Abstract
Architectural proteins mediate interactions between distant regions in the genome to bring together different regulatory elements while establishing a specific three-dimensional organization of the genetic material. Depletion of specific architectural proteins leads to miss regulation of gene expression and alterations in nuclear organization. The specificity of interactions mediated by architectural proteins depends on the nature, number, and orientation of their binding site at individual genomic locations. Knowledge of the mechanisms and rules governing interactions among architectural proteins may provide a code to predict the 3D organization of the genome.
Keywords: Transcription, nucleus, architectural proteins, CTCF
1. Introduction
The linear eukaryotic genome resides in the three dimensional nucleus in an organized manner [1]. Certain genomic regions are highly self-interactive, whereas interactions between these regions are much less frequent, thus organizing the genome into local chromatin interaction domains named topologically associating domains (TADs) [2–4]. Multiple lines of evidence suggest that this higher-order chromatin organization is functionally linked to genome function. TADs contain genes with coordinated expression [3], they overlap with DNA replication timing domains [2, 5], evolutionarily rearranged domains [6, 7], and oncogenic translocation-induced hyperacetylation domains [8], suggesting that TADs are a physical representation of the functional partitioning of the genome.
Architectural proteins are enriched at TAD borders and at regulatory elements interacting with each other within TADs [2–4, 9]. Binding sites for these proteins cluster at specific regions of the genome termed architectural proteins binding sites (APBSs) where they mediate chromatin interactions independent of their enhancer-blocking insulator function [10]. Here we first review the nature and genomic distribution of architectural proteins characterized in Drosophila and vertebrates. We then discuss results showing that loss of architectural protein function causes changes in chromatin interactions and alterations in transcription. Finally, we describe mechanistic models that aim to predict nuclear 3D organization from the linear information specified by the number, nature and binding site orientation of architectural proteins present at distinct sites in the genome.
2. Architectural proteins
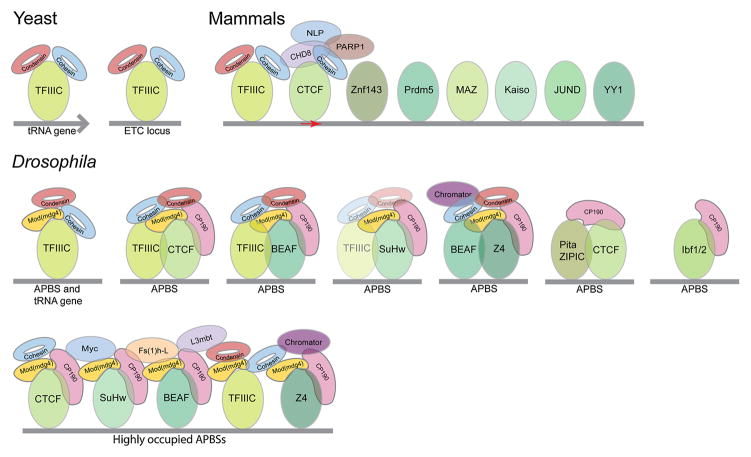
Contrary to vertebrates, in which CCCTC-binding Factor (CTCF) has been the main DNA-binding architectural protein studied in detail thus far, several DNA binding architectural proteins have been well characterized Drosophila. These include CTCF, Boundary Element Associated Factor 32 (BEAF-32), Suppressor of Hairy-wing [Su(Hw)], Transcription Factor IIIC (TFIIIC), Z4 (also called Putzig), Insulator binding factor 1 and 2 (Ibf1 and Ibf2), Pita, and Zn-finger Protein Interacting with CP190 (ZIPIC) [10–12]. These proteins bind to specific sites in the genome and recruit a series of associated factors, including Centrosomal Protein 190 (CP190), Modifier of mdg4 [Mod(mdg4)], Rad21, CAP-H2, L(3)mbt, Fs(1)h-L, and Chromator (also called Chriz). Although each DNA binding protein has a preference to interact with a specific subgroup of accessory proteins, this preference is not strict, and it is possible to find combinations of any of the architectural proteins described above at some genomic location (Figure 1). For example, TFIIIC, which is the main architectural protein found in yeast, is also present in Drosophila at tRNA genes together with Rad21 (a subunit of the cohesin complex) and CAP-H2 (a subunit of the condensin II complex) but it is also found at extra TFIIIC (ETC) sites with both DNA-binding and accessory architectural proteins, including CTCF, BEAF-32, and Su(Hw), CP190, and Mod(mdg4) [10]. BEAF-32 and Z4 colocalize at many promoter-proximal sites in the genome together with Chromator and CP190, whereas Su(Hw) colocalizes preferentially with CP190 and the Mod(mdg4)2.2 isoform. These genomic sites containing individual DNA-binding architectural proteins and several associated factors are called APBSs. Importantly, all or most DNA-binding and associated accessory architectural proteins colocalize in different numbers and combinations at distinct sites called high occupancy APBSs, which are preferentially located at the borders between TADs [10] (Figure 1). Additional candidate architectural proteins that possess canonical insulator function, including Early boundary activity (Elba), have been discovered in Drosophila but their genomic localization in the context of the ones described above has not been explored in detail [13].
