Abstract
Cellular innate immunity poses the first hurdle against invading viruses in their attempt to establish infection. This antiviral response is manifested with the detection of viral components by the host cell, followed by transduction of antiviral signals, transcription and translation of antiviral effectors and leads to the establishment of an antiviral state. These events occur in a rather branched and interconnected sequence than a linear path. Traditionally, these processes were studied in the context of a single virus and a host component. However, with the advent of rapid and affordable OMICS technologies it has become feasible to address such questions on a global scale. In the discipline of Systems Biology’, extensive omics datasets are assimilated using computational tools and mathematical models to acquire deeper understanding of complex biological processes. In this review we have catalogued and discussed the application of Systems Biology approaches in dissecting the antiviral innate immune responses.
Introduction
Antiviral innate immunity studies; performed extensively in recent years, revealed that it is executed through multiple overlapping pathways and involves multi-functional components which engage in elaborate cross talk (Schneider et al., 2014). The outcome of this response can be in favor of the host or the virus, depending on the dynamics of the interactions between virus and host factors. Complexity of the innate immune response arises, both, with the host where different cell types respond differently to viral infections and with the viruses, where differences in genetic makeup, cell tropism and replication kinetics elicit variable host responses (Zak et al., 2014). Such complex and dynamic nature of antiviral innate immunity makes it a perfect subject to tackle with the tools of systems biology. Here we have reviewed the current knowledge of anti-viral innate immune responses, described the system biology approaches to study these biological processes and compiled systems studies on anti-viral innate immunity, with special focus on influenza A viruses.
Anti-viral innate immunity
Viruses are obligate intracellular parasites and infect a broad range of living organisms, from prokaryotes to humans. In this review however, we will limit our discussion to human viral pathogens. The infecting viruses possess or generate pathogen associated molecular patterns (PAMPS) which can be viral genomic material, transcripts, replication intermediates or glycoproteins (Iwasaki, 2012). These PAMPs are detected by cellular pattern recognition receptors (PRRs), which then initiate a cascade of antiviral signaling. PRRs include Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I) like receptors (RLRs), AIM2 like receptors (ALRs), Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and a growing list of DNA sensors. TLRs are either present in endosomes where they detect viral nucleic acid (TLR 3/ 7/ 8/ 9) or on the cell membrane where they detect viral glycoproteins (TLR2/ TLR4) (O'Neill et al., 2013). The RLRs comprise of RIG-I, melanoma differentiation associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2), all of which are present in the cytosol and detect viral RNA or transcription intermediates (Yoneyama et al., 2015). There are several known DNA sensing mechanisms against viruses (DAI, AIM2, IFI16, DHX9, DDX41) and new sensors are being discovered regularly (Dempsey and Bowie, 2015). Most recent addition is cGAMP synthase (cGAS), which detects viral DNA and produces cGMP, which in turn binds to STING and activates anti-viral signaling (Ablasser et al., 2013). Viral DNA and RNA sensing mechanisms cross paths at several junctions, for example RNA Poll III is a DNA sensor, which detects and transcribes viral DNA and resulting RNA feeds back into the RIG-I pathway (Chiu et al., 2009). NLRs primarily detect bacterial pathogens, but can also detect viruses. Among NLRs, NOD2 has been reported to detect viral RNA whereas NLRP3 was shown to be activated by viral infection of dendritic cells (Allen et al., 2009; Sabbah et al., 2009). PAMP-PRR engagement leads to activation of interferon regulatory factors (IRF3/ IRF7/ IRF5) and NFkB transcription factors, which translocate to the nucleus and drive expression of secreted signaling molecules called interferons (IFN) and cytokines. This requires specific kinases (TBK1, IKKα/ IKKβ and MAPKs) and adaptor molecules MAVS (for RLRs), STING (for DNA sensors), TRIF and MYD88 (for TLRs) (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Kawai and Akira, 2010).
