Abstract
Developing safe and efficient non-viral gene delivery systems remains a major challenge. We present a new cationic poly(2-oxazoline) (CPOx) block copolymer for gene therapy that was synthesized by sequential polymerization of non-ionic 2-methyl-2-oxazoline and a new 2-oxazoline monomer, 2-(N-methyl, N-Boc-amino)-methyl-2-oxazoline, followed by deprotection of the pendant secondary amine groups. Upon mixing with plasmid DNA (pDNA), CPOx forms small (diameter ≈ 80 nm) and narrowly dispersed polyplexes (PDI < 0.2), which are stable upon dilution in saline and against thermal challenge. These polyplexes exhibited low plasma protein binding and very low cytotoxicity in vitro compared to the polyplexes of pDNA and poly(ethylene glycol)-b-poly(l-lysine) (PEG-b-PLL). CPOx/pDNA polyplexes at N/P = 5 bound considerably less plasma protein compared to polyplexes of PEG-b-PLL at the same N/P ratio. This is a unique aspect of the developed polyplexes emphasizing their potential for systemic delivery in vivo. The transfection efficiency of the polyplexes in B16 murine melanoma cells was low after 4 h but increased significantly for 10 h exposure time, indicative of slow internalization of polyplexes. Addition of Pluronic P85 boosted the transfection using CPOx/pDNA polyplexes considerably. The low protein binding of CPOx/pDNA polyplexes is particularly interesting for the future development of targeted gene delivery.
Keywords: gene delivery, click chemistry, biocompatibility, non-fouling, macrophage transfection
Introduction
Gene therapy is a promising strategy to treat devastating genetic disorders such as Leber's congenital amaurosis,[1, 2] X-linked adrenoleukodystrophy (ALD),[3] β-thalassemia,[4] severe combined immune deficiency (ADA-SCID),[5] hemophilia,[6, 7] as well as acquired diseases such as cancer[8-10] or neurodegenerative disorders.[11] Non-viral gene delivery vectors are intensively investigated as an alternative strategy to the viral vectors that exhibit high transfection efficiency but also often have high cost and serious safety issues.[12, 13] In contrast, non-viral vectors are typically less active but relatively safe, cost-efficient and easy-to-tailor systems, which are flexible to formulation design, amenable to modifications using target ligands, and capable of condensing large plasmid DNA (pDNA).[14-18] One major category of non-viral vectors is polycations – the positively charged macromolecules that are typically exemplified by poly(l-lysine) (PLL),[19, 20] polyethylenimine (PEI)[21-26] or chitosan.[26-30] Polycations condense pDNA via electrostatic interactions to form nano-sized polyelectrolyte complexes called polyplexes, which have been extensively studied as gene delivery vehicles.[31] Unfortunately, most polycations and polyplexes display high cytotoxicity.[19, 32-35] Grafting of poly(ethylene glycol) (PEG) to polycations is known to increase polyplexes stability. Thus, excess of polycations is avoided and toxicity may be reduced.[24, 36-41]
Although the use of PEG in drug and gene delivery systems has been well established to be relatively safe, this polymer also raises a few concerns. As a polyether, it is prone to oxidative degradation.[42-44] After administration, low molar mass PEG is predominantly cleared via kidney and considered safe,[45] yet several studies found evidence of persisting PEG in vivo.[46-48] Moreover, approximately 25% of patients have pre-existing anti-PEG antibodies, and do not respond to treatments employing PEGylated therapeutics because of accelerated blood clearance.[49, 50]
To overcome the limitations of PEG, poly(2-oxazolines) (POx) have been suggested as alternatives to PEG due to their highly tunable structure, versatile properties and favorable biological safety profiles.[51-53] POx are accessible via living cationic ring opening polymerization (LCROP) of 2-oxazolines, a robust and manageable synthetic approach, which allows incorporation of various functionalities in the polymer using functional initiation and termination reagents, or 2-substituted monomers.[54] LCROP can result in excellently control over POx length and dispersity (Mw/Mn = Đ < 1.2). The pendant amide moieties of POx are highly hydrated while the alkyl side chains can be fine-tuned by adjusting chain length to achieve amphiphilicity of each monomer unit or an overall hydrophobic character.[55] Both, poly(2-methyl-2-oxazoline) (PMeOx) and poly(2-ethyl-2-oxazoline) (PEtOx) exhibit “stealth” properties and long blood circulation times comparable or even superior to PEG when grafted onto liposomes[56, 57] or surfaces.[58, 59] Viegas et al. have reported that a conjugated of PEtOx and bovine serum albumin was less immunogenic as compared to the PEGylated protein in rats.[60] In addition, POx were investigated as biomaterials in form of surface-bound and soluble polymer brushes,[61-64] bactericidal materials,[65] polymeric micelles,[66-70] polymer drug conjugates,[71] polymer-protein conjugates,[72-74] polymersomes,[75, 76] polyplexes,[77, 78] and fibrous scaffolds[79] to date.
There have been attempts to develop gene delivery systems via partially hydrolyzing PEtOx into linear PEI-POx diblock polymers.[77, 80] One drawback of this approach is its complexity. First, the polymer has to be hydrolyzed, and second, coupled to an unhydrolyzed chain to obtain the desired block copolymers. Moreover, this approach does not allow introduction of other functional side chains. However, the direct synthesis of cationic POx by LCROP is also not straightforward, as the growing polymer chains are very sensitive to nucleophiles, which can terminate the propagating species.[51] Some kind of post-polymerization modification is therefore unavoidable if one desires well-defined polymers. Cationic 2-oxazoline monomers bearing tert-butyloxycarbonyl (Boc) protected primary amines were previously reported by one of us.[81] Polymerization of this monomer followed by deprotection of Boc groups generated POx with pendant cationic groups.[81] Hydrogel scaffolds produced with a very similar synthetic approach, were applied in bioanalytical systems such as gene chips to capture or enrich DNAs.[82] More recently, Rinkenauer et al. have explored a number of cationic 2-oxazoline polymers by polymer-analogue modification of POx side chains with primary or tertiary amines.[83] The researchers found that long hydrophobic side chain induced high cytotoxicity of the resulting polymer and primary amines with amine content of at least 40 mol% were required for efficient transfection. POx featuring pendant secondary amines were not reported previously.
Here, we describe a novel cationic POx (CPOx) copolymer comprising a non-ionic hydrophilic PMeOx block and a cationic poly[2-(N-methyl)aminomethyl-2-oxazoline] (PMAMeOx) block. We hypothesized that pendant secondary amine groups tethered by a short alkyl spacer, methylene, will reduce hydrophobicity and thus, toxicity while still retaining the ability to form polyplexes. A precursor polymer was synthesized by LCROP of 2-methyl-2-oxazoline (MeOx) and the novel 2-(N-methyl, N-Boc-amino)-methyl-2-oxazoline (Boc-MAMeOx) monomer. After deprotection, a polycationic polymer was obtained. We explored the potential of this block copolymer for gene delivery for macrophage transfection with potential applications in tumor immunotherapy.
2. Experimental Section
2.1. Materials and Methods
The monomer MeOx and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (Steinheim, Germany and St. Louis, US, respectively). Isobutylchloroformiate was purchased from Alfa Aesar (Karlsruhe, Germany), N-Boc-sarcosine from Bachem (Bubendorf, Switzerland). Dry solvents were stored under argon and over molecular sieve. PEG105-PLL51 was obtained from Alamanda Polymers (Huntsville, AL) with Mn = 13 kg/mol and dispersity Đ = 1.09. Pluronic P85 was kindly provided by BASF Corporation (North Mount Olive, NJ). Pre-cast Tris-HCl gels and Precision Plus Protein™ All Blue Standards were from Bio-Rad (Hercules, CA). SYPRO® Ruby protein gel stain and cell culture reagents including Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin (P/S) were purchased from Invitrogen (Carlsbad, CA). NAP desalting columns and LH-20 were from GE Healthcare (Piscataway, NJ). Luciferase (Luc) assay kit was from Promega (Madison, WI). gWIZ-Luc pDNA was from Genlantis (CA, USA). Carboxyrhodamine-Azide (AZ105) was purchased from Click Chemistry Tools (Scottsdale, AZ 85260). All other reagents and supplies were from Fisher Scientific (Pittsburgh, PA). Reagents and solvents were used as received, unless noted otherwise. The monomers and solvents used for the polymerization of POx were purified by distillation under reduced pressure and stored under dry argon or nitrogen. RAW264.7 murine macrophage and B16 murine melanoma cell lines were from American Type Culture Collection (ATCC, Manassas, VA) and cultured according to supplier's protocol.