Figure 1.
Organization of architectural proteins in different organisms. The main architectural protein in yeast is TFIIIC, which is able to recruit both cohesin and condensin. Drosophila has a large number of DNA-binding architectural proteins that bind to specific sequences in the genome and recruit a series of accessory proteins. Some of these DNA-binding proteins colocalize with other architectural proteins that recognize DNA sequences in close proximity, forming APBSs of varied occupancy. CTCF is the best characterized DNA-binding architectural protein in vertebrates but several other DNA-binding proteins have been shown to colocalize with CTCF and may also play an architectural role to ether enhancer or modify the ability of CTCF to establish interactions between distant sites in the genome.
CTCF and cohesin are the two main architectural proteins extensively characterized in vertebrates [14–17]. However, several other proteins have been shown to colocalize with CTCF at many genomic locations in mammals and to play a role in specific aspects of CTCF function, but their possible role in the establishment of 3D organization has not been explored in detail (Figure 1). For example, Yin Yang 1 (YY1) functions with CTCF during X-chromosome inactivation and both proteins colocalize extensively at evolutionarily conserved CTCF sites located preferentially at promoter-proximal regions [18]. YY1 interacts with cohesin and condensin, and has been shown to contribute to the 3D organization of the Igh locus [19, 20]. Furthermore, YY1 is enriched with CTCF at TAD borders [21]. As is the case in Drosophila, TFIIIC colocalizes with CTCF, cohesin, and the DNA-binding tumor suppressor protein Prdm5 at many locations of the mammalian genome [22–24]. The POZ-Zn finger transcription factor Kaiso interacts with CTCF but its genome-wide distribution or possible role in 3D organization has not been explored in detail [25]. Additional DNA binding proteins that colocalize extensively with CTCF include JunD, the Myc-associated zinc finger protein MAZ, and ZNF143 [26]. Finally, the nucleolar protein Nucleophosmin is required to recruit CTCF to the nucleolus in order to tether CTCF-mediated chromatin loops [27] (Figure 1).
It is striking that most DNA-binding architectural proteins characterized so far in both Drosophila and mammals are zinc finger proteins but it is unclear whether this conservation reflects a requirement for specific aspects of architectural protein function. In the case of CTCF, it has been shown that different combinations of zinc fingers can recognize different sequence motifs, possibly exposing other zinc fingers for protein-protein interaction. This may confer greater specificity to both its DNA- and protein-interacting capacity, while preserving the flexibility to relocate and mediate new chromatin interactions when a cell changes its fate [28, 29]. The degenerate consensus motif sequence of CTCF shows variable base content at many positions [30]. Indeed, three different types of CTCF motif sequences have been shown to be present at distinct genomic locations with respect to regulatory elements, different epigenetic features, and frequency of TSS-distal element interactions [31].