Interferons are the backbone of antiviral innate immune response. There are 3 classes of IFN among which Type I IFN and Type III IFN are primary contributors to antiviral innate immunity. Once secreted out of the infected cell, primary IFNs bind to IFN receptors on the same or bystander cells leading to activation of JAK-STAT pathway. In this pathway, transcription factors STAT1, STAT2 and IRF9 form interferon stimulated gene factor 3 complex (ISGF3), which translocates in to the nucleus and binds to interferon stimulated response elements (ISREs) and drives expression of hundreds of genes collectively known as interferon stimulated genes (ISGs). Many of these ISGs have specific or broad spectrum antiviral effectors functions such as Mx, IFITIMs, Viperin and Tetherin (see detailed review by Schneider et al 2014). Among ISGs are also RNA-sensing executors such as OAS and PKR, which can detect viral RNA and induce general RNA degradation mediated by OAS-RNaseL, and shut down mRNA translation (PKR-eIF2α) as a blanket strategy to restrict virus replication. ISGs also include IRFs, STATs and other positive regulators of IFN induction, which further potentiate the antiviral IFN response. Antiviral IFN signaling also leads to secretion of several cytokines and chemokines, which activate immune effector cells and prime the adaptive immune response against viral infections. Finally, IFN signaling is also reported to induce the expression of specific miRNAs, long non coding RNAs (lnc RNAs) and splice variants of specific transcripts which contribute to antiviral host response (Schneider et al., 2014). To keep the IFN signaling in check some ISGs also perform negative regulatory functions such as USP18 (Ritchie et al., 2004) and SOCS (Baetz et al., 2004).
In a parallel pathway, PRR activation (NLRs and ALRs) can lead to formation and activation of inflammasomes which activate caspase 1 to potentiate cytokine production and can eventually initiate cell death (Hornung et al., 2009; Martinon et al., 2009). The components of antiviral signaling are often regulated through, post translational modifications such as phosphorylation, ubiquitination, sumoylation, and ISGylation. These modifications are frequently carried out by ISGs, thus adding several activation and inhibition loops to the antiviral signaling cascades (Ivashkiv and Donlin, 2014).
Systems biology approach to study biological systems
Systems biology can be defined as the study of complex biological processes through orthogonal integration of varied ‘omics’ datasets obtained temporally under different conditions from the biological system in question (Ideker et al., 2001). The goal of a systems approach usually is to develop models which can predict the behavior of a biological system under specific conditions. It is a multidisciplinary field of science where biology, computation, mathematics and engineering come together to solve complex scientific problems. At the core of systems biology approaches are the high throughput methodologies for global data acquisition of different biological properties, computational tools for data analysis and mathematical algorithms to generate probabilistic models. The human genome project can be credited for bringing the systems biology approach to the research forefront (Aderem and Smith, 2004). It was the first major attempt to get global readouts of biological traits using high throughput sequencing technologies. This approach was further developed and employed to sequence the genomes of other important organisms. The vast amount of data generated through these projects required development of databases and software tools for analysis. This brought together researchers from biology and computer sciences and led to the emergence of the field of ‘Genomics’. With that ‘omics’ became the catch all phrase to describe all high throughput approaches to analyze biological systems. The knowledge of complete genome sequences of organisms allowed the design of oligonucleotide arrays to get genome level transcriptional readouts. With the advances in next generation sequencing, in addition to analyzing whole genome transcript levels, one can also detect splice variants, miRNAs and other non-coding RNAs under the discipline of ‘Transcriptomics’. Meanwhile the mass spectrometry based methods to study proteins saw major advances under the field of ‘Proteomics’. It became possible to study protein-protein and protein nucleic acid interactions, protein abundance and post translational modifications at whole genome level. The most complex and still in its infancy is the discipline of ‘Metabolomics’. The genomics, transcriptomics and proteomics datasets complement each other well, however metabolomics datasets are difficult to assimilate due to extremely complex range of metabolites produced in biological systems, which are unknown to large extent (Nicholson and Wilson, 2003). Nevertheless improved methods to study certain classes of metabolites such as lipids (Lipidomics) and sugars (Glycomics) have started contributing to systems studies.