NMR spectra were recorded on an Inova 400 (1H: 400 MHz, Varian) at 295 K. The critical micelle concentration (CMC) was determined by the Wilhelmy plate method using a Sigma 703D Tensiometer. The molar mass (Mn) and dispersity Đ (Mw/Mn) were measured by gel permeation chromatography (GPC) using a Polymer Laboratories GPC-120 (column setup: 1 × PSS GRAM analytical 1000 and 1 × PSS GRAM analytical 100 obtained from Polymer Standards Services, Mainz, Germany) using N,N-dimethyl acetamide (DMAc) (5 mmol/L LiBr, 1 wt% H2O, 70 °C, 1 mL/min) as eluent and poly(methyl methacrylate) standards.
2.2. Polymer synthesis
2.2.1. Synthesis of Boc-MAMeOx (3)
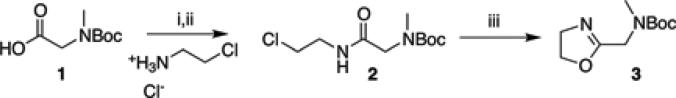
Boc-MAMeOx (3) monomer was synthesized by ring-closing reaction following the general procedure of Levy and Litt and a newer version of Weberskirch et al. (Scheme 1).[84, 85] Using a triple-necked flask with a thermometer and dropping funnel, 21.06 g (111.7 mmol) N-Boc-sarcosine (1) were dissolved in 350 mL tetrahydrofuran (THF) and 15.45 mL (111.7 mmol) triethylamine were added, and the mixture was cooled to 3 °C. Isobutylchloroformiate (14.5 mL, 111.7 mmol) was added dropwise under vigorous stirring. Subsequently, 12.95 g (111.7 mmol) chloroethylamine hydrochloride was dissolved in 50 mL dimethylformamide and added dropwise followed by 15.45 mL triethylamine. The yellowish solution was stirred at room temperature (RT) for 1 h. The reaction mixture was filtered and concentrated in vacuo. After dilution with dichloromethane, the solution was extracted thrice each with 10% aqueous soda and brine. The combined aqueous phases were extracted with dichloromethane. From the combined organic phases, all volatiles were removed, the residue (2) dissolved in 150 mL methanol and 22.505 g (162.8 mmol) of K2CO3 were added to the solution of 2, which was not isolated. The mixture was stirred under inert atmosphere overnight at RT and then refluxed for 5 h. Volatiles were removed at reduced pressure and the residue was dried at 0.002 mbar. From the yellowish solid the product (3) was obtained as a colorless liquid by fractionated distillation in vacuo (13.683 g, yield: 28.8%, bp (3.6 × 10−3 mbar) = 76-83 °C).
Scheme 1.
Synthesis of the monomer, Boc-MAMeOx (3). i) Triethylamine, isobutylchloroformate, 3 °C, THF, ii) Triethylamine, 2-chloroethylammonium chloride, dimethylformamide, iii) MeOH, K2CO3, reflux.
1H-NMR (d3-MeOD, 300 MHz, 295 K): 4.26 (t, 2H, N-CH2-CH2-O), 3.95 (s, 2H, C-CH2-N(Me)(Boc)), 3.74 (t, 2H, N-CH2-CH2-O), 2.82 (s, 3H, N-CH3), 1.35 ppm (d, 9H, O-C(CH3)3).
13C-NMR (d3-MeOD, 75 MHz, 295 K): 166.2/4 (C-CH2-N(Me)(Boc)), 156.1/155.7 (N-C(O)-O-C(CH3)3), 80.1 (N-C(O)-O-C(CH3)3), 68.0 (N-CH2-CH2-O), 53.2 (N-CH2-CH2-O), 45.7/44.9 (C-CH2-N(Me)(Boc)), 34.1 (N-CH3), 27.2 ppm (N-C(O)-O-C(CH3)3).
2.2.2. Synthesis of Propargyl-P(MeOx50-b-MAMeOx16), CPOx
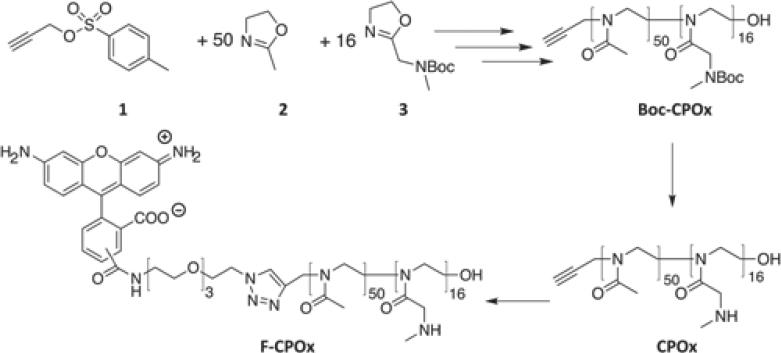
The polymer was synthesized following published methods (Scheme 2).[51, 68, 69] Briefly, under dry and inert condition 63.1 mg (0.3 mmol, 1 eq) of propargyl toluenesulfonate (tosylate) and 1276.5 mg (15 mmol, 50 eq) of MeOx were dissolved in 3 mL dry acetonitrile at RT. The mixture was heated (microwave-assisted synthesizer, 150W maximum, 130 °C) for 5 min. After cooling to RT, the cationic monomer for the second block, Boc-MAMeOx (1011.4 mg, 4.7 mmol, 16 eq) was added and the mixture was reacted at 70°C overnight. Finally the polymer was terminated by an excess of 5% aqueous K2CO3. The solvent was then removed and the residue was re-dissolved in 5 mL methanol/chloroform (3/1, v/v). After precipitation from cold diethylether, the product was isolated by centrifugation. The product was reprecipitated twice and the polymer (Boc-CPOx) was obtained as colorless powder after lyophilization (1.75 g, 68% yield). Removal of the Boc group was performed in trifluoroacetic acid (TFA):H2O:triisobutylsilane (TIBS) (95:2.5:2.5) and the mixture was dried and redissolved in water followed by dialysis against DI water for two days. Deprotected product CPOx (Scheme 2) was obtained by lyophilization of an aqueous solution. The yield of CPOx was 91.5% after deprotection of Boc-CPOx.
Scheme 2.
Block copolymerization of MeOx (2) and Boc-MAMeOx (3) using propargyl tosylate (1) as the initiator and 5% K2CO3 as the terminating agent. The deprotection of the pendant amino group of Boc-CPOx was performed with TFA to obtain CPOx. Fluorescently-labeled polymer (F-CPOx) was produced by click reaction of carboxyrhodamine 110-Azide with CPOx.
1H-NMR (CDCl3, 400 MHz, 298K): 4.1-3.9 (br, 37H, -N-CO-CH2-N-Boc, a), 3.47 (br, 249H, N-CH2-CH2, b), 2.87 (br, 63H, N-CH3, c), 2.05 (m, 147, -N-CO-CH3, d), 1.4 ppm (s, 180, N-C(O)-O-C(CH3)3, e).