3. Architectural proteins mediate interactions between distant sequences
Hi-C and Chromatin Interaction Analysis by Paired-End Tag sequencing (ChIA-PET) experiments conducted in Drosophila and in multiple mouse and human cell lines have shown enrichment of CTCF and cohesin at TAD borders [2, 3, 9], and at anchor regions of chromatin interaction within TADs [32, 33]. Contacts between cohesin-occupied anchor regions have significantly higher interaction frequency than those with cohesin at only one anchor or not mediated by cohesin [33], suggesting that cohesin facilitates the establishment or maintenance of contacts between the regulatory elements it occupies. However, multiple lines of evidence suggest CTCF and cohesin are not the only two architectural proteins involved in mediating contacts between distant sequences in mammals. In the human B-lymphoblastoid cell line GM12878, only 30% (2,857 out of 9,448) of all interactions are mediated by CTCF sites present at each of the anchor regions, whereas 54% (6,991 out of 12,903) of all chromatin interactions have CTCF at only one of the anchors [32]. Similarly, only 41% of 14,701 RAD21-mediated and 35% of 22,559 the Smc1a cohesin complex subunit mediated interactions occur between two CTCF-binding anchors in the human leukemia cell line K562 and in mouse ESCs, respectively [33, 34]. These results suggest that many chromatin interactions in mammals could be mediated by other architectural proteins or by the combination of cohesin with one or more of the possible candidate architectural proteins described above. For example, Znf143 and YY1 have been shown to be enriched at chromatin interaction anchors [21, 32, 33]. Although YY1 was found to extensively co-localize with CTCF in active genomic regions [18], TAD borders enriched with YY1 but not CTCF show higher enrichment for border-specific epigenomic features than borders enriched with both YY1 and CTCF [21], suggesting YY1 can function as an architectural protein independent of CTCF. Additionally, tRNA genes and SINE elements are also enriched at TAD borders [2], suggesting that TFIIIC, which binds to these sequences, may also be involved in mediating long range interactions [10, 35]. Furthermore, tRNA-like Mammalian-wide Interspersed Repeat (MIR) elements were recently characterized as a new group of sequences in the human genome that possess canonical insulator activity, are close to TAD borders, and appear to be CTCF-independent, suggesting the existence of proteins that bind these sequences with an architectural function [36]. These results suggest that Znf143, YY1, and TFIIIC along with its interaction partner the condensin II complex, may function as new architectural proteins in the mammalian genome but additional functional studies are required to verify their involvement in organizing chromatin contacts.
Although interactions that result in the formation of TADs are relatively stable during cell differentiation, contacts between architectural protein sites located within TADs are more variable [2, 37, 38]. It is possible that cell-type specific interactions result from the presence of architectural proteins in cell-type specific genomic locations. When the CTCF binding landscape was compared in 19 different cell types, 64% (50,149 out of 77,811) of all CTCF binding sites vary in at least one cell type [39], suggesting that CTCF binding is highly plastic across multiple cell types. Interestingly, CTCF sites whose genomic location is conserved across different cell types are enriched at TAD borders, perhaps explaining why TADs tend to be relatively maintained during development [10]. In addition to the existence of cell type-specific sites, it is also possible that architectural proteins collaborate with cell-type specific transcription factors in order to elicit distinct interactions in specific cell lineages. For example, the pluripotency transcription factors Oct4, Nanog, and Klf4 mediate embryonic stem cell (ESC)-specific promoter-enhancer interactions through binding to their cognate sequences and physically interacting with cohesin in ESCs [40–42]. Depletion of these transcription factors leads to decreased occupancy of cohesin and Mediator [40] and disruption of promoter-enhancer interactions [40, 41]. Depletion of cohesin or Mediator was also shown to abolish pluripotency-specific chromatin interactions [43, 44]. These results suggest that cell type-specific transcription factors may confer sequence specificity to cell type-specific chromatin interactions, and help recruit cohesin, which in turn, may stabilize these interactions.
4. Depletion of architectural proteins causes ectopic chromatin interactions and changes in transcription
The presence of CTCF and cohesin at TAD borders and at anchor regions within TADs suggests they may be responsible for the 3D organization of the genome visualized by Hi-C experiments. Indeed, depletion of CTCF by RNA interference (RNAi) in human embryonic kidney 293T (HEK293T) cells leads to reduced intra-TAD interactions and increased inter-TAD interactions, thus distorting the physical partitioning of the genome [45]. This result suggests that CTCF is responsible for mediating intra-TAD interactions and establishing boundaries between TADs. The later would be consistent with the classical enhancer-blocking insulator function if binding sites for architectural proteins inserted between enhancers and promoters of reporter genes cause the formation of a TAD border, which would hinder interactions between regulatory elements present on each side.
