Abstract
Collecting circulating tumor cells (CTCs) shed from solid tumor through a minimally invasive approach provides an opportunity to solve a long-standing oncology problem, the real-time monitoring of tumor state and analysis of tumor heterogeneity. However, efficient capture and detection of CTCs with diverse phenotypes is still challenging. In this work, a microfluidic assay is developed using the rationally-designed aptamer cocktails with synergistic effect. Enhanced and differential capture of CTCs for nonsmall cell lung cancer (NSCLC) patients is achieved. It is also demonstrated that the overall consideration of CTC counts obtained by multiple aptamer combinations can provide more comprehensive information in treatment monitoring.
1. Introduction
Tumor heterogeneity, which reflects the co-existence of cell clones with distinct phenotypes and behaviors in tumor(s), has been demonstrated for many cancers.[1] Such heterogeneity often consistently evolves, eventually leading to the uprising of cancer cells with resistant phenotypes.[2] As the sampling of tumor tissues by either surgery or needle biopsy is invasive and difficult to be repeatedly performed during the treatment, real-time classification of their heterogeneity through traditional methods is only theoretically possible.[3] In recent years, collecting circulating tumor cells (CTCs), which are cancer cells that detach from solid tumors and circulate in the peripheral blood, has emerged as a promising approach for tumor sampling.[4] As “liquid biopsy” of cancer, CTCs have the advantages of minimal invasiveness and convenient accessibility.[5] Besides the real-time monitoring of tumor burden, CTC detection also provides a chance to characterize the heterogeneity of tumor.
To enable the characterization of CTC heterogeneity, it is necessary to develop an enrichment process that meets the demands of sufficient capture efficiency and the ability to isolate cancer cells with different phenotypes. However, even after years of effort, it remains a technical challenge mainly for two reasons.[6,7] First, the abundance of CTCs is extremely low (approximately one CTC per billion normal hematopoietic cells in the peripheral blood of patients with advanced disease).[8] Second, few technology developments actually give consideration to cancer cells with different phenotypes. Anti-EpCAM (Anti-epithelial cell adhesion molecule) based CTC enrichments, to take an example, are so far the commonly-used strategy that has been validated in several types of cancer, including breast cancer, prostate cancer, and colon cancer.[9] However, it suffers from the loss of cancer cells with low EpCAM expression.[10] The combined use of additional antibodies (i.e., anti-HER2 and anti-EGFR) may offset the loss, but it is still insufficient for comprehensive characterization of CTC heterogeneity as the number of available antibodies against tumor-specific surface markers is very limited.[11] For the same reason, the antibody-based approach can only be applied to a few cancer types.
Using aptamers, the chemical antibodies, for the detection of CTCs has been recently explored with different mechanisms.[12] CTC enrichments using aptamer have been verified with artificial samples prepared with cancer cell lines (e.g., leukemia, colon cancer, gastric cancer) or cultivated mice tumors (orthotopic tumors, primary human glioblastoma cell).[13] Aside from comparable affinity and specificity, aptamers also possess advantages over natural made antibodies, such as long-term stability, synthetic reproducibility, and convenience for chemical modification.[14] More importantly, cell-specific aptamers can be generated through an in vitro process (cell-SELEX) even in the absence of knowledge about their molecular targets, making aptamers the ideal CTC targeting agent especially for cancer cells lacking available antibodies.[15] In contrast to fruitful results reported for artificial samples, the translational study of cell-SELEX generated aptamers as CTC enrichment and capture agents has been seldom addressed in clinic. One possibility is that in vitro cultured cancer cell line does not fully represent clinical situations. For example, it has been reported that patient CTCs could have a higher degree of heterogeneity comparing to dish-cultured cancer cell lines.[16] Therefore, CTC enrichment methods developed based on a single aptamer derived from cancer cell line might not function equally well in the detection of clinical samples.
Herein, to address the above issue, we develop an approach for rational design of aptamer cocktails with synergistic effect based on an existed aptamer panel. In the combined use of a silicon nanowire substrate (SiNS) embedded microfluidic chip,[17] the enhanced and differential capture of CTCs for nonsmall cell lung cancer (NSCLC) patients was achieved using cell-SELEX derived aptamers. Furthermore, we also explore the clinical value of this assay as well as its application potential in the characterization of CTC heterogeneity of cancer patients.
2. Results
2.1. Screening of Candidate Sequences
As reported earlier, we have already generated a panel of ten aptamers through cell-SELEX against representative NSCLC cells A549 (Table S1, Supporting Information).[18] These aptamers have shown different specificity to NSCLC subtypes. We expect that by modifying SiNS with multiple aptamers, it is possible to achieve enhanced cell affinity through a synergistic effect introduced by the concurrence of two or more independent receptor-ligand recognitions (Figure 1). Theoretically, we may create a large pool of aptamer cocktails based on this 10-aptamer panel. Rather than exhaustively examining each possibility, we designed a two-step process (Figure S1a, Supporting Information) to scale down the candidate pool by eliminating undesired sequences, which either have relatively low affinity to NSCLC cells or are inferior to other aptamers in the competition for target cells.
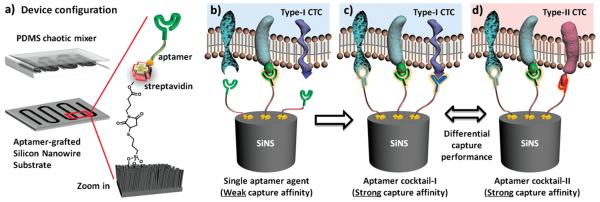
Figure 1.