2.2.3. Synthesis of carboxyrhodamine 110 fluorescence labeled polymers (F-CPOx)
Carboxyrhodamine110-Azide (4.15 mg, 3 eq) (Click Chemistry Tools) and CPOx (20 mg, 1 eq) were dissolved in 1 mL of water and methanol (1/1, v/v). Then 60 μg (0.1 eq) of copper (II) sulfate pentahydrate were added, supplemented with 208.6 μg (0.2 eq) Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA, 1 g/L stock solution in water/methanol) in water/methanol solution as the stabilizing ligand of copper. Oxygen was removed from the above mixture with a stream of argon gas and supplemented with 200 μL water/methanol solution containing sodium ascorbate (475.4 μg, from 2 g/L stock solution, 1 eq) drop-wise. The reaction was allowed to proceed for 24 h at RT, and reaction mixture was loaded onto LH-20 column to remove unreacted small molecules followed by addition of 10-fold molar excess of EDTA disodium salt and stirred at RT for 1 h. Finally, the reaction mixture was purified with NAP-10 desalting column, dialyzed against DI water for 2 days with MW-cutoff 3500 Da, (GE Healthcare, Piscataway, NJ) and lyophilized to give Carboxyrhodamine 110-labeled polymers F-CP (Scheme 2).[86] This was was repeated twice to completely remove free fluorescence dye. The fluorescence labeling efficiency was about 72% as determined from the calibration with free fluorescence dye.
2.3. Acid-Base Titration
The buffering capacity of the synthesized block copolymer was determined by acid-base titration.[87] Briefly, 2 mg CPOx was dissolved in 10 mL of 0.1 M NaCl to give a final concentration of 0.2 g/L, the pH of the polymer solution was adjusted to 10 with 1 N NaOH, and the solution was subsequently titrated with 0.1 M HCl. 0.1 M NaCl or 0.2 g/L PEG-PLL were also titrated as control and reference, respectively.
2.4 Formation and characterization of CP/pDNA polyplexes
For preparation of polyplexes, stock solutions of pDNA (1 g/L) and CPOx polymer (1 g/L) in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.4) were mixed at various N/P charge ratios (1 g CPOx polymer = (1/6130) mol giving the CPOx molar mass of 6130, and each mol of CPOx contains 16 mol secondary amine groups, therefore 1 g CPOx polymer = (1/6130) mol * 16 ≈ 2.6 mmol. The number of bases in 1 g of pDNA is 1.71 × 1021 ≈ 1.71× 1021 negative charges. Therefore, 1 g of pDNA = (1.71× 1021)/(6.022×1023) mol ≈ 3 mmol)[88] and vortexed immediately for 30 sec, then incubated for 30 min at RT before further use. The final concentration of pDNA was set to 20 mg/L. Hydrodynamic diameter (z-average hydrodynamic diameter), size distribution, polydispersity index (PDI) and ζ-potential of polyplexes were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano (Malvern Instruments Ltd., Malvern, UK). Results represent the average from three independently prepared polyplexes.
2.5. Ethidium bromide exclusion assay, agarose gel electrophoresis, and stability of polyplexes during storage, dilution and heating
The ability of CPOx and PEG-PLL to condense pDNA was assessed by a standard ethidium bromide (EtBr) exclusion assay via measuring the changes in EtBr/pDNA fluorescence.[89] pDNA (gWIZ-Luc) solutions in 10 mM sodium acetate buffer (pH 5.2) and 10 mM HEPES, pH 7.4 at concentration of 20 mg/L were mixed with EtBr (1 mg/L) and fluorescence measured using 545 nm excitation and 595 nm emission and set to 100 %. Background fluorescence was set to 0 % using EtBr (1 mg/L) solution alone. Fluorescence readings were taken following a stepwise addition of polymer solution.
For agarose gel electrophoresis, 20 μL of each polyplex solution was mixed with 4 μL 6x DNA gel loading dye and loaded on 1 % agarose gel containing EtBr (0.2 mg/L). The samples were electrophoresed in 1x Tris-Acetate-EDTA (TAE) running buffer for 1 h at 100 V, and pDNA bands were visualized under UV light (FluorChem E, ProteinSimple, Santa Clara, CA).
In order to analyze the stability upon storage at 4 °C, size and PDI of polyplexes were measured up to 7 days using DLS. Size and PDI of polyplexes were also monitored upon heating, where the temperature was increased step-wise from 25 °C to 70 °C. To ensure proper equilibration, the polyplexes were maintained for 5 min at the desired temperature before the measurement was started.[90]
2.6. Transmission electron microscopy (TEM)
The morphology of polyplexes was studied using a LEO EM910 TEM operating at 80 kV (Carl Zeiss SMT Inc., Peabody, MA) and digital images acquired using a Gatan Orius SC1000 CCD Digital Camera with Digital Micrograph 3.11.0 (Gatan Inc., Pleasanton, CA). Briefly, a drop of the polyplex solution was deposited on a copper grid/carbon film for 5 min after which the excess solution was wicked off with filter paper, and a drop of staining solution (1% uranyl acetate) was allowed to contact the sample for 10 seconds prior to the TEM imaging.
2.7. Plasma protein binding assay
To obtain rat plasma, blood was taken directly from ventricular dexter of male sprague-Dailey rats (5 weeks old, 270-300 g) into 15 mL tube containing anticoagulant EDTA 50 mg/mL. After centrifugation (2000 g, 5 min), plasma was separated from blood cells and was placed on ice for later protein binding experimental use. Pooled plasma (150 μL) was incubated with 50 μL PEG-PLL/pDNA, PLL/pDNA, and CPOx/pDNA polyplexes ([pDNA] = 20 mg/L, N/P ratios as indicated) for 30 min at 37 °C. Mixtures were centrifuged at 16,000 g for 40 min to precipitate polyplex-bound plasma proteins from unbound plasma proteins. Unbound components were removed by 3 cycles of washing/centrifugation in 10 mM HEPES buffer (pH 7.4). The plasma protein-bound polyplexes pellets were then washed with cold acetone to remove any lipids that were co-precipitated. Plasma proteins bound to polyplexes were ultimately desorbed from the pellets using 50 μL 2% SDS (in 0.5 M Tris-HCl containing 10% glycerol, pH 6.8) at 100 °C for 5 min. For comparison, the rat plasma proteins were collected from pooled rat plasma by direct precipitation with cold acetone. The amount of protein bound to polyplexes was determined using MicroBCA Assay™. Equal volumes of samples (8 μL each) were loaded on a SDS-PAGE gel, electrophoresed and stained using SYPRO Ruby. Selected protein bands were assigned using mass spectrometry (AB Sciex 4800 Plus MALDi-ToF/ToF).
2.8. Cell culture and in vitro cell viability assay
RAW264.7 murine macrophages and murine melanoma B16 cell lines were cultured in DMEM supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin and 100 mg/L streptomycin (complete medium). For the cytotoxicity assay, cells were seeded at a density of 2 × 104 cells per well in 96-well plates and allowed to adhere for 24 h prior to the addition of the polymer at various concentrations ranging from 1 mg/L up to 1 g/L in full media. PEG-PLL was used as a positive control. Cells were incubated with polymer solutions for 48 h and cell viability was determined by MTT assay (100 μg/well, 3h). Data represents the average of six separate wells for each polymer concentration.[68] RAW264.7 murine macrophages were incubated with CPOx/pDNA or PEG-PLL/pDNA polyplexes prepared at N/P = 5 or 10 under the same conditions as in the transfection experiments (polyplexes containing 0.1 μg pDNA in 100 μL medium per well of 96-well plate) for 4 h or 10 h, and then incubated with complete medium for another 24 h before measuring the cell viability using MTT assay. Data represent average ± S.D. (n = 6).