Several studies have investigated the effect of reducing cohesin levels on chromatin interactions in both Drosophila and human cells. As is the case for CTCF, intra-TAD contacts were greatly reduced [45–48], consistent with the role of cohesin in mediating promoter-enhancer interactions within TADs [49]. Two of the studies showed that inter-TAD interactions were generally unchanged [45, 47], whereas the other two studies showed enhanced inter-TAD contacts [46, 48], consistent with the interdependent relationship between CTCF and cohesin [14, 15]. The discrepancy could be related to the use of different cell types or methodology to deplete cohesin, resulting in incomplete depletion under some conditions. In all cases, establishment of ectopic chromatin interactions are accompanied by changes in expression of genes whose promoter regions are significantly enriched in CTCF/cohesin [45, 47].
The causal relationship between the presence of architectural proteins and establishment of borders between TADs is supported by experiments showing that a 58-kb deletion of a CTCF-containing border region between two TADs in the X chromosome of mouse ESCs leads to ectopic chromatin interactions across the previous border [3], suggesting that TAD border regions contain information, perhaps in the form of CTCF sites or other functional elements present in the deleted region, that have the ability to establish TADs. Several studies have used the CRISPR-Cas9 genome-editing technique to explore the function of specific CTCF sites in the genome. For example, deletion of a CTCF binding site located between the active Hoxa6 and silenced Hoxa7 genes in mouse ESC-derived motor neurons results in the establishment of new interactions detected by 4C-seq and activation of Hoxa7 [50].
Similarly, deletion of CTCF sites present at a TAD border results in ectopic chromatin interactions and changes in gene expression causing limb malformations [51]. These results suggest that architectural proteins present at TAD borders are responsible for the formation of the two adjacent TADs. This could be the result of an inhibitory effect on cross-border interactions or a positive directional effect that favors interactions towards the two adjacent TADs.
5. Occupancy of genomic sites may determine architectural protein function in the establishment of a specific 3D organization
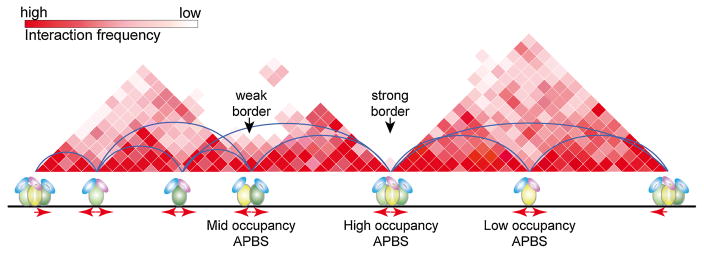
Although architectural proteins such as CTCF are enriched at TAD borders, they are also present at sites inside TADs, including sub-TAD borders, raising the question of what causes architectural protein sites to have functionally different properties. The definition of TAD versus sub-TAD is relative and it is ultimately based on the number of interactions present between each structure with respect to adjacent regions (Figure 2). TADs show frequent interactions within TADs and few interactions between TADs, whereas the number of interactions between sub-TADs is higher than between TADs. Therefore, whether a specific region of the genome is defined as a TAD or a sub-TAD is a consequence of the approaches and algorithms used to define these structures. To avoid this somewhat arbitrary distinction between TADs and sub-TADs, it is possible to define an insulation index or TAD border strength as the ratio between the number of interactions within certain window on both sides of a TAD border and the number of interactions across the border [46, 48]. This concept can then be applied not only to TAD borders but to any DNA fragment in the genome. Borders between TADs will have a high insulation index or border strength whereas sub-TADs will have a lower one. When one then compares the border strength of any DNA fragment in the genome with the number of architectural proteins present at sites in the same fragment, there is a striking correlation between APBS occupancy and border strength in Drosophila and, to a lesser extent, in mammals [10]. High occupancy APBSs preferentially reside at Drosophila TAD borders, whereas medium and low occupancy APBSs are mainly located within TADs (Figure 2). Additionally, the occupancy level of APBSs also correlates with the canonical enhancer-blocking activity of architectural proteins as determined by classical reporter assays in Drosophila [10]. Thus, high occupancy APBSs possess strong insulator activity and preclude interactions between adjacent TADs, establishing the TAD-based partitioning of the genome. Medium and low occupancy APBSs within TADs form sub-TADs and mediate interaction between intra-TAD regulatory elements without posing a strong restriction on the formation of loops required for proper gene expression [48]. These results underscore the importance of viewing TAD border strength as a continuous instead of a binary variable [10, 52]. Furthermore, the correlation between APBS occupancy and border strength suggests that cells may use the recruitment of different numbers of architectural proteins to specific genomic locations as a mechanism to achieve a rich spectrum of biological regulatory control, explaining the gradient of changes observed at TAD borders when cells are subject to differentiation cues or environmental stress [38, 48, 52].