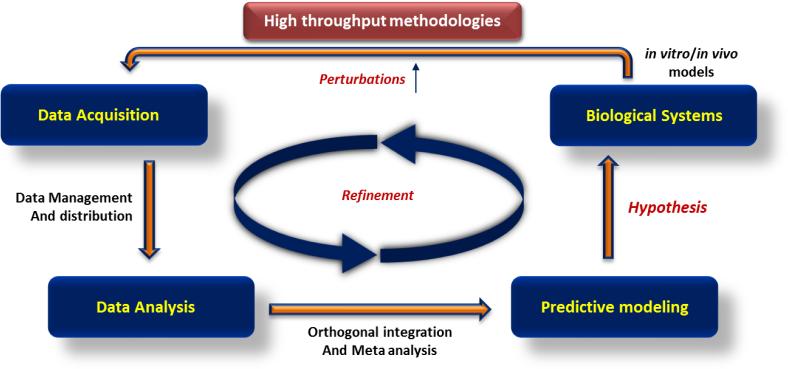
Typically, a systems biology experiment involves measurement of different biological properties such as transcript and protein levels at global scale over a period of time under different conditions. During the experiment, the biological system is perturbed by different means to induce differences in behavior of the system. So far RNAi has been the preferred technology for introducing gene specific or genome level perturbations. With emergence of CRISPR technology scientists have a powerful off the shelf tool for studying the role of genetic elements in biological processes (Sternberg and Doudna, 2015). The advantage of CRISPR technology over RNAi lies in its ability to abolish target gene expression completely, modify parts of it or target non coding regulatory regions of the genome. With emerging applications and variants of CRISPR technology, it may replace RNAi in future systems level studies. The data generated during systems experiments is analyzed using computational methods to identify peculiar enrichment of any class of molecules. A network analysis of the enriched components is usually carried out to identify direct and indirect molecular interactions which may contribute to the change in the behavior of the biological system (Ng et al., 2006). There are several open source and commercial computational tools available to study large scale datasets, which are compiled in Table 1 according to their specific applications. Further, different classes of omics datasets are subjected to Meta-analysis where they are overlapped, integrated and assimilated. This allows identification of key regulators of biological processes, which behave in similar way across varied datasets and are more likely to be true hits. The true power of a systems approach lies however, in its ability to observe ‘emergent properties’ of a system which arise due to combinatorial action of its components (Zak et al., 2014). A familiar example of an emergent property is the sound of music, which emerges when several instruments are played together and it cannot be appreciated by listening to the sound of individual instruments. Once the key components of a biological process are identified, mathematical algorithms are applied to predict a functioning model for the system under study (Azhar and Vodovotz, 2014). The hypothesis put forward through this model is tested again on the biological system through selective perturbations, data acquisition and analysis. This cycle of experiments is reiterated to refine and validate the model until it can predict behavior of the biological system robustly (Fig 2).
Table 1.
Systems Biology and Innate immunity Resources
Figure 2. Systems biology approach to study biological systems.
Systems biology studies involve parallel measurement of varied properties of the biological system using high throughput methods. The acquired data is then analyzed using computational tools to predict key regulators of the biological process being studied. Next, a mathematical model is generated to predict interaction between key regulators under different conditions and how it will affect the phenotype. This model is tested by selected perturbations and repetitive cycle of experimentation, until a robust model for the system is generated
Systems studies on anti-viral innate immunity
In addition to being complex and dynamic, there are other characteristics of the innate immune system which make it amenable to be studied using systems approaches. It is possible to isolate different immune effector cell populations and study their specific contribution to innate immunity. Advances in viral reverse genetics allow introduction of desired mutations in the viral genome and suitable animal models are available for many viral pathogens (Ye et al., 2014). Traditionally the antiviral innate immunity has been studied in a reductionist approach focusing on small set of host and virus factors at a time. With advances in high-throughput omics technologies it became feasible to study cellular processes during viral infection at global scale. The genetically tractable invertebrate model organisms such as C. elegans and Drosophila melanogaster were used first to study innate immunity and contributed significantly to the knowledge, such as the discovery of TLRs (Irazoqui et al., 2010). In mammalian systems RNAi has been the mainstay for genome wide perturbations in systems analysis of innate immunity. It has been used in combination with transcriptional, proteomic and recently with lipidomic profiling to get systems level understanding of anti-viral innate immune responses. One of the early applications of systems approaches to mammalian systems was done by Bouwmeester et al to map the components of the TNF-a/NF-kB pathway (Bouwmeester et al., 2004). Extensive work has been done at the Institute for systems biology (ISB, Seattle), initially to understand TLR signaling in macrophages and dendritic cells in response to bacterial pathogens (Gilchrist et al., 2006; Litvak et al., 2009). Later, in the context of anti-viral response they used systems approach to discover a negative regulatory action of FOXO3 on IRF7-Type I IFN antiviral signaling pathway (Litvak et al., 2012). Chevrier et al used transcriptional profiling, genetic and small-molecule perturbations, and phosphoproteomics to identify novel regulators of anti-viral TLR signaling in dendritic cells (Chevrier et al., 2011). In a similar study, the same group identified mediators of antiviral signaling in