2.9. Cellular internalization of polyplexes and in vitro transfection
Cellular internalization of polyplexes was studied using confocal microscopy. Briefly, cells were seeded onto sterile Lab-Tek II chambered coverglass. After overnight attachment, the cells were treated with polyplexes [F-CP (115 mg/L)/ TOTO-3 labeled pDNA (20 mg/L), N/P = 5] for various time points from 2, 4, 6, 8, 10, 12 to 24 h. pDNA labeling with TOTO-3 was performed according to manufacturer's protocol (Life Technologies, Grand Island, NY). At the end of each time point, cells were washed thrice with 1xPBS and fixed with 4% paraformaldehyde for 10 min followed by imaging using confocal microscopy (LSM 510 laser scaning microscope, Zeiss, Thornwood, NY, USA) with argon ion laser and corresponding filter set. Digital images were obtained using a CCD camera (Photometrics). All imaging conditions, including laser power, photomultiplier tube, and offset settings, remained constant for each comparison set. For in vitro transfection study, the RAW264.7 macrophage or B16 melanoma cells were split one day prior to transfection and plated in 48-well plates at cell density of 3×104 per well for RAW264.7 cells and 1×104 for B16 cells. Before transfection, the cell culture medium was replaced with fresh serum-free DMEM or complete DMEM media. Transfection solutions were prepared in 1.5 mL Eppendorf tube by mixing (i) 4 μg gWIZ-Luc plasmid DNA, (ii) 150 μL HEPES, and (iii) CP at predetermined volume to give desired N/P ratio. The cells in each well were transfected with polyplexes containing 0.5 μg of plasmid DNA in 200 μL volume at 37°C for 4 h or 10 h (n=3). Then the complexes were removed and cells were incubated for additional 24 h in fresh complete DMEM medium or with Pluronic P85 excipient (in complete media) for extra 3 h before changing to complete medium. Cells were washed with 1xPBS and then lysed with 120 μL 1x cell culture lysis buffer (Promega). The luciferase activity in each sample was measured using a Promega Luminometer after adding luciferin substrate.
2.10. Statistical Analysis
Unless otherwise specified, all statistical comparisons were made using Student's t test. A result with p < 0.05 was considered statistically significant.
3. Results
3.1. Synthesis and characterization of Boc-MAMeOx (3)
A new monomer was designed to yield polymers containing pendant secondary amine groups after polymerization and deprotection. The synthesis was performed by a two-step reaction from readily available reagents following a standard route for 2-oxazoline synthesis (Scheme 1). First, Boc-Sarcosine (1) was coupled with 2-chloroethylammonium chloride in THF to yield 2-chloroethyl(N-Boc)sarcosinamide (2). Ring closure under basic conditions yields the monomer Boc-MAMeOx (3). The required purity for polymerization was obtained after fractionated distillation under reduced pressure.
The chemical structure of purified 3 as outlined in Scheme 1 was verified via 1H-NMR spectroscopy (Figure 1).
Figure 1.
1H-NMR spectrum (300 MHz, d3-MeOD, 295 K) of the protected monomer 3 (Boc-MAMeOx) along with the assignment of all observed signals.
3.2. Polymer synthesis and characterization
The living polymerization was initiated using propargyl tosylate in order to incorporate a functional alkyne group at the hydrophilic non-ionic PMeOx block end for consecutive “click” chemistry reactions (Scheme 2). In the current work, this was used to introduce an azide-bearing fluorescent dye with good efficiency (72 %). The molar mass of the product, Boc-CPOx, as determined using GPC (Mn,GPC) was close to the desired value (Mcalc) and the dispersity was reasonably low. Since the Boc-protected PMAMeOx block is hydrophobic, the copolymer is amphiphilic and self-assemble in aqueous media into micelles with a CMC value around 19 mg/L (2.1 μM) at 20°C as determined using tensiometry (Wilhelmy plate). Boc-CP exhibited a characteristic signal of Boc-protection group (tert.-butyl protons) at 1.4 ppm (Figure 2A). After deprotection under standard acidic condition, the signal intensity assigned to these protons was strongly reduced (CPOx, Figure 2B). However, it is noteworthy that complete removal of the Boc-protection groups was apparently not achieved using a standard protocol. Approx. 10 % of the protection groups remained on the polymer. Even after a second deprotection attempt, approx. 5% of the Boc groups remained visible in the NMR spectrum (CPOx′, Figure 2C). For further experiments, we hypothesized that the remaining hydrophobic groups may actually stabilize the complexes. Thus, we did not conduct further attempts for quantitative removal of the Boc groups.
Figure 2.
1H-NMR spectra (400 MHz, d3-MeOD, 295 K) of A) Boc-CPOx and the deprotected polymer B) after single (CPOx) and C) repeated deprotection procedures (CPOx′), along with the respective structures and the peak assignments. The signal assignable to protons of the Boc-protection group was greatly attenuated but remained visible in the CPOx spectrum (estimated 10% Boc residual). This peak nearly disappeared (estimated <5% Boc residual in C) after repeated deprotection procedure to further remove residual Boc groups (CPOx′).
3.3. Formation and physicochemical characterization of CPOx/pDNA polyplexes
This study investigated the potential use of a POx block copolymer with pendant hydrophilic cationic moieties for non-viral gene delivery. In contrast to a recently published polymer analogue addition approach by Schubert and co-workers,[83] we introduced the positive charges via a monomer-deprotection route. The buffering capacity of the pendant amino groups at low pH may be important for transfection efficiency of polyplexes since buffering effects are believed to facilitate endosomal escape of polyplexes and thereby protect DNA from nuclease degradation in the endosomal compartment (proton sponge effect).[91-93] Therefore, we first measured the buffering capacity of the block copolymer by acid-base titration in 0.1 M NaCl aqueous solution (Figure 3 A).[87] At the same mass concentration, both CPOx and PEG-PLL showed similar titration curves, however, CPOx appears to buffer slightly better between pH 7.4 and 5.2, relevant for endosomal escape (Figure 3 B).
Figure 3.
A). Titration curves for CPOx copolymer and PEG-PLL reference in 0.1M aqueous NaCl (pH 10, adjusted with NaOH) using 0.1M HCl. As a control, the titration curve of NaCl is also presented. The concentration of polymers was 0.2 g/L. B). Enlarged inset of rectangular region in A), pH range from 5.2 to 7.4.
The ability of CPOx to condense pDNA and form polyplexes was assessed by EtBr exclusion (Figure 4A) and gel retardation (Figure 4B) assays. CPOx was mixed with gWIZ-Luc pDNA at various N/P ratios. Condensation curves for polyplexes at pH 5.2 or pH 7.4 both showed typical transition between N/P ratios of 1 and 2.
Figure 4.
A) pDNA condensation by CPOx assayed by EtBr exclusion. Change in fluorescence intensity after addition of CPOx to pDNA (20 mg/L)/EtBr (1 mg/L) solution at various N/P ratios in 10 mM HEPES (■, pH 7.4) or 10 mM sodium acetate buffer (□, pH 5.2). A relative fluorescence of 100 RFU represents the fluorescence of pDNA/EtBr solution in the absence of CPOx while 0 RFU indicates background fluorescence of EtBr not intercalated into DNA. B) Gel retardation assay of pDNA and polyplexes (prepared in 10 mM HEPES at pH 7.4) at various N/P ratios of 1/3, 2, 5, 10 and 20, loaded on 1% agarose gel, stained with EtBr, and visualized under UV light.
Gel retardation assay suggested that CPOx is able to condense DNAs and produce polyplexes. When N/P ratio exceeded 2, CPOx/pDNA polyplexes were unable to migrate on the gel and remained immobilized in the vicinity of the gel loading sites.
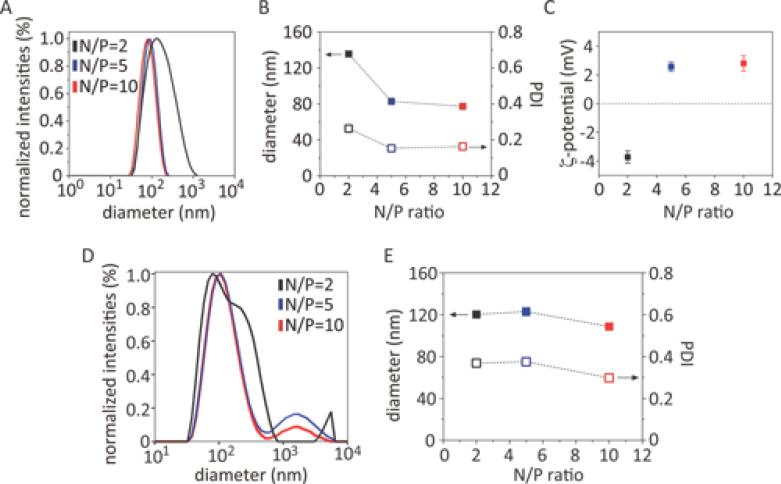
Further investigations of the polyplexes by dynamic light scattering (DLS) (Figure 5A) revealed that CPOx indeed effectively condensed DNA into nanoparticles. Polyplexes at N/P = 2 exhibited a relatively large particle size about d = 132 nm and a high polydispersity (PDI ≈ 0.3). Interestingly, the measured ζ-potential was essentially neutral (Figure 5B). The formation of uniform and smaller polyplex nanoparticles could be realized by an increase of the N/P ratios to N/P = 5 or N/P = 10 (d = 83 and 78 nm, respectively, with PDIs ≈ 0.17). Again, these particles display an essentially neutral surface charge (Figure 5B and C). The DLS results were in good agreement with our observations in agarose gel electrophoresis.