Figure 2.
The role of architectural proteins in the formation of TADs. The figure shows a cartoon representation of a Hi-C heatmap showing 3 TADs. Each TAD has a variable number of sub-TADs indicated by darker red tones inside the TADs. Architectural proteins (shown as spheroids or tori of different colors) are present at APBSs located at the borders of TADs and sub-TADs. Based on results obtained in Drosophila, APBSs with high architectural protein occupancy are located at strong TAD borders, whereas lower occupancy APBSs are found at weak TAD or sub-TAD borders. Although this correlation is also found in mammals, it is not as strong as in Drosophila. In mammals the orientation of CTCF binding sites (shown by red arrowheads) also contributes to the establishment of interactions leading to the formation of TADs and sub-TADs. Although divergently-oriented CTCF sites are enriched at TAD borders, it is unclear whether this is also the case at sub-TADs. It is not currently understood what distinguishes TAD and sub-TAD borders in vertebrate cells.
In addition to APBSs containing several DNA-binding architectural proteins, mammals may also utilize clusters of CTCF sites for a similar purpose. For instance, a cluster of ten CTCF motifs with the same orientation adjacent to the 3’RR enhancer in the mouse Igh locus has been suggested to be important to form long-range contacts responsible for the translocations involving this locus found in various types of cancers [53]. The existence and possible functional significance of clusters of CTCF sites in the genome is underscored by recent studies of the distribution of the CTCF homolog BORIS (also called CTCFL). This protein is expressed at very low levels in most mouse and human cells but it is present in the male germ line and in tumor tissues. Analysis of the distribution of BORIS in these cell types shows that this protein is present at sites containing two CTCF motifs separated by 33–58 bp. These doublets of CTCF motifs are preferentially found at promoters and enhancers and are normally occupied by CTCF, but are co-occupied by CTCF and BORIS in germ and tumor cells [54]. Whether multiple adjacent CTCF sites play a different organizational role in 3D genome architecture than single sites merits further analyses.
6. Orientation of architectural protein sites guides directional looping in mammals
The formation of a specific 3D organization of the chromatin fiber in the nucleus requires the establishment of a specific pattern of genome-wide interactions between APBSs that is determined, at least in part, by their relative occupancy level. This appears to be the case in both Drosophila and mammals [10]. However, cells from vertebrate organisms possess an additional level of control to regulate the nature of the interactions that can take place in the genome that relies on the orientation of the CTCF binding site, which does not appear to occur in Drosophila (M. H. Nichols and V. G. Corces, unpublished observations). Indeed, the vast majority of chromatin interactions with a unique CTCF motif at each anchor (2,574 out of 2,857, 90%) in GM12878 cells show the two motifs in a convergent orientation. This preference for interactions between convergent CTCF motifs has also been observed in several other human cell lines [32]. Interactions between binding sites arranged in the same orientation still occur, although less frequently, and interactions between CTCF sites in a divergent orientation rarely take place. In the mouse Igh locus, the intergenic control region 1 (IGCR1) located in the VH-to-D intergenic region regulates V(D)J recombination by balancing proximal and distal V(H) use. IGCR1 contains two divergently oriented CTCF binding sites that are required for proper IGCR1 function by forming loops with other convergently-oriented CTCF sites on either side of the Igh locus in mouse B cells [12]. The ability of CTCF sites to preferentially interact with sequences located downstream from the site explains the finding that TAD borders tend to contain two closely linked CTCF sites located in divergent orientation. This pattern of divergent sites at TAD borders has been conserved during mammalian evolution in syntenic regions of the mouse, dog, and macaque genomes [6]. Interestingly, the TAD organization surrounding the clusters of Six homeobox genes, which has been evolutionarily conserved since the Cambrian explosion, is established by two divergent CTCF sites conserved in the mouse, zebrafish and sea urchin genomes [7].