Schematic description of aptamer cocktail based CTC assay. a) Microfluidic CTC chip is composed of an aptamer-grafted silicon nanowire substrate (SiNS) and an overlaid PDMS chaotic mixer. b) When a single aptamer capture agent was employed, the capture affinity of the device is relatively weak for the lack of synergistic binding. c) By using cocktail capture agents, the synergistic effects among individual aptamers lead to an enhanced capture affinity. d) Different cocktail capture agents are expected to have differential capture performance for CTC subpopulation recognition.
In the first step, the affinities of ten aptamers to five different NSCLC cell lines (i.e., A549 human lung adenocarcinoma, H460 human large-cell carcinoma, H292 human pulmonary mucoepidermoid carcinoma, H1299 human lung Adenocarcinoma-lymph node metastasis, and SK-MES-1 human lung squamous carcinoma) were evaluated using flow cytometry. As high binding affinity is a prerequisite for efficient capture of rare CTC cells, the best two aptamers with highest affinities to each NSCLC cell line (i.e., Ap1/Ap5 for A549, Ap1/Ap4 for H460, Ap1/Ap2 for H292, Ap1/Ap5 for H1299, and Ap3/Ap5 for SK-MES-1) were selected for further evaluation (Figure S1b, Supporting Information). Thus, the library of candidate aptamers was narrowed down to five sequences, which were, respectively, Ap1, Ap2, Ap3, Ap4, and Ap5. To maximize the synergy among individual aptamers, a competition assay was employed to eliminate the pairing of aptamers which may compete for the same binding site (Figure S1c, Supporting Information). As a result, four sequences (Ap1, Ap2, Ap3, and Ap4) were picked out as candidate aptamers for the formulation of aptamer cocktails.
2.2. Verification of Aptamers’ Synergistic Effect
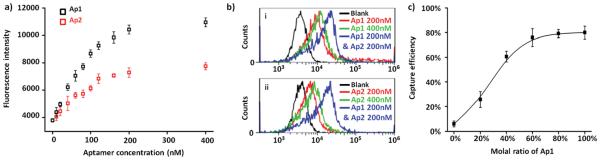
The idea of using aptamer cocktails is built on a precondition that two or more aptamers may yield synergistic effect and leads to increased cell affinity (Figure 1b,c). Therefore, we carried out the following experiments to verify the feasibility. Aptamers Ap1 and Ap2 with fluorescent reporter group were employed for the labeling of A549 cells at various concentrations (0–400 × 10−9 m). After removing unbound aptamers, A549 cells were examined by flow cytometry. As shown in Figure 2a, the fluorescence intensity of cells reached a platform when the concentration of Ap1 or Ap2 was higher than 200 × 10−9 m. We then side by side compared the fluorescent signal of A549 cells incubated, respectively, with 200 × 10−9 m Ap1, 400 × 10−9 m Ap1, and a mixture solution containing 200 × 10−9 m Ap1 and 200 × 10−9 m Ap2. The results showed that increasing the concentration of Ap1 from 200 × 10−9 m to 400 × 10−9 m hardly led to signal increase (Figure 2b). In contrast, the fluorescence became notably enhanced by adding 200 × 10−9 m Ap2 to 200 × 10−9 m Ap1. Similar results were observed after switching the positions of Ap1 and Ap2, indicating occurrence of synergistic effect on aptamer binding.
Figure 2.
a) Fluorescence intensity of A549 cells measured after incubation with Ap1 or Ap2 at a series of concentrations (e.g., 0–400 × 10−9 M). b) Comparisons of flow cytometry results of A549 cells after incubation with different aptamer solutions containing: (i) Ap1 200 × 10−9 M, Ap1 400 × 10−9 M, or Ap1 200 × 10−9 M plus Ap2 200 × 10−9 M; (ii) Ap2 200 × 10−9 M, Ap2 400 × 10−9 M, or Ap1 200 × 10−9 M plus Ap2 200 × 10−9 M. c) The capture efficiency of A549 cells of SiNS with different aptamer densities, which were achieved by mixing a scramble DNA sequence Rc and aptamer Ap1 at various proportions during surface modification.
To ensure that this synergistic effect can play a role in the existing platform (i.e., SiNS embedded microfluidic chips), we also examined the cell capture efficiency as a function of aptamer density. In this experiment, we selected aptamer molecule Ap1, which has the highest overall affinity to 5 NSCLC cell lines, and a scramble DNA sequence Rc (Table S1, Supporting Information), which can compete with Ap1 for the binding site on SiNS but has no interaction with target cells. This scramble DNA was mixed with aptamer Ap1 in different molar ratios but with a fixed total concentration (10 × 10−6 m) during modification to prepare SiNS with different densities of cell-binding aptamer. Following the previously reported procedures of evaluating cell capture yield,[17] we found that the efficiency gradually increased with aptamer density and reaches a maximum when the ratio of Ap1 rises to ≈60% (Figure 2c). This result suggested that there was a surplus of binding sites for aptamer molecules due to the increased surface area and the topographic interactions introduced by nanostructured substrate. [19] Therefore, it is possible to improve the cell capture performance by introducing other synergistic aptamers.