response to cytosolic DNA and retroviruses (Lee et al., 2013). In another study, Cho et al used genome wide RNAi approach to discover regulators of virus induced necrotic cell death (Martinon et al., 2009). Using a more proteomics inclined approach Li et al characterized the human innate immunity interactome, and its genetic gain and loss of function validation led to identification of mind bomb (MIB) E3 ligases which controlled ubiquitination of TBK1(Li et al., 2011). Using proteomic methods to screen DNA binding proteins which are also induced by IFN, Bürckstümmer et al identified AIM2 as cytosolic sensor of double-stranded DNA viruses, which recruits the inflammasome and triggers IL-1b production (Burckstummer et al., 2009). At the genome level innate immunity is regulated by epigenetic modifications and interaction between transcription regulators (Stender and Glass, 2013). In a comprehensive unbiased study Amit et al revealed a transcriptional circuit involving 125 factors (transcription factors, chromatin modifiers, RNA binding proteins) which regulated or fine-tuned anti-viral transcription (Amit et al., 2009). In a similar approach Zaslavsky et al identified a temporal cascade of transcriptional factors regulating antiviral transcriptional response to Newcastle disease virus in DCs (dendritic cells) (Zaslavsky et al., 2010). Transcriptional profiling of non-coding host RNAs during virus infection has revealed their regulatory roles in innate immunity. In case of influenza infection, specific changes in levels of miRNAs and other small non-coding RNAs have been reported to regulate the early innate immune response and cell death induction (Li et al., 2010; Peng et al., 2011).
An additional layer of complexity is added at single cell level, where in a homogenous population, individual cells may exhibit different phenotypes based on genetic and proteomic status as well as in stochastic events. Indeed, Shalek et al discovered a bimodal variation in messenger RNA abundance and splicing patterns by performing single cell RNAseq based profiling of DCs in response to LPS. They also identified a module of 137 highly variable genes which regulated antiviral signaling through STAT2 and IRF7 (Shalek et al., 2013). In terms of metabolic profiling, a limited number of Lipidomics studies have been conducted to understand lipid biomarkers associated with viral pathogenicity and anti-viral immune responses (Diamond et al., 2010; Morita et al., 2013; Tam et al., 2013). In an important study, Luber et al compared the lipid profile of CD8+ and CD4+ DCs in response to influenza virus infection and discovered differential expression of PRRs and sensitivity of viral infection (Luber et al., 2010). This study underscored the molecular basis of variable response of different immune effector cells to viral infections. The IFN signaling culminates with expression of hundreds of ISGs, which have varied role in anti-viral response (de Veer et al., 2001). Overexpression of selected set of genes to discover novel anti-viral molecules has been a fruitful approach. Charlie Rice's group has conducted screens of over 400 ISGs, using a panel of reporter viruses and discovered several anti-viral effectors, many of them having broad spectrum effect such as cGAS (Schoggins et al., 2014; Schoggins et al., 2011). Tripartite motif (TRIM) family of proteins has been reported to perform protein modifications and have antimicrobial activity. In an overexpression accompanied with knockdown screen of 75 TRIM family proteins, several member proteins were found to regulate IFN signaling in response to viral infections (Versteeg et al., 2013). Using a cDNA overexpression approach additional regulators of antiviral signaling have been discovered, such as STING in DNA sensing and MafB in IRF3 regulation (Ishikawa and Barber, 2008; Kim and Seed, 2010). Finally, genetic polymorphisms associated with innate immunity genes in humans can yield a lot of information about regulation of innate immune response and viral disease pathogenesis. Genome Wide association (GWA) studies, Exome sequencing, global profiling of single nucleotide polymorphisms (SNPs) and gene copy number variations (CNV) can be used to identify genes responsible for susceptibility to enhanced viral disease or predisposition to autoimmune disorders. Jean-Laurent Casanova's group has successfully used this approach to identify the role of ISG15 in regulation of innate immune signaling and to associate IRF7 and TLR3 deficiencies with severe influenza pathogenesis and herpes encephalitis, respectively, in humans (Ciancanelli et al., 2015; Zhang et al., 2015; Guo et al., 2011). Integrative analysis of published genome-wide datasets at large scale using uniform statistical methods can help visualize unique and diverse molecular patterns in antiviral immunity. On those lines, recently Gorenshteyn et al performed a tour de force comprehensive computational analysis of 38,088 diverse genome scale immunological experimental datasets. This allowed them to create an extensive molecular interaction network capable of predicting antiviral responses (Gorenshteyn et al., 2015). Along similar lines, in context of influenza A viruses, we have performed a meta-analysis of genome scale RNAi datasets and integrated it with global protein interaction data to uncover a functional biochemical landscape for influenza-host interactions, which allowed us to predict several novel antiviral host factors (unpublished). These data and tools are accessible at http://www.metascape.org/IAV.