The role of CTCF binding site orientation in the establishment of specific interactions by this protein has been analyzed in detail by deleting or inverting specific CTCF sites in the genome [55–57]. At the human protocadherin (Pcdh) gene cluster, CTCF plays an important role in the regulation of enhancer-promoter choice responsible for the stochastic expression of specific Pcdh isoforms [56]. Each Pcdh isoform is transcribed from a specific promoter containing a binding site for CTCF oriented in the direction of transcription. Transcription from a specific promoter requires interactions with downstream enhancers, which contain CTCF sites arranged in a convergent orientation with respect to those present in the promoter regions. Each variable exon and enhancer has a CTCF binding site. Using the CRISPR-Cas9 technique, inversions of key CTCF binding sites were created, switching their orientation. 4C analysis shows that the inverted CTCF binding sites now have an inverted interaction bias. This confirms the causal relationship between DNA binding site orientation and the direction of looping. Furthermore, the change in looping directionality is accompanied by changes in transcription, indicating a functional role for the CTCF mediated interactions in regulating gene expression [56]. De Laat and collaborators have performed a similar analysis in mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs), using 4C to map contacts mediated by 86 CTCF sites present in the genome of these cells [55]. Interactions between CTCF sites, even over distances of 5.8 Mb, show a preference in directionality, with 65% of loops forming between CTCF sites in convergent, 1% divergent, and 34% in the same orientation. Deletion of specific CTCF sites results in disruption of loops between convergent sites, but inversion of a site does not result in the formation of new loops with sites now arranged in a convergent orientation [55]. Ruan and colleagues have used ChIA-PET to examine the role of CTCF binding site orientation on the establishment of loops genome-wide at a resolution of a few hundred bp [58]. They find approximately 54,000 CTCF-mediated loops, 99% of which also contain cohesin in one or both anchors. Of these, 65% are formed between CTCF sites in the forward-reverse orientation whereas 33% are between sites in tandem orientation. Loops formed between convergent CTCF sites are associated with high contact frequency, suggesting that they represent stronger interactions or interactions present in most cells. Importantly, the two types of loop appear to have different functions, with loops between tandemly arranged CTCF sites preferentially enriched in enhancer-promoter interactions in housekeeping gens [58].
7. Extrusion models explain CTCF site directionality in the formation of loops
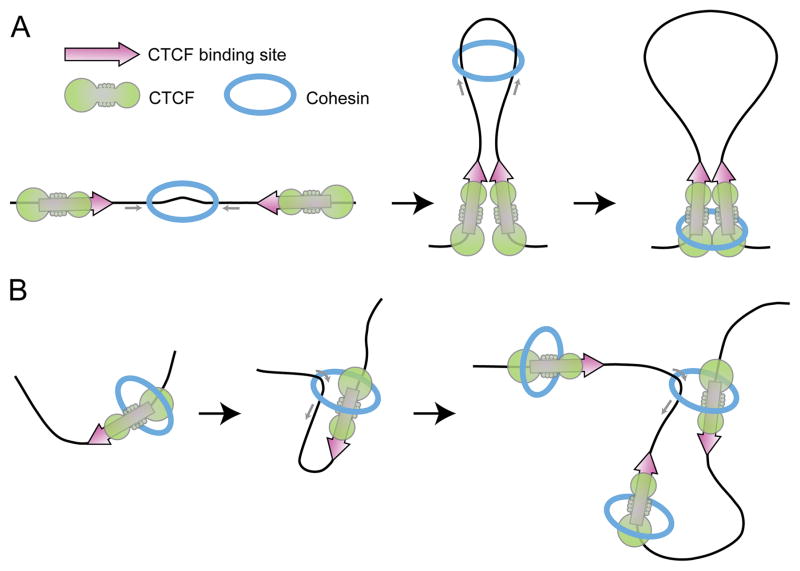
An intriguing question arising from these observations is how the two CTCF proteins encounter each other and form a loop in the 3-D nucleus preferentially when the binding sites are arranged in a convergent versus divergent orientation. A model of random collision and stabilization is inconsistent with the fact that CTCF bound to a pair of convergent motifs represents the same molecular entity as CTCF bound to a pair of divergent motifs, since head-to-head and tail-to-tail configurations are equivalent in the 3D space [59]. Therefore, the preferential recognition of convergently versus divergently oriented CTCF molecules requires constrains in the dimensions of the interacting space. These constrains may be imposed by the cohesin ring, which is thought to form a circle around the two interacting chromatin 10 nm fibers. A model for the generation of self-organized chromosome loops by proteins that can bind to two DNA sites and translocate along the DNA has been proposed by Alipour and Marko. This model is applicable to any DNA-loop-extruding enzyme, such as condensin or cohesin, which would form loops whose length can be regulated by the presence of architectural proteins [60]. The loop extrusion model has been recently extended to explain the formation of loops in interphase chromatin by modeling a 10 Mb region as a polymer bound by loop extruding proteins such as cohesin, and is able to predict the consequence of depletion of CTCF or cohesin in agreement with experimental observations [57, 61] (Figure 3A). This model is agnostic about how the cohesin or condensin are recruited to chromatin, critical for this model is halting of