Figure 5.
DLS measurements of the A, D) size distributions; B, E) particle size and PDI; and C) ζ-potential of polyplexes as a function of N/P ratio of 2 (black), 5 (blue), and 10 (red). Polyplexes were prepared using the CPOx (A-C) or CPOx′ (D, E), respectively.
Following standard deprotection procedure, we were unable to completely remove Boc protection group and about 10% of Boc groups remained on the polymer. However, this particular polymer formed small and defined (relatively uniform) complexes with pDNA. In contrast, the polymer produced after repeated deprotection that still contained a trace amount of Boc groups, <5%, formed less defined complexes with pDNA (Figure 5 D and E). This is very interesting and we hypothesize that CPOx containing some Boc groups bearing hydrophobic properties may promote the core formation of CPOx/pDNA complexes. Examples for such hydrophobic stabilization of polyplexes can be indeed found in the literature.[94-97]
We also investigated morphology of the polyplexes via TEM (Figure 6). A representative TEM image of the polyplexes at N/P ratio 10 indicated essentially spherical particle morphology and we estimated a diameter of 50 nm for these particles. It should be noted that the TEM sizes are usually smaller than the hydrodynamic diameters obtained by DLS.
Figure 6.
Representative TEM image of CPOx/pDNA polyplex at N/P ratio of 10 indicated a spherical morphology. Ten μL of polyplex solution (10 μg/mL pDNA) were deposited onto copper grid with carbon film for 5 min and then negatively stained with 1% uranyl acetate. (Scale bar, 100 nm).
3.4. Stability of CPOx/pDNA polyplexes
The stability and shelf-life of polycation/DNA polyplexes is a critical factor to facilitate the translation into in vivo applications. We assessed the stability of CPOx/pDNA polyplexes upon storage at 4 °C for up to one week in 10 mM HEPES buffer (pH 7.4) and observed no considerable changes of the particle size or PDI for polyplexes formed at N/P = 2 (d ≈ 130 nm, PDI ≈ 0.25) and N/P = 5 (d ≈ 83 nm and PDI ≈ 0.16) (Figure 7). The CPOx/pDNA at N/P = 10 showed good stability for up to 5 days but at day 7 the average particle diameter increased from 78 nm to almost 100 nm and their PDI from 0.17 to about 0.3 (Figure 7).
Figure 7.
A) Diameter and B) PDI of CPOx/pDNA polyplexes at various N/P = 2, 5 and 10 as a function of storage time. Each data point represents the average (± S.D.) of independently prepared samples (n = 3)
CPOx/pDNA polyplexes formed at N/P = 1 disintegrated upon 10-fold dilution with 10 mM HEPES buffer (pH 7.4), however, at N/P = 5 polyplexes were stable and no change in size (d = 82 nm) and PDI (0.16) was detected upon 10-fold dilution (Figure 8A).
Figure 8.
Stability of CPOx/pDNA polyplexes upon dilution and heating. A) Average particle diameter and PDI of polyplexes formed at N/P = 5 (I), upon 10-fold dilutions with 10 mM HEPES buffer (pH 7.4) (II), or 150 mM NaCl (III) and of polyplexes (N/P = 5) produced directly in 150 mM NaCl (IV). B) Stability of polypexes in 10 mM HEPES buffer (pH 7.4) at N/P = 5 (■) and N/P = 10 (□) upon heating from 25 °C to 70 °C.
In contrast, when the CPOx/pDNA polyplex at N/P = 5 was diluted 10-fold in isotonic saline solution (0.9% NaCl), both the size and PDI increased almost two fold to about 150 nm and 0.4, respectively. Despite the significant increase, this result is a first indication that such polyplexes may be applicable for in vivo studies as the nanoparticles are stable in isotonic solutions. However, it would be beneficial to obtain polyplexes of significantly lower dispersity. In fact, when we prepared polyplexes at N/P = 5 directly in 0.9% NaCl instead of 10 mM HEPES buffer, CPOx/pDNA polyplexes with an average particle diameter of d = 135 ± 1 nm and narrow PDI of 0.275 ± 0.008 (n = 3) were obtained (Figure 8A). The thermal stability of CPOx/pDNA polyplexes prepared at N/P = 5 and 10 was also investigated. The development of nanoparticle size and PDI were determined as a function of the temperature from 25 °C up to 70 °C (Figure 8B). Interestingly, the average hydrodynamic particle diameter exhibited only a small but steady increase while the increase and strong variation of the PDI at 70 °C may indicate an onset of particle aggregation for both N/P = 5 and 10.
3.5 Polyplex plasma protein binding
Plasma proteins bound to nanoparticles are directly linked to the uptake and further clearance of these nanoparticles by the mononuclear phagocyte system (MPS) in vivo.[35, 98-100] Hydrophilic POx, i.e. PMeOx and PEtOx homopolymers, have been long known to exhibit anti-fouling or so called “stealth” properties, similar to or superior to PEG.[56, 59, 101, 102] This prompted us to evaluate the plasma protein binding of CPOx/pDNA polyplexes and compare it with PEGylated polyplexes. The polyplexes formed at different N/P ratios (5, 10, and 20) were exposed to pooled rat plasma for 30 min at 37 °C. The protein binding to the polyplexes was quantified using MicroBCA assay. Interestingly, very little plasma protein bound to CPOx/pDNA polyplexes as compared to the PEGylated control polyplexes PEG-PLL/pDNA or PLL/pDNA (Figure 9A). Analysis of the bound proteins by SDS-PAGE (Figure 9B) confirmed the low protein binding to CPOx/pDNA prepared at any N/P ratios. Analysis by mass spectrometry identified a few of the major proteins bound to PLL/pDNA polyplexes, which included IgG light and heavy chains, C3, C4, and C5 proteins of the complement system, and β and γ chain of fibrinogen isoforms (Figure 9B, small red box from bottom to top respectively). The low protein binding of the present polyplexes is particularly promising for targeted gene delivery.
Figure 9.
A) Total protein adsorption in isolated polyplexes as determined by MicroBCA assay. ** Comparison with PEG-PLL/pDNA polyplexes, p < 0.05. B) SDS-PAGE of adsorbed plasma proteins on PEG-PLL/pDNA, PLL/pDNA, and CPOx/pDNA polyplexes at N/P ratio of 5, 10 and 20 (8 μL each sample). Rat plasma proteins were separated on the gel as a reference. Protein bands on the SDS-PAGE gel were visualized by SYPRO Ruby staining.
