, Maniezzo N, et al. MGMT and MLH1 methylation in Helicobacter pylori-infected children and adults. World J Gastroenterol. 2013;19:3043–3051. doi: 10.3748/wjg.v19.i20.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagland HR, Berg M, Jolma IW, Carlsen A, Soreide K. Molecular Pathways and Cellular Metabolism in Colorectal Cancer. Dig Surg. 2013;30:12–25. doi: 10.1159/000347166. [DOI] [PubMed] [Google Scholar]
- 31.Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, Smans K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res. 2013;52:585–589. doi: 10.1016/j.plipres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbenhardt C, Poole EM, Kulmacz RJ, et al. Phospholipase A2G1B polymorphisms and risk of colorectal neoplasia. Int J Mol Epidemiol Genet. 2013;4:140–149. [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes LA, Melotte V, de Schrijver J, et al. The CpG island methylator phenotype: what’s in a name? Cancer Res. 2013;73:5858–5868. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 34.Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109:1004–1012. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amirian ES, Petrosino JF, Ajami NJ, Liu Y, Mims MP, Scheurer ME. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infect Agent Cancer. 2013;8:42. doi: 10.1186/1750-9378-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmeister M, Blaker H, Kloor M, et al. Body mass index and microsatellite instability in colorectal cancer: a population-based study. Cancer Epidemiol Biomarkers Prev. 2013;22:2303–2311. doi: 10.1158/1055-9965.EPI-13-0239. [DOI] [PubMed] [Google Scholar]
- 37.Araujo RF, Jr, Lira GA, Guedes HG, et al. Lifestyle and family history influence cancer prognosis in Brazilian individuals. Pathol Res Pract. 2013;209:753–757. doi: 10.1016/j.prp.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Esterhuyse MM, Kaufmann SH. Diagnostic biomarkers are hidden in the infected host’s epigenome. Expert Rev Mol Diagn. 2013;13:625–637. doi: 10.1586/14737159.2013.811897. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y, Yang SR, Wang PP, et al. Influence of pre-diagnostic cigarette smoking on colorectal cancer survival: overall and by tumour molecular phenotype. Br J Cancer. 2014;110:1359–1366. doi: 10.1038/bjc.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagland HR, Soreide K. Cellular metabolism in colorectal carcinogenesis: Influence of lifestyle, gut microbiome and metabolic pathways. Cancer Lett. 2015;356:273–280. doi: 10.1016/j.canlet.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 41.Shaheen NJ. Editorial: what is behind the remarkable increase in esophageal adenocarcinoma? Am J Gastroenterol. 2014;109:345–347. doi: 10.1038/ajg.2014.35. [DOI] [PubMed] [Google Scholar]
- 42.Brandstedt J, Wangefjord S, Nodin B, Eberhard J, Jirstrom K, Manjer J. Associations of hormone replacement therapy and oral contraceptives with risk of colorectal cancer defined by clinicopathological factors, beta-catenin alterations, expression of cyclin D1, p53, and microsatellite-instability. BMC Cancer. 2014;14:371. doi: 10.1186/1471-2407-14-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coppede F. The role of epigenetics in colorectal cancer. Expert Rev Gastroenterol Hepatol. 2014;8:935–948. doi: 10.1586/17474124.2014.924397. [DOI] [PubMed] [Google Scholar]
- 44.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross AJ, Moore SC, Boca S, et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120:3049–3057. doi: 10.1002/cncr.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simons CC, van den Brandt PA, Stehouwer C, van Engeland M, Weijenberg MP. Body size, physical activity, early life energy restriction, and associations with methylated insulin-like growth factor binding protein genes in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:1852–1862. doi: 10.1158/1055-9965.EPI-13-1285. [DOI] [PubMed] [Google Scholar]
- 47.Haque TR, Bradshaw PT, Crockett SD. Risk Factors for Serrated Polyps of the Colorectum. Dig Dis Sci. 2014;59:2874–2889. doi: 10.1007/s10620-014-3277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan BM, Wolff RK, Valeri N, et al. An analysis of genetic factors related to risk of inflammatory bowel disease and colon cancer. Cancer Epidemiol. 2014;38:583–590. doi: 10.1016/j.canep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huser V, Sincan M, Cimino JJ. Developing genomic knowledge bases and databases to support clinical management: current perspectives. Pharmgenomics Pers Med. 2014;7:275–283. doi: 10.2147/PGPM.S49904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wennersten C, Andersson G, Boman K, Nodin B, Gaber A, Jirstrom K. Incident urothelial cancer in the Malmo Diet and Cancer Study: cohort characteristics and further validation of ezrin as a prognostic biomarker. Diagn Pathol. 2014;9:189. doi: 10.1186/s13000-014-0189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikeska T, Craig JM. DNA methylation biomarkers: cancer and beyond. Genes. 2014;5:821–864. doi: 10.3390/genes5030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell PT, Deka A, Briggs P, et al. Establishment of the Cancer Prevention Study II Nutrition Cohort Colorectal Tissue Repository. Cancer Epidemiol Biomarkers Prev. 2014;23:2694–2702. doi: 10.1158/1055-9965.EPI-14-0541. [DOI] [PubMed] [Google Scholar]