2.3. Formulation of Aptamer Capture Agents
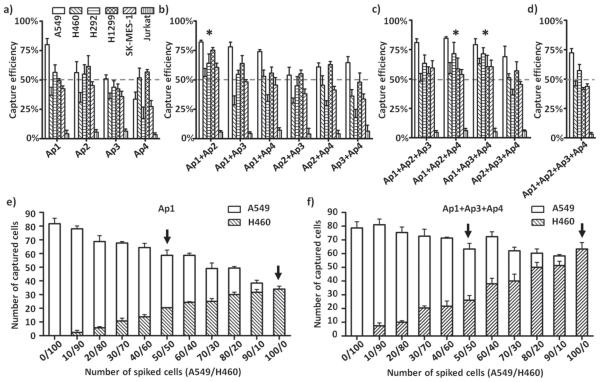
With the four candidate sequences (Ap1, Ap2, Ap3, and Ap4) we selected, different categories of aptamer combinations, including single aptamer, dual-, tri-, and quad-aptamer cocktails, were prepared by mixing individual aptamers in equal molar ratio with a total concentration of 10 × 10−6 m. Cell suspensions (100 cells mL−1) of the five NSCLC cell lines (A549, H460, H292, H1299, and SK-MES-1) and a control (Jurkat human T lymphocytes) were applied to test the capture performance of different aptamer(s). The results summarized in Figure 3a–d supported our earlier finding—the combined use of SiNS embedded microfluidic chips and NSCLC-specific aptamer(s) conferred specificity to the devices for capturing NSCLC cells instead of control cells (Jurkat cells). Moreover, comparing to single aptamers, aptamer cocktails showed generally enhanced cell capture efficiencies. Specifically, the average efficiencies observed for dual (Figure 3b) and tri-aptamer (Figure 3c) cocktails outperform those for single aptamers (Figure 3a). The quad-aptamer combination (Figure 3d), however, does not exhibit the highest efficiencies, possibly due to excessively reduced density of each aptamer.
Figure 3.
a–d) Cell capture studies of SiNS modified with four different categories of capture agents, which were, respectively, a) single aptamer, b) dual-, c) tri-, and d) quad-aptamer cocktails using five NSCLC cell lines (A549, H460, H292, H1299, and SK-MES-1) and a control (Jurkat). Three aptamer cocktails (i.e., Ap1+Ap2, Ap1+Ap2+Ap4, and Ap1+Ap3+Ap4, marked with asterisk) showed >50% capture performance across all five NSCLC cell lines. e,f) Cells capture experiments using heterogeneous artificial samples prepared by mixing H460 and A549 cells at different ratios (e.g., H460/A549 ranges from 0/100 to 100/0). After processing through SiNS modified with e) aptamer Ap1, f) aptamer cocktail Ap1+Ap3+Ap4, the numbers of immobilized A549 cells (blank column) and H460 cells (diagonal pattern) were counted separately.
It was also worthy to note that the same combination of aptamers displayed nonequivalent cell capture capacity for different NSCLC cell lines. Such differential capture performance indicated that assays based on one specific aptamer or aptamer cocktail may lead to a biased enrichment of CTCs. Therefore, we designed an experiment to study how this differential performance will affect the capture of cancer cells in a more heterogeneous sample. First, artificial samples were prepared by mixing Dio prestained A549 and Dil prestained H460 cell lines at a series of proportions while the total number of spiked cells remained unchanged (100 cells per 1 mL). These samples were then processed through SiNS embedded microfluidic chips modified with single aptamer Ap1 or aptamer cocktail Ap1+Ap3+Ap4. According to Figure 3e,f, though the spiked total cell numbers were 100 for all conditions, the enumeration results of Ap1 modified chips roughly dropped from 80 to 40 (up to 50%) as the proportion of H460 gradually increased. This was because the capture efficiency of Ap1 toward H460 was much lower than A549. So the number of captured cells was mainly determined by the amount of A549, the dominant subpopulation in cell suspension. On the other hand, the cocktail Ap1+Ap3+Ap4 had comparable capture yields for H460 and A549, so its detection result was in better accordance with the total number of cancer cells. To take an example, if we assume two samples 50 A549/50 H460 and 100 H460 (Figure 3e,f, marked with arrow) are collected from the same patient at two different time points to evaluate the therapeutic effect of ongoing treatment, the falling of one tumor subpopulation (e.g., A549) and the rise of the other (e.g., H460) does not actually change the total number of CTCs. In this case, the number variations of CTC monitored by cocktail Ap1+Ap3+Ap4 were apparently more accurate than Ap1.
Based on the above cell line experiments, we finally picked out three cocktail combinations exhibiting >50% capture performance across all of the five NSCLC cell lines, which were, respectively, Cocktail A (Ap1+Ap2), Cocktail B (Ap1+Ap2+Ap4), and Cocktail C (Ap1+Ap3+Ap4). These three aptamer cocktails together with single aptamer Ap1, which had the best performance among its own category, were selected for the translational studies using blood samples collected from advanced NSCLC patients.