3.6. Determination of in vitro cytotoxicity of CP polymer
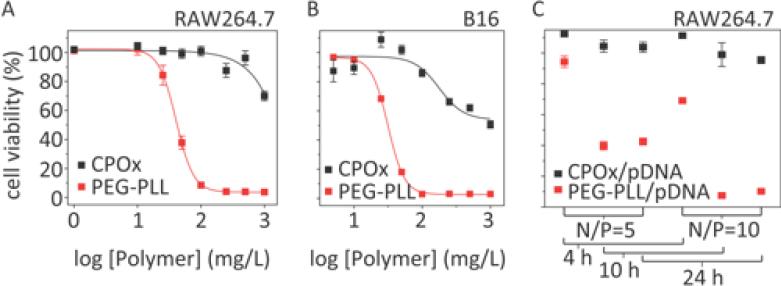
Many polycationic delivery systems have cellular toxicity issues. In order to form stable polycation/pDNA complexes, a large excess of cationic polymer is often required.[14, 33, 38] In turn, this may induce significant cytotoxicity due to the interaction of polycations with the cell membranes. In this respect, we studied the in vitro cytotoxicity of the CPOx in the cell lines we were interested to transfect, RAW264.7 murine macrophage and B16 melanoma cells using PEG-PLL as control. Interestingly, CPOx showed little if any toxicity in RAW264.7 cells at concentrations up to 0.5 g/L (IC50 > 1 g/L) (Figure 10A, black line) while PEG-PLL induced cytotoxicity with an IC50 of 41 mg/L. Similar results were observed in B16 melanoma cells where the IC50 value of CPOx was about 1 g/L while that of PEG-PLL was close to 32 mg/L (Figure 10B). Moreover, the CPOx/pDNA polyplexes were significantly less toxic in RAW264.7 cells than the PEG-PLL/pDNA polyplexes at the same N/P ratios of 5 or 10 following 4 h, 10 h and 24 h exposures, conditions used subsequently for transfection studies (Figure 10C).
Figure 10.
In vitro cytotoxicity of polymers and polyplexes. Cell viability was assessed using MTT assay after 48 h incubation with polymers. Conditions for polyplexes were the same as in the transfection experiments (polyplexes containing 0.1 μg pDNA in 100 μL medium per well of 96-well plate) for 4 h, 10 h and 24 h, and then incubated with complete medium for another 24 h before measuring the cell viability using MTT assay. Data represent average ± S.D. (n = 6).
3.7. Analysis of cellular uptake of CPOx/pDNA polyplex in macrophages
Cellular uptake of CPOx/pDNA polyplex (N/P = 5) was studied in RAW264.7 cells using confocal microscopy. For this experiment, CPOx was fluorescently labeled with carboxyrhodamine 110 using click chemistry to give F-CPOx, while pDNA was visualized using TOTO-3 (Figure 11). Cells were imaged after incubation (N/P = 5) for 0.5, 2, 4, 6, 8, 10 and 24 h, respectively. We found that the uptake process in RAW264.7 cells was rather slow. Up to 4 h exposure, no fluorescence in the cells could be detected. After 6 and 8 h exposures the green fluorescence of F-CPOx and red fluorescence of pDNA was detectable in the cells, while after 10 h exposure the majority of cells were positive for both F-CPOx and pDNA fluorescence. Furthermore, after 24 h incubation both the green and red label fluorescence concentrated in the perinuclear region of the cells (Figure 11). However, significant loss of cell viability was also observed after 24 h of continuous polyplex exposure. Similar results were obtained for the polyplex at N/P 10 (data not shown).
Figure 11.
Cellular uptake of F-CPOx/pDNA polyplex (N/P = 5) in RAW264.7 macrophages studied by confocal fluorescence microscopy. Blue, nuclear staining (Hoechst); Green, F-CPOx; Red, TOTO-3 labeled pDNA. Cells were incubated with polyplex (20 mg/L pDNA) for 0, 0.5, 2, 4, 6, 8, 10 and 24 h, then washed with PBS and fixed in 4 % paraformaldehyde before confocal imaging.
3.8. In vitro transfection
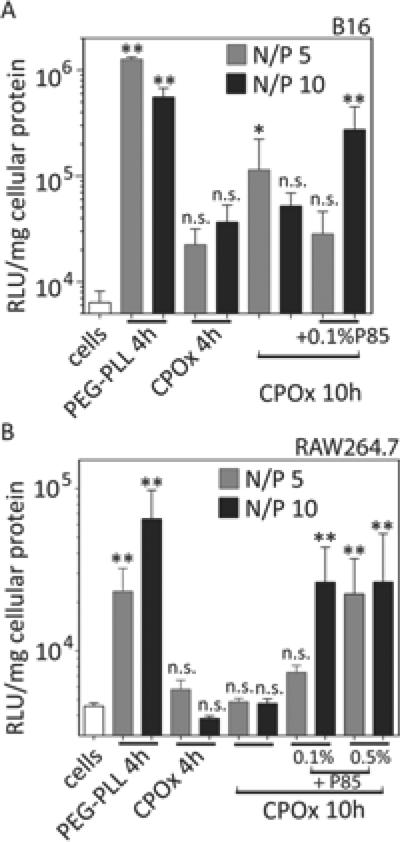
Gene delivery was evaluated with CPOx/DNA polyplexes at N/P = 5 and 10 in B16 murine melanoma cells and RAW264.7 murine macrophages, which are known to be difficult to transfect.[103, 104] Again, the PEG-PLL/pDNA polyplexes were used as a reference in the transfection experiments. All transfections were performed in complete DMEM medium containing 10% FBS for 4 h and 10 h. The 10 h time point was selected given the fact that the CPOx/pDNA polyplexes internalized relatively slow into RAW264.7 but still showed acceptable levels of cytotoxicty (Figure 10 and 11).
The transfection observed in the B16 cells with the control PEG-PLL/pDNA polyplexes was significant although relatively low. The transfection observed after exposure of these cells to CPOx/pDNA polyplexes at N/P = 5 or 10 for 4 h was not significantly different from the baseline (Figure 12A). When the exposure time was increased to 10 h the CPOx/pDNA polyplexes at N/P = 5 produced significant transgene expression in these cells, albeit several fold lower as compared to the control. Since the transfection levels were low for all experiments, we explored the effect of a gene expression adjuvant, an amphiphilic block copolymer, poly(ethylene oxide)-poly(propylene oxide)-poly(ethylene oxide) triblock copolymer (Pluronic P85) that was previously shown to increase transgene expression both in vitro and in vivo.[105, 106] In particular, using different polyplexes this block copolymer was shown to increase cellular uptake of pDNA in vitro, nuclear transport of the pDNA and the transcription of intranuclear pDNA.[105, 107] Indeed, after adding P85 to CPOx/pDNA polyplexes at N/P 10 we observed a significant increase of the cell transfection although the gene expression levels in this treatment group were still less than those observed for PEG-PLL/pDNA.
Figure 12.
In vitro transfection after incubation with PEG-PLL/pDNA and CPOx/pDNA at N/P = 5 or 10 in DMEM + 10% FBS for 4 h or 10 h. Alternatively, cells were transfected with CPOx/pDNA polyplex supplemented with 0.1% or 0.5% Pluronic P85. After transfection, cells were incubated with fresh complete medium for another 24 h and lysed subsequently for luciferase measurement. Statistical comparison with cells only, * p < 0.05, ** p < 0.01. n.s., not significant (n = 3).
In RAW264.7 macrophages, PEG-PLL/pDNA polyplexes produced significant albeit low levels of transfection (Figure 12B). In contrast, we saw no transfection after either 4 or 10 h incubation of the cells with CPOx/pDNA polyplexes. Again, addition of either 0.1 % or 0.5 % Pluronic P85 boosted the transfection for CPOx/pDNA polyplexes to significant levels. In these cases the gene expression reached comparable levels to the PEG-PLL/pDNA control.
4. Discussion
It is well known that macrophages play a key role in immune responses through direct and indirect mechanisms.[108, 109] A significant population of macrophages derived from monocytic precursors are believed to be associated with malignant tissues in the tumor microenvironment. This special phenotype of macrophages, often termed tumor-associated macrophages (TAMs), act as pro-tumoral components. Previous reports demonstrated a number of chemoattractants such as CCL2 and CCL5 secreted by tumor or stromal cells are responsible for the accumulation of TAMs at the tumor sites.[110, 111] Similar roles may also be played by the vascular endothelial growth factor (VEGF), the platelet derived growth factor (PDGF), the transforming growth factor beta (TGFβ) and the macrophage colony stimulating factor (M-CSF), of which the latter may promote macrophage survival and differentiation. TAMs also facilitate tumor cells producing matrix proteases to degrade the extracellular matrix (ECM) and therefore promote tumor invasion and metastasis.[112] Gene delivery targeted at macrophages either by re-educating macrophages towards tumoricidal phenotype or directly wiping out these populations offers great opportunities for a long-term cancer immunotherapy. Interestingly, it was recently discovered that macrophages can transfect neighboring cells simply by transporting DNAs via excreted lipid vesicles termed exosomes.[113, 114] In this regard, we proposed that transfecting TAMs can provide for possibilities to infiltrate and impact deep tissues of tumors. The transfection efficiency in macrophages has proven to be extremely low and although a number of non-viral gene transfer methods attempted to improve macrophage transfection,[115-117] all these methods have the drawback of a rather low transfection efficiency and/or high cellular toxicity. This study was designed as a first step towards development of a platform to deliver genes to TAMs and to reduce the notorious cytotoxicity of polyplex gene delivery systems. For targeted delivery, a low unspecific uptake is important, which can be correleated with unspecific protein binding. Therefore, protein binding of the presented polyplexes was studied.