Systems Studies on Influenza A viruses
Influenza viruses have negative sense segmented RNA genome, which can acquire mutations rapidly through genetic shift and drift. This allows the virus to change its host specificity, virulence and escape the restriction posed by drugs or vaccines (Medina and Garcia-Sastre, 2011). Factors contributing to emergence of novel influenza strains are poorly understood and new strains often cause intermittent pandemics at global scale, case in point the 2009 H1N1 pandemic. The seasonal influenza strains usually have low pathogenicity; however the 1918 H1N1, H5N1 ‘Bird Flu’ and recently emerged H7N9 viruses have shown severe pathogenesis and increased mortality in humans. Orthogonal integration of transcriptional profiling, proteomic and RNAi data to generate detailed virus-host interaction network has been done with many viral pathogens including Influenza A viruses (Konig et al., 2008; Watanabe et al., 2010). These studies are contributing to emergence of ‘Systems Virology’ as a discipline in its own (Law et al., 2013); along the way these studies are also yielding knowledge about anti-viral immune responses (Baas et al., 2006; Shapira et al., 2009). For instance Brass et al conducted a genome wide RNAi screen to identify host dependency factors for IAV, and in the process they also discovered IFITM proteins as broad spectrum anti-viral effectors (Brass et al., 2009). Apart from revealing antiviral signaling components and effectors, system level studies have also pointed to a major role of early innate immune response in the viral pathogenesis. In the case of Influenza A virus infections, transcriptional and proteomic profiling of infected animal models has shown co-operative action among IAV genome segments and host innate immunity and apoptosis genes in severe pathogenesis (Brown et al., 2010; Cheung et al., 2012; Kash et al., 2006; Kroeker et al., 2012). In another such study, Brandes et al applied transcriptional and chemokine profiling during IAV infection to differentiate the contribution of virus vs host in IAV associated pathogenesis and discovered an innate immune chemokine loop as important contributor (Brandes et al., 2013). Michael G. Katze's group has done extensive application of systems methods to understand influenza biology, specially the factors regulating viral pathogenesis (Korth et al., 2013). We, in collaboration with other groups have undertaken the ‘Fluomics’ initiative to achieve systems level understanding of influenza biology (http://www.fluomics.org/). A much desired application of the system based knowledge of antiviral immunity to viruses, is the development of more effective vaccines. This goal is compounded by additional variables such as age, immune status of the individuals, vaccine formulation and administration regimen (Hagan et al., 2015). Scientists have started analyzing the innate and adaptive immune responses in humans post vaccination in comprehensive manner in order to discover biomarkers of efficient vaccine response to influenza and other viruses (Andersen-Nissen et al., 2012; Nakaya et al., 2011; Querec et al., 2009; Tsang et al., 2014). These studies are contributing to the emergence of ‘Systems Vaccinology’ as a new research discipline (Nakaya et al., 2012).
Conclusion and perspective
As High throughput methods of data acquisition evolve, their precision and accuracy will increase and related costs will come down. This will make systems approaches to biological problems more popular and common practice. In order to make novel discoveries and avoid rediscovering known facts, it is crucial to integrate biologically diverse data sets. More powerful and universal algorithms are desired for this purpose. As increasing number of research groups generate omics datasets, data management is a crucial issue to avoid getting lost in translation. It's important that omics datasets are made available to the research community through easy to access databases where different studies can be compared in useful ways. Systems level understanding of antiviral innate immunity holds the key to the design of more effective and broad spectrum antivirals and vaccines. With advances in pharmacogenomics, system level understanding of innate immunity in context of individual patient genetics could be translated into personalized medicine for treating viral infections and inflammatory disorders. However, to realize this goal, active collaborations are warranted for knowledge and resource sharing between clinical and systems biology researchers. In conclusion, systems biology approaches to understand functioning of antiviral immunity is poised to change paradigms, introduce new concepts in virus-host interactions and usher the anti-viral modalities into the era of ‘Systems medicine’.