2.4. CTC Capture and Reproducibility Evaluation
The protocol for CTC enumeration study with clinic samples included three general steps: (1) blood collection/process, (2) CTC capture, and (3) immunostaining and microscopy imaging (see Figure S2, Supporting Information). To evaluate the reproducibility of our method, 12 NSCLC patients (P1–P12) and 12 healthy donors (H1–H12) were divided into four groups. Each of these groups corresponds to one combination of aptamers (see study design in Figure S3, Supporting Information). For each individual, 3 mL blood was collected, split into three aliquots, and subjected to parallel CTC enumeration studies using microfluidic devices modified with the same aptamer(s). The results are summarized in Table 1, in which statistically significant differences between NSCLC patients and healthy donors were observed (t-test, P < 0.05 for all four combinations). Among 36 tests of healthy donors, only in two cases, one false positive cell was detected. We set the threshold value for CTC detection as one cell count to ensure the reliability of our method. For all 36 tests of the patients, no false negative result was observed, indicating excellent sensitivity and selectivity of our platform. We also found the values of standard deviation (Std) and coefficient of variation (CV) for all 12 patients are quite low (Table 1), which suggested our method has good reproducibility.
Table 1.
The reproducibility study of aptamer-modified microfluidic chips.
| Object | ID | Test 1 | Test 2 | Test 3 | Std | CV | |
|---|---|---|---|---|---|---|---|
| Ap1 | Patient | P1 | 4 | 4 | 5 | 0.58 | 13.32% |
| P2 | 7 | 6 | 4 | 1.53 | 26.96% | ||
| P3 | 4 | 5 | 6 | 1.00 | 20.00% | ||
| Average | – | – | – | 1.04 | 20.1% | ||
| Healthy donor | H1 | 0 | 0 | 0 | – | – | |
| H2 | 0 | 0 | 0 | – | – | ||
| H3 | 0 | 0 | 0 | – | – | ||
| Cocktail A | Patient | P4 | 10 | 11 | 8 | 1.53 | 15.80% |
| P5 | 4 | 7 | 6 | 1.53 | 26.96% | ||
| P6 | 8 | 10 | 7 | 1.53 | 18.33% | ||
| Average | – | – | – | 1.53 | 20.4% | ||
| Healthy donor | H4 | 0 | 0 | 0 | – | – | |
| H5 | 0 | 0 | 0 | – | – | ||
| H6 | 1 | 0 | 0 | – | – | ||
| Cocktail B | Patient | P10 | 16 | 14 | 19 | 2.52 | 15.41% |
| P11 | 11 | 8 | 10 | 1.53 | 15.80% | ||
| P12 | 12 | 9 | 13 | 2.08 | 18.37% | ||
| Average | – | – | – | 2.04 | 16.5% | ||
| Healthy donor | H10 | 0 | 0 | 0 | – | – | |
| H11 | 0 | 0 | 0 | – | – | ||
| H12 | 0 | 0 | 0 | – | – | ||
| Cocktail C | Patient | P7 | 7 | 7 | 8 | 0.58 | 7.87% |
| P8 | 14 | 17 | 16 | 1.53 | 9.75% | ||
| P9 | 12 | 13 | 14 | 1.00 | 7.69% | ||
| Average | – | – | – | 1.04 | 8.4% | ||
| Healthy donor | H7 | 0 | 0 | 0 | – | – | |
| H8 | 0 | 0 | 1 | – | – | ||
| H9 | 0 | 0 | 0 | – | – |
Statistically significant differences were observed between CTC counts of NSCLC patients and healthy donors, i.e., Ap1 (t-test, P = 0.006), Cocktail A (t-test, P = 0.021), Cocktail B (t-test, P = 0.025), and Cocktail C (t-test, P = 0.040).
Abbreviations: Std, standard deviation; CV, coefficient variation.
2.5. Parallel CTC Enumeration
To further evaluate the performance of aptamer and aptamer cocktails in clinical practice, we performed the parallel CTC enumeration study in which each patient was checked using four different combinations of aptamers. A total of 11 stage-IV NSCLC patients (Table S2, Supporting Information) were enrolled for this experiment. Following the same procedure as above, 4 mL blood sample was collected from the same patient and subjected to parallel CTC enumeration studies using four devices modified with Ap1, Cocktail A, B, and C, respectively (see Figure S4, Supporting Information).
The enumeration results are summarized in Table 2. A certain number of CTCs were detected successfully for all 11 patients, with only three out of 44 tests the CTC number fell on the threshold value (CTC = 1). Similar to the cell line experiment, aptamer cocktails, particularly tri-aptamer combinations Cocktail B and C, captured more CTCs than single aptamer Ap1 in general. It supported the idea that aptamer cocktails may improve the capture efficiency in detection of patient samples. In addition to the improved capture efficiency, we noticed that CTC counts obtained by different aptamer combinations formed a distinct profile for each patient (i.e., CTC numbers vs aptamer capture agents). To illustrate this point, we highlighted the numbers in Table 2 according to the range of cell counts (gray 1 ≤ CTC < 5, blue 5 ≤ CTC < 10, yellow 10 ≤ CTC < 20, and red CTC ≥ 20).
Table 2.
Parallel CTC enumeration study using four different combinations of aptamers.
| Object | ID | Ap1 | Cocktail A | Cocktail B | Cocktail C |
|---|---|---|---|---|---|
| Adenocarcinoma | P13 | 2 | 1 | 2 | 2 |
| P14 | 12 | 25 | 16 | 11 | |
| P15 | 26 | 28 | 58 | 71 | |
| P16 | 3 | 2 | 12 | 4 | |
| P17 | 7 | 6 | 11 | 23 | |
| P18 | 4 | 3 | 6 | 2 | |
| P19 | 2 | 2 | 3 | 1 | |
| P20 | 2 | 6 | 7 | 9 | |
| SqCC | P21 | 2 | 8 | 7 | 6 |
| P22 | 2 | 1 | 3 | 5 | |
| P23 | 5 | 3 | 16 | 8 |
All CTC counts are divided into five different categories and highlighted with different colors, which are, respectively, gray (0 ≤ CTC < 5), blue (5 ≤ CTC < 10), yellow (10 ≤ CTC < 20), and red (CTC ≥ 20).