Various polycations and polyplexes formed thereof have been extensively explored as carriers for non-viral gene delivery although their applications in vivo have been hindered by their low transfection efficiency, their cytotoxicity as well as the rapid clearance from the plasma circulation by the immune system. Although polyplexes have been investigated for quite some time, numerous research groups are still actively investigating new polymers with the goal to reduce toxicity, non-specific cell uptake and increase the transfection efficacy at the same time.[83] We designed a new alternative system based on a protected secondary amine-functionalized 2-oxazoline monomer, and synthesized polycationic POx block copolymer, CPOx, containing pendant secondary amine groups by co-polymerization of this new monomer with MeOx. This new polycationic block copolymer exhibit promising properties such as a low cytotoxicity, and formation of stable polyplexes with pDNA. While in low and medium ionic strength media (DI water and 10 mM HEPES) polyplex diameters were below 100 nm, size increased to around 150 nm when in transferred to or prepared in high ionic strength media (150 mM NaCl). Important to note, the polyplexes were essentially neutral in terms of ζ-potential. In a recent study on polycationic POx with primary and tertiary amines, the polyplex particle size was reported only at N/P = 20.[83] The reported particle diameter varied between 67 and 255 nm but were mostly > 100 nm with significantly positive ζ-potential and variable dispersities.[83] In addition, CPOx/pDNA polyplexes show very low plasma protein binding as compared to the polyplexes based on a conventional cationic copolymer, PEG-PLL.
The finding of the significantly decreased protein binding for CPOx/pDNA may be attributed to a stealth-effect enabled by the hydrophilic PMeOx block of CPOx, forming a hydrophilic protective shell around the CPOx/pDNA nanoparticles. This PMeOx shell may be more efficient as compard to the PEG shell of PEG-PLL/pDNA polyplexes because PMeOx is reported to be more hydrophilic than the PEG.[60, 118] However, the low protein binding may be also attributed to the nature of the polycationic block, which is designed to be more hydrophilic as compared to PLL or previously described polycationic POx. In summary, the difference between the CPOx and PEG-PLL compared in this study is the overall higher hydrophilicity of CPOx and thus, either the hydrophilicity of shell or the core or the lower amphiphilic contrast between both may explain the favorable stealth property of polyplex particles formed with CPOx.
Stability against environmental challenges such as temperature, ionic strength and protein load are important criteria of polyplexes.[90] CPOx/pDNA polyplexes were found to be stable upon dilution in low ionic strength buffers and saline as well as upon storage at 4°C for at least 1 week. We also found that the CPOx/pDNA polyplexes are stable even at elevated temperatures (70 °C). While studying interactions of the CPOx/pDNA polyplexes with cells, we found slow internalization into RAW264.7 macrophages. Eventually, after 10 h, appreciable amounts were detected. The slow internalization of the CPOx/pDNA might be accociated to the hydrophilic shell formed by the PMeOx block as the stealth-effect also diminishes interactions of the polyplex particles with the cell membrane. Interestingly though, amphiphilic POx, which also exhibit very low protein binding[119] were shown to be internalized very rapidly in a variety of cells.[120] This marked difference may be attributed to the hydrophilic character of the cationic block forming the polyplex core and/or the good stability of the overall polyplex construct as it do not allow disintegration of the particles upon cell membrane contact. As a general rule, internalization and toxicity of polyplexes formed by PEG-modified polycations (e.g. PEG-PLL) are lower than internalization and toxicity of polyplexes formed by the same polycations that do not carry PEG chains (e.g. PLL).[121, 122] The newly designed CPOx/pDNA polyplexes are significantly less toxic (IC50 ≥ 1 g/L) than the PEG-containing ones. A direct comparison with recent work by Rinkenauer et al.[83] is difficult since in this paper the cytotoxicity was tested using a different assay to a maximal concentration of 200 mg/L for 24 h while in the present work, incubation was performed for 48 h. The toxicity of the presented polymers is in the same order of magnitude as reported before by Hsiue et al. for POx-PEI copolymers.[77]
To optimize the in vitro transfection using CPOx/pDNA polyplexes the time of the exposure of the cells to these polyplexes was increased to 10 h compared to conventional 4 h.[123] Even at prolonged exposure the CPOx/pDNA polyplexes revealed very low transfection activity that was same or less than the transfection activity of the PEG-PLL/pDNA control. We performed multiple transfection experiments and found that for hard-to-transfect RAW264.7 macrophages the transfection efficiency of the CPOx/pDNA polyplexes was negligible, however, it could be boosted to significant levels by coadministration of P85, known to increase transfection due to, in part, the enhancement of polyplex uptake and the activation of the nuclear factor-κB (NF-κB) signaling pathways.[124, 125] We also noted considerable increase for the transfection of B16 murine melanoma cells with CPOx/pDNA polyplexes and Pluronic P85 systems, as compared to the polyplexes alone. This is a very promising observation especially in view of our prior findings that Pluronic block copolymers, and in particular, Pluronic P85 were shown to greatly improve non-viral gene transfer in vivo, including increased gene expression in macrophages and other immune response cells.[126]
Although the transfection levels observed with the CPOx/pDNA polyplexes without the block copolymer adjuvant were lower as compared to PEG-PLL/pDNA, the presented results provide a good starting point for further developments because the POx system is highly tuneable in its properties and allows precise adjustments of the polymer architecture, amphiphilic contrast and charge distribution. All of these aspects will be subject of future work. On the other hand, the slow CPOx/pDNA polyplex uptake by macrophages also offers an opportunity to minimize non-specific uptake in vivo and provides a sufficient time window for active targeting of specific cell populations using selective cell surface binding motifs. These can readily be introduced into the CPOx by click chemistry with the alkyne group at the hydrophilic polymer terminus or by introduction of a suitable monomer.[120] At the same time, this might also boost selective endocytosis only for recognized cells. The ‘clickable’ property of the developed CPOx polymer was evidenced by successfully attachment of a fluorescence dye.
5. Conclusions
We prepared the first cationic POx with pendant secondary amine groups from a new protected 2-oxazoline monomer (Boc-MAMeOx). The secondary amine is linked to the polymer backbone by a short methylene linker in order to minimize the hydrophobic character of the monomer unit. Living cationic polymerization of Boc-MAMeOx with the hydrophilic MeOx and deprotection gave a block copolymer (CPOx) that effectively complexed pDNA to small and narrow distributed polyplex nanoparticles of excellent stability upon storage and dilution in physiological saline as well as upon thermal challenge. The CPOx/pDNA polyplexes show low to no cytotoxicity and very low plasma protein binding as compared to conventional PEG-PLL-based polyplexes. The cell uptake of CPOx/pDNA was found to be slow and the transfection efficiency into macrophages or melanoma cells was not satisfactory, but could be significantly improved using Pluronic P85 as a gene transfer adjuvant. The CPOx polymer is functionalized with a “clickable” terminus allowing for attachment of targeting ligands, fluorescent dyes and other functional molecules to the resulting polyplexes. In future studies stability and targeting of the new polyplexes in complex biological media can be further explored both in vitro and in vivo.