- 53.Witvliet MI. World health survey: a useful yet underutilized global health data source. Austin J Public Health Epidemiol. 2014;1:id1012. [Google Scholar]
- 54.Li P, Wu H, Zhang H, et al. Aspirin use after diagnosis but not prediagnosis improves established colorectal cancer survival: a meta-analysis. Gut. 2015;64:1419–1425. doi: 10.1136/gutjnl-2014-308260. [DOI] [PubMed] [Google Scholar]
- 55.Wild CP, Bucher JR, de Jong BW, et al. Translational cancer research: balancing prevention and treatment to combat cancer globally. J Natl Cancer Inst. 2015;107:353. doi: 10.1093/jnci/dju353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caiazza F, Ryan EJ, Doherty G, Winter DC, Sheahan K. Estrogen receptors and their implications in colorecal carcinogenesis. Front Oncol. 2015;5:Article 19. doi: 10.3389/fonc.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015;16:2472–2496. doi: 10.3390/ijms16022472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tillmans LS, Vierkant RA, Wang AH, et al. Associations between Environmental Exposures and Incident Colorectal Cancer by ESR2 Protein Expression Level in a Population-Based Cohort of Older Women. Cancer Epidemiol Biomarkers Prev. 2015;24:713–719. doi: 10.1158/1055-9965.EPI-14-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cisyk AL, Penner-Goeke S, Lichtensztejn Z, et al. Characterizing the prevalence of chromosome instability in interval colorectal cancer. Neoplasia. 2015;17:306–316. doi: 10.1016/j.neo.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weisenberger DJ, Levine AJ, Long TI, et al. Association of the Colorectal CpG Island Methylator Phenotype with Molecular Features, Risk Factors and Family History. Cancer Epidemiol Biomarkers Prev. 2015;24:512–519. doi: 10.1158/1055-9965.EPI-14-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao C. Molecular pathological epidemiology: an interdisciplinary field for study of hepatocellular carcinoma. Austin J Gastroenterol. 2015;2:1040. [Google Scholar]
- 62.Szylberg L, Janiczek M, Popiel A, Marszalek A. Serrated polyps and their alternative pathway to the colorectal cancer: a systematic review. Gastroenterol Res Pract. 2015;2015 doi: 10.1155/2015/573814. ID 573814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong EY, Chua C, Beh SY, Koh D, Chong D, Tan IB. Addressing the needs of colorectal cancer survivors: current strategies and future directions. Expert Rev Anticancer Ther. 2015;15:639–648. doi: 10.1586/14737140.2015.1038248. [DOI] [PubMed] [Google Scholar]
- 64.Begg CB, Orlow I, Zabor EC, et al. Identifying Etiologically Distinct Sub-Types of Cancer: A Demonstration Project Involving Breast Cancer. Cancer Med. 2015;4:1432–1439. doi: 10.1002/cam4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell PT, Newton CC, Newcomb PA, et al. Association between body mass index and mortality for colorectal cancer survivors: overall and by tumor molecular phenotype. Cancer Epidemiol Biomarkers Prev. 2015;24:1229–1238. doi: 10.1158/1055-9965.EPI-15-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta AM, Osse M, Kolkman-Uljee S, Fleuren GJ, Jordanova ES. Molecular background of ERAP1 downregulation in cervical carcinoma. Analytical Cellular Pathology. 2015;2015:article ID 367837. doi: 10.1155/2015/367837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishi A, Kawachi I, Koenen KC, Wu K, Nishihara R, Ogino S. Lifecourse Epidemiology and Molecular Pathological Epidemiology. Am J Prev Med. 2015;48:116–119. doi: 10.1016/j.amepre.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogino S, Campbell PT, Nishihara R, et al. Proceedings of The Second International Molecular Pathological Epidemiology (MPE) Meeting. Cancer Causes Control. 2015;26:959–972. doi: 10.1007/s10552-015-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogino S, King EE, Beck AH, Sherman ME, Milner DA, Giovannucci E. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am J Epidemiol. 2012;176:659–667. doi: 10.1093/aje/kws226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: Emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26:465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki H, Yamamoto E, Maruyama R, Niinuma T, Kai M. Biological significance of the CpG island methylator phenotype. Biochem Biophys Res Commun. 2014;455:35–42. doi: 10.1016/j.bbrc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Song M, Nishihara R, Wu K, et al. Marine omega-3 Polyunsaturated Fatty Acids and Risk of Colorectal Cancer According to Microsatellite Instability. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv007. pii: djv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin JH, Giovannucci E. Environmental exposure and tumor heterogeneity in colorectal cancer risk and outcomes. Curr Colorectal Cancer Rep. 2014;10:94–104. [Google Scholar]