Abbreviation: SqCC: squamous cell carcinoma.
We also carried out a side by side comparison between anti-EpCAM and aptamers. As summarized in Table S3 (Supporting Information), microfluidic devices coated with aptamer cocktails generally captured more CTCs than anti-EpCAM for the enrolled patients (n = 3), further demonstrating the advantages of our aptamer-based CTC detection strategy.
2.6. Treatment Monitoring
It is known that the heterogeneity of a patient’s tumor will not remain steady during his or her treatment. The number, proportion, and even surface phenotypes of CTC subpopulations may vary independently. So the accuracy of treatment monitoring based on the number of CTCs counted by one specific capture agent cannot be guaranteed. Therefore, we proposed to perform parallel CTC enumeration study and evaluate its clinical significance in treatment monitoring.
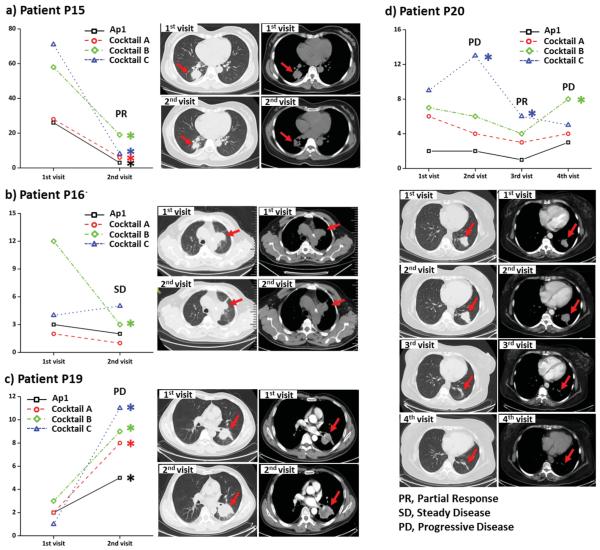
In this study, the profile variations of four patients (i.e., P15, P16, P19, and P20) before and after treatment of Pemetrexed-disodium and Cisplatin were examined and compared with their computed tomography (CT) images. Based on the result of our reproducibility study for each device (Table 1), we set two criteria to ensure the reliable comparison of CTC counts from two separate tests (e.g., N and N′ in the test before and after treatment) for the same patient: (1) ΔN = |N −N′ | ≥2•Std and (2) ΔN = |N − N′ | ≥2•CV•N. N and N′ cannot be regarded as significantly different (marked with *) unless both two criteria can be satisfied.
As given in Figure 4, the parallel enumeration tests for patients P15 (Figure 4a) and P19 (Figure 4c) all showed consistent CTC number change after treatment and were highly correlated with patients’ CT images and disease status. However, for patient P16, for example, the CTC counts obtained by three combinations (Ap1, Cocktail A, and Cocktail C) remained unchanged according to our criteria, while a significant decrease was observed in the presence of capture agent Cocktail B (Figure 4b). Associated with the fact that there was no detectable change in tumor size, this patient should be regarded as stable disease, which was in accordance with the number changes of Ap1, Cocktail A, and Cocktail C. The difference in CTC change in parallel enumeration tests indicated the co-existence of CTCs with different sensitivities to ongoing therapy. The result also supported the assumption that CTC counting from one specific capture agent may not fully reflected disease status, while the CTC enumeration profile acquired from parallel enumeration study can provide more comprehensive information.
Figure 4.
Monitoring the treatment response of patients, a) P15, b) P16, c) P19, and d) P20, by comparing the parallel CTC enumeration results before and after therapy, as well as the correlation between their CTC number variations and CT images. CTC counts significantly higher or lower than last visit are marked with asterisks.
After receiving the first therapy of Pemetrexeddisodium and Cisplatin treatment, the tumor size of patient P20 showed notably increase (Figure 4d). At the same time, comparing the results of CTC detection in the first (before first therapy) and second visits (after first therapy), the CTCs obtained from one combination (Cocktail C) showed significant increase while the rest three remained unchanged, indicating this patient was insensitive to the first therapy. Both imaging and CTC enumeration results agreed with his clinical assessment of progressive disease (PD). Since then, this patient was switched to the second treatment with Gefitinib. At the third visit, reduced tumor size was observed, which suggested the clinical assessment was partial response. Accordingly, CTC counts for Cocktail C showed remarkable decrease. The rest three combinations also exhibited CTC number decrease in a certain degree, but they were too small to be regarded as significant based on our criteria. At the forth visit after a period of Gefitinib treatment, patient P20's disease was regarded as progressing (PD) according to his clinic symptoms even though there was no detectable size increase of tumor. The CTC enumeration profile showed the decrease of CTC counts for Cocktail C became insignificant. Meanwhile, the CTC counts for Cocktail B became notably higher. It indicated the uprising of CTCs (captured by Cocktail B) that were insensitive of the second therapy. This growing tumor group might be too small to be caught, thus it was not noted with CT imaging, but was successfully detected with the parallel CTC enumeration tests with aptamer cocktails.