the loop extrusion process by properly oriented CTCF at a domain boundary. A second model based on the loop extrusion concept suggests that bending of the DNA as a consequence of CTCF binding is the initial step in the formation of the loop, which is then extruded by CTCF-bound cohesin [59] (Figure 3B). In either case, the constrain of two CTCF interacting sites to a 1D space would facilitate recognition of structural features in the CTCF protein molecule to allow interaction between convergently versus divergently oriented CTCF sites. These structural features must be determined by the topology of the architectural protein complex. CTCF-cohesin interactions take place between the C-terminal domain of the former and the SA2 subunit of the latter [62]. This is in agreement with results from ChIP-seq experiments showing that the location of Rad21 is shifted with respect to that of CTCF and towards the downstream of the CTCF motif sequence [63]. This arrangement may be important to halt the processivity of cohesin during loop extrusion or to initiate the extrusion process in the proper direction [57, 59, 61].
Figure 3.
Models to explain the preferential establishment of interactions between divergent CTCF sites. A. Cohesin associates with a randomly located genomic site, and initiates the formation of a loop. Cohesin then extrudes a progressively larger loop until it becomes halted at a boundary element, potentially formed by interactions between cohesin and CTCFs with a particular orientation [61]. B. CTCF binds to its recognition sequence and bends the DNA, thus initiating the formation of a loop. CTCF-bound cohesin is then able to extrude a loop by pulling one of the DNA strands. The process stops when a CTCF-bound site in a convergent orientation interacts with the site where the loop was initiated. This interaction is mediated by the two CTCF proteins, which cannot interact when in divergent orientation.
8. Conclusions and perspectives
At first sight there seems to be a difference between the mechanisms Drosophila and mammals employ to establish interactions between specific sites in the genome. Drosophila appears to rely on using a variety of proteins that can colocalize at genomic sites of different occupancy. The degree of occupancy then determines the frequency, and perhaps specificity and functionality, of interactions between sites, with highly occupied sites forming strong TAD borders. Mammals, on the other hand seem to rely preferentially on the orientation of CTCF sites in the genome to choose the interacting partner. However, mammalian cells have a large number of potential architectural proteins that have been shown to interact or colocalize in the genome with CTCF but whose function in 3D organization has not been explored in detail. Therefore, it is possible that, in addition to CTCF site orientation, mammals also rely on APBS occupancy as an additional mechanism to establish specificity of interactions. In addition, some of these potential architectural proteins in mammals could have more specific functions in the process of loop formation between convergent CTCF sites. For example, binding of YY1 to the c-fos promoter induces bending of the underlying DNA [64], and, therefore, YY1 could cooperate with CTCF in the initial steps of loop formation and extrusion. It is possible that, in addition to other architectural proteins, the presence of multiple adjacent CTCF sites play a more deterministic role than single sites in the establishment of specific interactions by stopping the progression of cohesin or favoring the formation of the initial bend during the process of loop extrusion. The specific role of clustered versus single CTCF sites in the 3D organization of the genome remains an important issue for future studies. Finally, it is currently unclear whether contacts mediated by architectural proteins are disrupted during the cell cycle. The TAD organization visible by Hi-C during interphase cannot be observed in cells during mitosis [65]. It is possible that all or most interactions responsible for building the characteristic TAD organization during interphase are disrupted and arranged in a different manner in order to build the highly condensed mitotic chromosomes. However, given the apparent energy-dependent needs to establish orientation-specific interactions between CTCF sites, it is likely that mitotic chromosomes are formed through additional levels of condensation on top of the normal interphase structures.