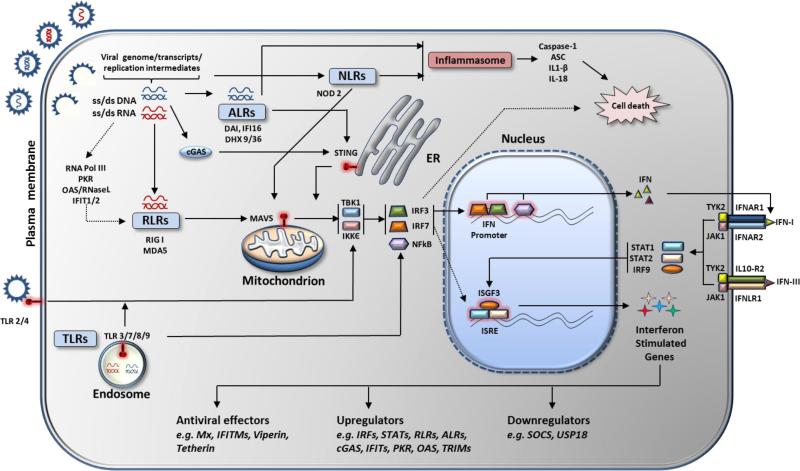
Figure 1. Scheme of antiviral innate immune signaling in mammalian cells.
Upon infection viral ligands (ss/ds RNA/DNA, RNA/DNA hybrids, glycoproteins) are detected by host viral sensors (PRRs, TLRs, ALRs, NLRs) which signal through STING and MAVS to activate IRFs and NFkB, which in turn move in to nucleus, bind to IFN promoter and drive IFN expression. Primary IFNs are secreted out of the cell where they bind to IFNAR and initiate JAK-STAT pathway. This involves formation of ISGF3 with STATs and IRF9, which translocate into the nucleus and bind to ISRE elements to drive expression of ISGs. Among ISGs are antiviral effectors, IFN signaling enhancers (IFNs, IRFs, STATs, PRRs) and inhibitors (SOCS, USP18). PRRs can also lead to formation of inflammasome complex which activates caspase 1 and initiates cell death response upon viral infection.
Highlights.
Cellular anti-viral innate immune response is highly dynamic and complex.
Systems biology approach is well suited to study anti-viral innate immunity.
Systems studies are explaining Influenza pathogenesis and vaccine efficacy.
Acknowledgements
We thank Ekta Tripathi, Gowthamee Thangavel and Michael Schotsaert for critically reviewing the manuscript. Research in Adolfo García-Sastre's lab is supported by NIH grants U19AI106754, U19AI117873, R01DA033773, U19AI118610, R21AI119304 and by CRIP (Center for Research in Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, Clark HF. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6(4):319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aderem A, Smith KD. A systems approach to dissecting immunity and inflammation. Semin Immunol. 2004;16(1):55–67. doi: 10.1016/j.smim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326(5950):257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Nissen E, Heit A, McElrath MJ. Profiling immunity to HIV vaccines with systems biology. Curr Opin HIV AIDS. 2012;7(1):32–37. doi: 10.1097/COH.0b013e32834ddcd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen LL, Mork N, Reinert LS, Kofod-Olsen E, Narita R, Jorgensen SE, Skipper KA, Honing K, Gad HH, Ostergaard L, Orntoft TF, Hornung V, Paludan SR, Mikkelsen JG, Fujita T, Christiansen M, Hartmann R, Mogensen TH. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212(9):1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar N, Vodovotz Y. Innate immunity in disease: insights from mathematical modeling and analysis. Adv Exp Med Biol. 2014;844:227–243. doi: 10.1007/978-1-4939-2095-2_11. [DOI] [PubMed] [Google Scholar]
- Baas T, Baskin CR, Diamond DL, Garcia-Sastre A, Bielefeldt-Ohmann H, Tumpey TM, Thomas MJ, Carter VS, Teal TH, Van Hoeven N, Proll S, Jacobs JM, Caldwell ZR, Gritsenko MA, Hukkanen RR, Camp DG, 2nd, Smith RD, Katze MG. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80(21):10813–10828. doi: 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34(Database issue):D504–506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz A, Frey M, Heeg K, Dalpke AH. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J Biol Chem. 2004;279(52):54708–54715. doi: 10.1074/jbc.M410992200. [DOI] [PubMed] [Google Scholar]
- Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007;23(8):1040–1042. doi: 10.1093/bioinformatics/btm057. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2-3):234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4(3):R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS, Lynn DJ. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, Deville Y, van Helden J. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36(Web Server issue):W444–451. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JN, Palermo RE, Baskin CR, Gritsenko M, Sabourin PJ, Long JP, Sabourin CL, Bielefeldt-Ohmann H, Garcia-Sastre A, Albrecht R, Tumpey TM, Jacobs JM, Smith RD, Katze MG. Macaque proteome response to highly pathogenic avian influenza and 1918 reassortant influenza virus infections. J Virol. 