3. Discussion
The two-step in vitro process we used in this work allows for the acquisition of aptamer cocktails without knowing their molecular target in advance. Benefited from this property, aptamer cocktails with synergistic effect show a better overall capture efficiency comparing to single aptamer, which had been demonstrated not only in cell line experiment but also in the detection of patient samples. Of course, although the aptamers we used here were all selected against cancer cell lines, this strategy is open to a variety of capture agents, such as aptamers or even antibodies targeting peptide, protein, or tumor tissue.
In the parallel enumeration study, we noticed that CTC counts obtained by different aptamer combinations formed a distinct profile for each patient. This cannot be simply attributed to the generally improved capture efficiency as they also differ among patients of the same pathology. Associated with the study using heterogeneous cell line samples, we believe this phenomenon reflected the individualized CTC heterogeneity of patients, since a specific aptamer or aptamer cocktail might differentially enrich CTC subpopulations. The result of treatment monitoring supported this idea to a certain extent. As for the same patient (e.g., P20), CTCs captured by different aptamer(s) showed different treatment sensitivities.
In our opinion, parallel CTC enumeration assays involving multiple combinations of aptamers can possibly provide a more-inclusive coverage in terms of CTC subpopulations comparing to that from one aptamer or one aptamer cocktail, thus allowing more comprehensive and accurate monitoring of treatment responses during cancer therapy. Even without validation in the molecular analysis of the captured CTCs, the real-time analysis of CTC enumeration profile already demonstrates its potential in treatment monitoring of cancer patients.
4. Conclusion
In summary, for the first time as we know, the enrichment and capture of CTCs from NSCLC patients was successfully performed using aptamers based microfluidic assay. We also demonstrated that aptamer cocktails showed enhanced and differential performance in CTC capture. The dynamic monitoring of CTC counts using multiple combinations of aptamers provides additional information for patients’ prognosis and treatment response, serving as a valuable tool in the clinical management of patients. Considering the easy access of aptamer panels from cell-SELEX, this method can be replicated in other cancers. We also envision that elaborately formulated aptamer cocktails may offer a potential solution for the selectively enrichment of CTC subpopulations, which once realized will be an important step toward the real-time characterization of CTC heterogeneity.
5. Experimental Section
Fabrication of SiNS Embedded Microfluidic Chips
The SiNS embedded microfluidic chips composed of two separate components, a patterned SiNS and a PDMS (polydimethylsiloxane) chaotic mixer. The fabrication process has been reported elsewhere.[7] Briefly the patterned SiNS was fabricated by a standard photolithography and a chemical wet etching process. The SiNS was then functionalized with streptavidin according to the previously established method. The PDMS chaotic mixer was prepared following a standard soft lithography method. These two components were aligned and sandwiched with a holder made of stainless steel to form a complete device. Aptamer (Invitrogen) or anti-EpCAM (R&D) solution was loaded into the channel and incubated at room temperature before use.
Cell Culture
Six cell lines, i.e., A549 (adenocarcinoma), H460 (large cell carcinoma), H292 (pulmonary mucoepidermoid carcinoma), H1299 (large cell carcinoma), SK-MES-1 (squamous carcinoma), and Jurkat T (immortalized human T lymphocyte cells), were all obtained from National Platform of Experimental Cell Resources for Sci-Tech. A549 cells were maintained in DMEM medium (Dulbecco’s modified eagle medium) with 10% fetal bovine serum (FBS) and 100 units mL−1 penicillin-streptomycin. The rest five cell lines were cultured in RPMI 1640 medium with 10% FBS (HyClone) and 100 units mL−1 penicillin-streptomycin (Sigma-Aldrich).
Flow Cytometry Experiment
For flow cytometry experiment, cultured cell lines harvested with 10 × 10−3 M EDTA (ethylenediamine tetraacetic acid) were resuspended into working buffer (DPBS (Dulbecco phosphate-buffered saline) with 0.1 mg mL−1 yeast tRNA/4.5 g L−1 glucose/1 mg mL−1 BSA (Albumin from bovine serum) /5 × 10−3 M MgCl2) and split into equal aliquots, which contained about 2 × 105 cells. FAM (6-carboxy-fluorescein) labeled aptamer was added into separate aliquot forming a final concentration of 500 × 10−9 M. After 30 min incubation at 37 °C, aptamer labeled cells were centrifugal washed three times with DPBS containing 5 × 10−3 M MgCl2 to remove unbound DNA molecules. Fluorescent intensity of the cells labeled with different aptamers was measured by a flow cytometry (BDAccuri C6, USA).
Cell Capture Experiments Using Artificial Samples
For the preparation of artificial samples, EDTA harvested cells were resuspended into culture medium containing 5% (v/v) lipophilic tracers with fluorescence (Dio or Dil, Invitrogen) for 30 min. Fluorescent labeled cells were washed with PBS (phosphate-buffered saline) and serially diluted. To achieve precise spiking number, at least five aliquots of cell suspension were transferred to a microplate and enumerated under a fluorescent microscope (Olympus IX71, Japan). Finally, artificial samples (100 cells mL−1) were prepared by spiking a given volume of cell suspension into working buffer, DMEM with 5 × 10−3 M MgCl2. For a heterogeneous sample, the cell suspension of two cancer cell lines (i.e., H460 and A549) labeled with different fluorescence tracers were mixed at different ratios.