Acknowledgments
Work in the authors’ lab is supported by US Public Health Service Award R01GM035463 from the National Institutes of Health. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–9. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Marin C, Tena JJ, Acemel RD, Lopez-Mayorga M, Naranjo S, de la Calle-Mustienes E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci U S A. 2015;112:7542–7. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015;29:1507–23. doi: 10.1101/gad.267583.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–84. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuartero S, Fresan U, Reina O, Planet E, Espinas ML. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014;33:637–47. doi: 10.1002/embj.201386001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maksimenko O, Bartkuhn M, Stakhov V, Herold M, Zolotarev N, Jox T, et al. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015;25:89–99. doi: 10.1101/gr.174169.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki T, Sarkeshik A, Yates J, Schedl P. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife. 2012;1:e00171. doi: 10.7554/eLife.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 15.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–66. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwalie PC, Ward MC, Cain CE, Faure AJ, Gilad Y, Odom DT, et al. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 2013;14:R148. doi: 10.1186/gb-2013-14-12-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atchison ML. Function of YY1 in Long-Distance DNA Interactions. Front Immunol. 2014;5:45. doi: 10.3389/fimmu.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerasimova T, Guo C, Ghosh A, Qiu X, Montefiori L, Verma-Gaur J, et al. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev. 2015;29:1683–95. doi: 10.1101/gad.263871.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore BL, Aitken S, Semple CA. Integrative modeling reveals the principles of multi-scale chromatin boundary formation in human nuclear organization. Genome Biol. 2015;16:110. doi: 10.1186/s13059-015-0661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galli GG, Carrara M, Francavilla C, de Lichtenberg KH, Olsen JV, Calogero RA, et al. Genomic and proteomic analyses of Prdm5 reveal interactions with insulator binding proteins in embryonic stem cells. Mol Cell Biol. 2013;33:4504–16. doi: 10.1128/MCB.00545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carriere L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:270–83. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Defossez PA, Kelly KF, Filion GJ, Perez-Torrado R, Magdinier F, Menoni H, et al. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J Biol Chem. 2005;280:43017–23. doi: 10.1074/jbc.M510802200. [DOI] [PubMed] [Google Scholar]
- 26.Xie D, Boyle AP, Wu L, Zhai J, Kawli T, Snyder M. Dynamic trans-acting factor colocalization in human cells. Cell. 2013;155:713–24. doi: 10.1016/j.cell.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 28.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–89. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang R, Wang C, Skogerbo G, Zhang Z. Functional diversity of CTCFs is encoded in their binding motifs. BMC Genomics. 2015;16:649. doi: 10.1186/s12864-015-1824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–17. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crepaldi L, Policarpi C, Coatti A, Sherlock WT, Jongbloets BC, Down TA, et al. Binding of TFIIIC to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet. 2013;9:e1003699. doi: 10.1371/journal.pgen.1003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Vicente-Garcia C, Seruggia D, Molto E, Fernandez-Minan A, Neto A, et al. MIR retrotransposon sequences provide insulators to the human genome. Proc Natl Acad Sci U S A. 2015;112:E4428–37. doi: 10.1073/pnas.1507253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 41.de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–31. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 42.Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–16. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13:30–5. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–77. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell. 2015;58:216–31. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, et al. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–21. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandra T, Ewels PA, Schoenfelder S, Furlan-Magaril M, Wingett SW, Kirschner K, et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10:471–83. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aiden EL, Casellas R. Somatic Rearrangement in B Cells: It's (Mostly) Nuclear Physics. Cell. 2015;162:708–11. doi: 10.1016/j.cell.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Mendez-Catala CF, Kang S, Suzuki T, et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015;16:161. doi: 10.1186/s13059-015-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 56.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–10. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, PS, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015 doi: 10.1016/j.cell.2015.11.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols MH, Corces VG. A CTCF Code for 3D Genome Architecture. Cell. 2015;162:703–5. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–12. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. 2015 doi: 10.1016/j.celrep.2016.04.085. bioRxiv. doi: http://dx.doi.org/10.1101/024620. [DOI] [PMC free article] [PubMed]
- 62.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–83. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natesan S, Gilman MZ. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 65.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, et al. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]