2010;84(22):12058–12068. doi: 10.1128/JVI.01129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Cheung CY, Chan EY, Krasnoselsky A, Purdy D, Navare AT, Bryan JT, Leung CK, Hui KP, Peiris JS, Katze MG. H5N1 virus causes significant perturbations in host proteome very early in influenza virus-infected primary human monocyte-derived macrophages. J Infect Dis. 2012;206(5):640–645. doi: 10.1093/infdis/jis423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, Garber M, Eisenhaure TM, Yosef N, Robinson J, Sutton A, Andersen MS, Root DE, von Andrian U, Jones RB, Park H, Carr SA, Regev A, Amit I, Hacohen N. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147(4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, Lafaille FG, Trouillet C, Schmolke M, Albrecht RA, Israelsson E, Lim HK, Casadio M, Hermesh T, Lorenzo L, Leung LW, Pedergnana V, Boisson B, Okada S, Picard C, Ringuier B, Troussier F, Chaussabel D, Abel L, Pellier I, Notarangelo LD, Garcia-Sastre A, Basler CF, Geissmann F, Zhang SY, Snoeck HW, Casanova JL. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348(6233):448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- Dempsey A, Bowie AG. Innate immune recognition of DNA: A recent history. Virology. 2015;479-480C:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG, 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6(1):e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35(Web Server issue):W606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441(7090):173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Gorenshteyn D, Zaslavsky E, Fribourg M, Park CY, Wong AK, Tadych A, Hartmann BM, Albrecht RA, Garcia-Sastre A, Kleinstein SH, Troyanskaya OG, Sealfon SC. Interactive Big Data Resource to Elucidate Human Immune Pathways and Diseases. Immunity. 2015;43(3):605–614. doi: 10.1016/j.immuni.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Audry M, Ciancanelli M, Alsina L, Azevedo J, Herman M, Anguiano E, Sancho-Shimizu V, Lorenzo L, Pauwels E, Philippe PB, Perez de Diego R, Cardon A, Vogt G, Picard C, Andrianirina ZZ, Rozenberg F, Lebon P, Plancoulaine S, Tardieu M, Valerie D, Jouanguy E, Chaussabel D, Geissmann F, Abel L, Casanova JL, Zhang SY. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208(10):2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan T, Nakaya HI, Subramaniam S, Pulendran B. Systems vaccinology: Enabling rational vaccine design with systems biological approaches. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW, Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, Kanagawa O, Kawamoto H, Kawano K, Koseki H, Kubo M, Kurita-Miki A, Kurosaki T, Masuda K, Nakata M, Oboki K, Ohno H, Okamoto M, Okayama Y, J OW, Saito H, Saito T, Sakuma M, Sato K, Sato K, Seino K, Setoguchi R, Tamura Y, Tanaka M, Taniguchi M, Taniuchi I, Teng A, Watanabe T, Watarai H, Yamasaki S, Ohara O. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23(21):2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chang YC, Wang Y, Huang CL, Liu Y, Tian F, Granger B, Delisi C. VisANT 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res. 2013;41(Web Server issue):W225–231. doi: 10.1093/nar/gkt401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J, Forum S. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- International HapMap, C. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10(1):47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, Garcia-Sastre A, Swayne DE, Katze MG. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443(7111):578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kim H, Seed B. The transcription factor MafB antagonizes antiviral responses by blocking recruitment of coactivators to the transcription factor IRF3. Nat Immunol. 2010;11(8):743–750. doi: 10.1038/ni.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth MJ, Tchitchek N, Benecke AG, Katze MG. Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses. Semin Immunol. 2013;25(3):228–239. doi: 10.1016/j.smim.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker AL, Ezzati P, Halayko AJ, Coombs KM. Response of primary human airway epithelial cells to influenza infection: a quantitative proteomic study. J Proteome Res. 2012;11(8):4132–4146. doi: 10.1021/pr300239r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Korth MJ, Benecke AG, Katze MG. Systems virology: host-directed approaches to viral pathogenesis and drug targeting. Nat Rev Microbiol. 