1 mL artificial sample was loaded into a disposable syringe and slowly injected into a SiNS embedded microfluidic device modified with aptamer(s) using a syringe pump (LongerPump, Baoding, China). After sample processing, 200 µL 4% paraformaldehyde (PFA) was introduced into the channel for the fixation of captured cells. The device was then disassembled. The SiNS was rinsed with PBS and mounted to a cover slide using mounting solution (Fisher). The number of captured cells was enumerated under a fluorescent microscope (IX71, Olympus).
Formulation of Aptamer Capture Agents
In the first step, each aptamer’ (Table S1, Supporting Information) cell binding affinity was evaluated with 5 NSCLC cell lines (A549, H292, H1299, H460 and SK-MES-1). 10 NSCLC specific aptamers were added into separate aliquots of cell suspension forming a final concentration of 500 × 10−9 M. After incubating at room temperature for 30 min, the fluorescent intensity of aptamer labeled cells were measured by flow cytometry. The two aptamers with the highest binding affinity for each individual cell line were selected for following experiment (Figure S1b, Supporting Information). After binding affinity study, 5 NSCLC specific aptamers were chosen for further evaluation (i.e. Ap1, Ap2, Ap3, Ap4 and Ap5).
In the second step, a competition assay (Figure S1c, Supporting Information) was employed to eliminate the pairing of aptamers with mutual competition in order to maximize the synergy among them. Similar to the procedure above, A549 cells were split into equal aliquots (400 µl containing 2×105 cells). A549 cells in control group were incubated with 500 × 10−9 M FAM labeled aptamer A at room temperature for 30 min. Meanwhile, A 549 cells in experiment group were incubated with 500 × 10−9 M FAM labeled aptamer and 5 µM non labeled competing aptamer. By comparing the fluorescent intensity of cell in both groups, we were able to understand whether competition occurred between two aptamers (Figure S1d,e, Supporting Information). Finally, the aptamer Ap1, Ap2, Ap3 and Ap4 were selected for the formulation of cocktail capture agents.
General Protocol for CTC Analysis with Clinical Samples
For clinical analysis, a general protocol was applied (Figure S2, Supporting Information). The blood sample was collected from each individual using a vacutainer (BD Vacutainer EDTA Blood collection tubes), and subjected to red blood cell lysis using a commercial buffer (BioLegend) following the standard protocol. After centrifugation, the cell pellet was resuspended into working buffer (DMEM containing 5 × 10−3 M MgCl2) depending on the original sample volume. 1-mL cell sample was then loaded into a disposable syringe and process through an aptamer conjugated microfluidic chip. For the fixation of captured cells, 200 µL 4% PFA was introduced into the channel. After disassembling of the device, 300 µL of 0.5% Triton X-100 was added onto the SiNS for 10 min for cell permeabilization. Then, unconjugated mouse anti-cytokeratin antibody (BD) and unconjugated rabbit anti-CD45 (AbCAM) diluted with 2% BSA in PBS were loaded onto the SiNS for 2 h at room temperature. After washing with PBS, a second antibody cocktail composed of Alexa 488 conjugated Donkey anti-Mouse and Alexa 555 conjugated Donkey anti-Rabbit diluted in 2% BSA was added and incubated for 30 min at room temperature prior to DAPI (4′,6-diamidino-2-phenylindole) staining. The SiNS was then thoroughly rinsed with PBS and mounted to a glass cover slide using mounting solution. Finally, the SiNS was imaged under a fluorescence microscope (IX71, Olympus) to identify nanosubstrate-immobilized CTCs (DAPI+/CK+/CD45−) from nonspecifically captured white blood cells (WBCs) (DAPI+/CK−/CD45+).
Reproducibility Study
Twenty four individuals including 12 NSCLC patients and 12 healthy donors were enrolled for this study. 3 mL peripheral blood was collected from each individual and treated following the same protocol described above. After that, three equal aliquots of resultant cell suspension were processed through identical microfluidic chips modified with one specific formula of aptamer agent (Figure S3, Supporting Information). Captured cells were then stained with fluorescent antibodies as well as DAPI and enumerated under microscope. To evaluate the selectivity of our method, an independent sample t-test was used to compare CTC counts of cancer patients and healthy donors. Ps < 0.05 was considered statistically significant. The Std and CV for each aptamer capture agent are also summarized in Table 1.
Parallel CTC Enumeration Study
11 NSCLC patients (Table S2, Supporting Information) were enrolled for the parallel CTC enumeration study. 4 mL peripheral blood was collected and subjected to sample pre-treatment following general protocol. Four aliquots of resultant cell suspension were processed through microfluidic chips modified with different aptamer agents (i.e. Ap1, Cocktail A, Cocktail B and Cocktail C) in parallel (see Figure S4, Supporting Information). Captured cells were stained with fluorescent antibodies and enumerated under microscope.