- 74.Nishihara R, Wu K, Lochhead P, et al. Long-term Colorectal Cancer Incidence and Mortality after Lower Endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular Pathways Involved in Colorectal Cancer: Implications for Disease Behavior and Prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
- 77.Nosho K, Kure S, Irahara N, et al. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620. e1603. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song M, Nishihara R, Wang M, et al. Plasma 25-hydroxyvitamin D and colorectal cancer risk according to tumour immunity status. Gut. 2015;65:296–304. doi: 10.1136/gutjnl-2014-308852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatterjee N, Sinha S, Diver WR, Feigelson HS. Analysis of cohort studies with multivariate and partially observed disease classification data. Biometrika. 2010;97:683–698. doi: 10.1093/biomet/asq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosner B, Glynn RJ, Tamimi RM, et al. Breast cancer risk prediction with heterogeneous risk profiles according to breast cancer tumor markers. Am J Epidemiol. 2013;178:296–308. doi: 10.1093/aje/kws457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang M, Kuchiba A, Ogino S. A meta-regression method for studying etiologic heterogeneity across disease subtypes classified by multiple biomarkers. Am J Epidemiol. 2015;182:263–270. doi: 10.1093/aje/kwv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogino S, Giovannucci E. Commentary: Lifestyle factors and colorectal cancer microsatellite instability - molecular pathological epidemiology science, based on unique tumour principle. In J Epidemiol. 2012;41:1072–1074. doi: 10.1093/ije/dys076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Field AE, Camargo CA, Ogino S. The merits of subtyping obestity: one size does not fit all. JAMA. 2013;310:2147–2148. doi: 10.1001/jama.2013.281501. [DOI] [PubMed] [Google Scholar]
- 84.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61:794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T-cells in colorectal carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ogino S, Lochhead P, Giovannucci E, Meyerhardt JA, Fuchs CS, Chan AT. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2014;33:2949–2955. doi: 10.1038/onc.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikramuddin S, Livingston EH. New Insights on Bariatric Surgery Outcomes. JAMA. 2013;310:2401–2402. doi: 10.1001/jama.2013.280927. [DOI] [PubMed] [Google Scholar]
- 89.van Winkel R. Aetiological stratification as a conceptual framework for gene-by-environment interaction research in psychiatry. Epidemiology and Psychiatric Sciences. 2015;24:6–11. doi: 10.1017/S2045796014000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lochhead P, Chan AT, Giovannucci E, et al. Progress and opportunities in molecular pathological epidemiology of colorectal premalignant lesions. Am J Gastroenterol. 2014;109:1205–1214. doi: 10.1038/ajg.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lochhead P, Chan AT, Nishihara R, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;28:14–29. doi: 10.1038/modpathol.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation status, and colorectal cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Domingo E, Church DN, Sieber O, et al. Evaluation of PIK3CA mutation as a predictor of benefit from NSAID therapy in colorectal cancer. J Clin Oncol. 2013;31:4297–4305. doi: 10.1200/JCO.2013.50.0322. [DOI] [PubMed] [Google Scholar]
- 95.Fink SP, Yamauchi M, Nishihara R, et al. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of 15-Hydroxyprostaglandin Dehydrogenase (HPGD) Sci Transl Med. 2014;6:233re232. doi: 10.1126/scitranslmed.3008481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chan AT, Ogino S, Fuchs CS. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 97.Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tougeron D, Sha D, Manthravadi S, Sinicrope FA. Aspirin and colorectal cancer: Back to the Future. Clin Cancer Res. 2014;20:1087–1094. doi: 10.1158/1078-0432.CCR-13-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng K, Ogino S, Meyerhardt JA, et al. Relationship Between Statin Use and Colon Cancer Recurrence and Survival: Results From CALGB 89803. J Natl Cancer Inst. 2011;103:1540–1551. doi: 10.1093/jnci/djr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee JE, Baba Y, Ng K, et al. Statin use and colorectal cancer risk according to molecular subtypes in two large prospective cohort studies. Cancer Prev Res (Phila) 2011;4:1808–1815. doi: 10.1158/1940-6207.CAPR-11-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]