2013;11(7):455–466. doi: 10.1038/nrmicro3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, Dotiwala F, Sen J, Doench JG, Orzalli MH, Kramnik I, Knipe DM, Lieberman J, Carr SA, Hacohen N. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14(2):179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, Kasturi S, Carlone GM, Quinn C, Chaussabel D, Palucka AK, Mulligan MJ, Ahmed R, Stephens DS, Nakaya HI, Pulendran B. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35(3):426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol. 2010;84(6):3023–3032. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10(4):437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, Rust AG, Bergthaler A, Aitchison JD, Aderem A. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012;490(7420):421–425. doi: 10.1038/nature11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32(2):279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. PRIDE: the proteomics identifications database. Proteomics. 2005;5(13):3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Kuba K, Ichikawa A, Nakayama M, Katahira J, Iwamoto R, Watanebe T, Sakabe S, Daidoji T, Nakamura S, Kadowaki A, Ohto T, Nakanishi H, Taguchi R, Nakaya T, Murakami M, Yoneda Y, Arai H, Kawaoka Y, Penninger JM, Arita M, Imai Y. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153(1):112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;4(2):193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of crosstalk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8(6):571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- Ng A, Bursteinas B, Gao Q, Mollison E, Zvelebil M. Resources for integrative systems biology: from data through databases to networks and dynamic system models. Brief Bioinform. 2006;7(4):318–330. doi: 10.1093/bib/bbl036. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID. Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, Schroth GP, Tumpey TM, Li C, Kawaoka Y, Baric RS, Katze MG. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio. 2011;2(6) doi: 10.1128/mBio.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, de la Torre JC, Zhang DE. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10(12):1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10(10):1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Glass CK. Epigenomic control of the innate immune response. Curr Opin Pharmacol. 2013;13(4):582–587. doi: 10.1016/j.coph.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Doudna JA. Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol Cell. 2015;58(4):568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- Tam VC, Quehenberger O, Oshansky CM, Suen R, Armando AM, Treuting PM, Thomas PG, Dennis EA, Aderem A. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154(1):213–227. doi: 10.1016/j.cell.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, Moir S, Dickler HB, Perl S, Cheung F, Baylor HC, Consortium CHI. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38(2):384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7(6):427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Evans JG, Stambas J. Influenza reverse genetics: dissecting immunity and pathogenesis. Expert Rev Mol Med. 2014;16:e2. doi: 10.1017/erm.2014.4. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Zak DE, Tam VC, Aderem A. Systems-level analysis of innate immunity. Annu Rev Immunol. 2014;32:547–577. doi: 10.1146/annurev-immunol-032713-120254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavsky E, Hershberg U, Seto J, Pham AM, Marquez S, Duke JL, Wetmur JG, Tenoever BR, Sealfon SC, Kleinstein SH. Antiviral response dictated by choreographed cascade of transcription factors. J Immunol. 2010;184(6):2908–2917. doi: 10.4049/jimmunol.0903453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, Rice GI, Chen C, Mansouri N, Mahdaviani SA, Itan Y, Boisson B, Okada S, Zeng L, Wang X, Jiang H, Liu W, Han T, Liu D, Ma T, Wang B, Liu M, Liu JY, Wang QK, Yalnizoglu D, Radoshevich L, Uze G, Gros P, Rozenberg F, Zhang SY, Jouanguy E, Bustamante J, Garcia-Sastre A, Abel L, Lebon P, Notarangelo LD, Crow YJ, Boisson-Dupuis S, Casanova JL, Pellegrini S. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517(7532):89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]