Supplementary Material
Acknowledgements
L.Z. and C.T. contributed equally to this work. This work was supported by the National Basic Research Program of China (Grant Nos. 2013CB933701 and 2011CB91100), the National Natural Science Foundation of China (Grant Nos. 21127901, 21402211, and 81472186), Chinese Academy of Sciences (Grant No. XDA09030308), and Beijing Municipal Natural Science Foundation (Grant No. 7142122).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Libo Zhao, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China
Dr. Chuanhao Tang, Department of Lung Cancer, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China
Dr. Li Xu, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China
Dr. Zhen Zhang, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China
Dr. Xiaoyan Li, Department of Lung Cancer, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China
Haixu Hu, Laboratory of Oncology, Translational Medicine Center, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China.
Si Cheng, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China.
Wei Zhou, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China.
Mengfei Huang, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China.
Anna Fong, Department of Molecular and Medical Pharmacology, California NanoSystems Institute, University of California, Los Angeles, CA 90095, USA.
Prof. Bing Liu, Laboratory of Oncology, Translational Medicine Center, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China
Prof. Hsian-Rong Tseng, Department of Molecular and Medical Pharmacology, California NanoSystems Institute, University of California, Los Angeles, CA 90095, USA.
Prof. Hongjun Gao, Department of Lung Cancer, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China.
Dr. Yi Liu, Laboratory of Oncology, Translational Medicine Center, Affiliated Hospital of Academy of Military Medical Sciences, Beijing 100071, P. R. China.
Prof. Xiaohong Fang, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Molecular Nanostructure and Nanotechnology Institute of Chemistry Chinese Academy of Sciences, Beiyi Street 2#, Zhongguancun, Beijing 100190, P. R. China.
References
- [1].Marusyk A, Almendro V, Polyak K. Nat. Rev. Cancer. 2012;12:323. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- [2].Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Nature. 2011;472:90. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Marusyk A, Polyak K. Biochim. Biophys. Acta. 2010;1805:105. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alix-Panabieres C, Pantel K. Clin. Chem. 2013;59:110. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- [5].Hayes DF, Paoletti C. J. Intern. Med. 2013;274:137. doi: 10.1111/joim.12047. [DOI] [PubMed] [Google Scholar]
- [6].a) Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yu M, Stott S, Toner M, Maheswaran S, Haber DA. J. Cell Biol. 2011;192:373. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang S, Liu K, Liu J, Yu Z, Xu X, Zhao L, Lee T, Lee E, Reiss J, Lee Y, Chung L, Huang J, Rettig M, Seligson D, Duraiswamy K, Shen C, Tseng H. Angew. Chem. Int. Ed. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qian W, Zhang Y, Chen W. Small. 2015;11:3850. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- [9].Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI, Sastre J, Maestro ML, Puente J, Veganzones S, Alfonso R, Rafael S, Garcia-Saenz JA, Vidaurreta M, Martin M, Arroyo M, Sanz-Casla MT, Diaz-Rubio E. Clin. Cancer Res. Ann. Oncol. 2007;2008;1319:7053, 935. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- [10].Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yu M, Bardia A, Wittner B, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339:580. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Dharmasiri U, Balamurugan S, Adams AA, Okagbare PI, Obubuafo A, Soper SA. Electrophoresis. 2009;30:3289. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Herr JK, Smith JE, Medley CD, Shangguan DH, Tan WH. Anal. Chem. 2006;78:2918. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]; c) Sheng WA, Chen T, Tan WH, Fan ZH. ACS Nano. 2013;7:7067. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zamay GS, Kolovskaya OS, Zamay TN, Glazyrin YE, Krat AV, Zubkova O, Spivak E, Wehbe M, Gargaun A, Muharemagic D, Komarova M, Grigorieva V, Savchenko A, Modestov AA, Berezovski MV, Zamay AS. Mol. Ther. 2015;23:1486. doi: 10.1038/mt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Song Y, Zhu Z, An Y, Zhang W, Zhang H, Liu D, Yu C, Duan W, Yang C. Anal. Chem. 2013;85:4141. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]; b) Wan YA, Kim YT, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Cancer Res. 2010;70:9371. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- [14].Jayasena SD. Clin. Chem. 1999;45:1628. [PubMed] [Google Scholar]
- [15].a) Tuerk C, Gold L. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; b) Ellington AD, Szostak JW. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; c) Zeng ZH, Tung CH, Zu YL. Mol. Ther.—Nucleic Acids. 2014;3:e184. doi: 10.1038/mtna.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zamay GS, Kolovskaya OS, Zamay TN, Glazyrin YE, Krat AV, Zubkova O, Spivak E, Wehbe M, Gargaun A, Muharemagic D, Komarova M, Grigorieva V, Savchenko A, Modestov AA, Berezovski MV, Zamay AS. Mol. Ther. 2015;23:1486. doi: 10.1038/mt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Powell AA, Talasaz AH, Zhang HY, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. Plos One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, OuYang W-H, Xu X, Zhang Z, Song M, Lee T, Garcia MA, Xiong B, Hou S, Tseng H-R, Fang X. Adv. Mater. 2013;25:2368. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Xu L, Zhang Z, Zhao Z, Liu Q, Tan W, Fang X. Am. J. Biomed. Sci. 2013;5:47. [Google Scholar]; b) Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Analyst. 2009;134:1808. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- [19].a) Wang ST, Wang H, Jiao J, Chen KJ, Owens GE, Kamei KI, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew. Chem. Int. Ed. 2009;48:8970. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mitchell MJ, Castellanos CA, King MR. J. Biomed. Mater. Res. A. 2015;103:3407. doi: 10.1002/jbm.a.35445. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hughes AD, King MR. Langmuir. 2010;26:12155. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.