Acknowledgments
This study was supported by the Eshelman Gift Trust funds (to A.V.K), by Award No. KUK-F1-029-32, made by King Abdullah University of Science and Technology (KAUST), by the Free State of Bavaria, the Fonds der Chemischen Industrie through a junior faculty support grant and start-up funding from the German Plastics Center SKZ and the University of Würzburg (all to R. L.) and partially by the Cancer Nanotechnology Platform Partnership grant (U01 CA116591, to A.V.K) of the National Cancer Institute Alliance for Cancer Nanotechnology. Z. H. is also grateful to GlaxoSmithKline Clinical Research and Drug Development Fellowship support. We would like to thank the Chapel Hill Analytical and Nanofabrication Laboratory (CHANL) and Proteomics Core Facility at UNC-CH. We also acknowledge technical support from Mr. Matthew Haney for the confocal microscopy, Mr. Yuhang Jiang for the gel electrophoresis, Mr. Vivek Mahajan for the supply of pDNA and Mr. Jonas F. Nawroth for the monomer synthesis.
References
- 1.Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, Windsor EA, Conlon TJ, Sumaroka A, Roman AJ, Byrne BJ, Jacobson SG, Engl N. J. Med. 2009;361:725. doi: 10.1056/NEJMc0903652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying GS, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, Auricchio A. Mol. Ther. 2010;18:643. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, Mahlaoui N, Kiermer V, Mittelstaedt D, Bellesme C, Lahlou N, Lefrère F, Blanche S, Audit M, Payen E, Leboulch P, l'Homme B, Bougneres P, Von Kalle C, Fischer A, Cavazzana-Calvo M, Aubourg P. Science. 2009;326:818. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser J. Science. 2009;326:1468. doi: 10.1126/science.326.5959.1468-b. [DOI] [PubMed] [Google Scholar]
- 5.Candotti F, Shaw KL, Muul L, Carbonaro D, Sokolic R, Choi C, Schurman SH, Garabedian E, Kesserwan C, Jagadeesh GJ, Fu PY, Gschweng E, Cooper A, Tisdale JF, Weinberg KI, Crooks GM, Kapoor N, Shah A, Abdel-Azim H, Yu XJ, Smogorzewska M, Wayne AS, Rosenblatt HM, Davis CM, Hanson C, Rishi RG, Wang X, Gjertson D, Yang OO, Balamurugan A, Bauer G, Ireland JA, Engel BC, Podsakoff GM, Hershfield MS, Blaese RM, Parkman R, Kohn DB. Blood. 2012;120:3635. doi: 10.1182/blood-2012-02-400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monahan PE, Gui T. Curr. Opin. Hematol. 2013 doi: 10.1097/MOH.0b013e328363c1a1. [DOI] [PubMed] [Google Scholar]
- 7.Skinner MW. Mol. Ther. 2013;21:1. doi: 10.1038/mt.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kershaw MH, Westwood JA, Darcy PK. Nat. Rev. Cancer. 2013;13:525. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 9.Tuppurainen L, Sallinen H, Kokki E, Koponen J, Anttila M, Pulkkinen K, Heikura T, Toivanen P, Hamalainen K, Kosma VM, Heinonen S, Alitalo K, Yla-Herttuala S. Hum. Gene Ther. Clin. Dev. 2013;24:29. doi: 10.1089/humc.2013.006. [DOI] [PubMed] [Google Scholar]
- 10.Yao X, Yoshioka Y, Morishige T, Eto Y, Narimatsu S, Kawai Y, Mizuguchi H, Gao JQ, Mukai Y, Okada N, Nakagawa S. Mol. Ther. 2011;19:1619. doi: 10.1038/mt.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan C. Nat. Biotechnol. 2011;29:121. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- 12.Auman JT. Curr. Opin. Mol. Ther. 2010;12:637. [PubMed] [Google Scholar]
- 13.Wu TL, Zhou D. Adv. Drug Deliv. Rev. 2011;63:671. doi: 10.1016/j.addr.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Akhtar S. Gene Ther. 2006;13:739. doi: 10.1038/sj.gt.3302692. [DOI] [PubMed] [Google Scholar]
- 15.Pringle IA, Hyde SC, Gill DR. Expert Opin. Biol. Ther. 2009;9:991. doi: 10.1517/14712590903055029. [DOI] [PubMed] [Google Scholar]
- 16.Rogers ML, Rush RA. J. Control. Release. 2012;157:183. doi: 10.1016/j.jconrel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Zibert JR, Wallbrecht K, Schön M, Mir LM, Jacobsen GK, Trochon-Joseph V, Bouquet C, Villadsen LS, Cadossi R, Skov L, Schön MP. J. Clin. Invest. 2011;121:410. doi: 10.1172/JCI41295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchelmeister HY, Karczewski S, Gutschmidt A, Knauer S, Schmuck C. Angew. Chem. Int. Ed. Engl. 2013;52:14016. doi: 10.1002/anie.201306929. [DOI] [PubMed] [Google Scholar]
- 19.Lollo CP, Banaszczyk MG, Mullen PM, Coffin CC, Wu D, Carlo AT, Bassett DL, Gouveia EK, Carlo DJ. Methods Mol. Med. 2002;69:1. doi: 10.1385/1-59259-141-8:001. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Oulad-Abdelghani M, Zelkin AN, Wang Y, Haikel Y, Mainard D, Voegel JC, Caruso F, Benkirane-Jessel N. Biomaterials. 2010;31:1699. doi: 10.1016/j.biomaterials.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 21.Boletta A, Benigni A, Lutz J, Remuzzi G, Soria MR, Monaco L. Hum. Gene Ther. 1997;8:1243. doi: 10.1089/hum.1997.8.10-1243. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Tian H, Kano A, Maruyama A, Chen X, Park TG. J. Control. Release. 2011;152(Suppl. 1):e134. doi: 10.1016/j.jconrel.2011.08.045. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Wang Z, Wang R, Lu T, Wang W, Drug Target J. 2007;15:714. doi: 10.1080/10611860701637974. [DOI] [PubMed] [Google Scholar]
- 24.Hong JW, Park JH, Huh KM, Chung H, Kwon IC, Jeong SY. J. Control. Release. 2004;99:167. doi: 10.1016/j.jconrel.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Gene Ther. 2005;12:1023. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 26.Wong K, Sun G, Zhang X, Dai H, Liu Y, He C, Leong KW. Bioconjugate Chem. 2006;17:152. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 27.Duceppe N, Tabrizian M. Expert. Opin. Drug Deliv. 2010;7:1191. doi: 10.1517/17425247.2010.514604. [DOI] [PubMed] [Google Scholar]
- 28.Jean M, Alameh M, Buschmann MD, Merzouki A. Gene Ther. 2011;18:807. doi: 10.1038/gt.2011.25. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Guo T, Zhou D, Hu Y, Zhou H, Wang S, Chen J, Zhang Z. J. Control. Release. 2011;154:177. doi: 10.1016/j.jconrel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Malmo J, Sørgård H, Vårum KM, Strand SP. J. Control. Release. 2012;158:261. doi: 10.1016/j.jconrel.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Van Kuringen HP, Lenoir J, Adriaens E, Bender J, De Geest BG, Hoogenboom R. Macromol. Biosci. 2012;12:1114. doi: 10.1002/mabi.201200080. [DOI] [PubMed] [Google Scholar]
- 32.Ballarín-González B, Howard KA. Adv. Drug Deliv. Rev. 2012;64:1717. doi: 10.1016/j.addr.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Kim SW. Cold Spring Harb. Protoc. 2012;2012:433. doi: 10.1101/pdb.ip068619. [DOI] [PubMed] [Google Scholar]
- 34.Prevette LE, Mullen DG, Holl MM. Mol. Pharm. 2010;7:870. doi: 10.1021/mp100027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeil SE. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009;1:264. doi: 10.1002/wnan.6. [DOI] [PubMed] [Google Scholar]
- 36.Deng J, Gao N, Wang Y, Yi H, Fang S, Ma Y, Cai L. Biomacromolecules. 2012;13:3795. doi: 10.1021/bm3012538. [DOI] [PubMed] [Google Scholar]
- 37.Lin YL, Liu YK, Tsai NM, Hsieh JH, Chen CH, Lin CM, Liao KW. Nanomedicine. 2012;8:318. doi: 10.1016/j.nano.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Lutz GJ, Sirsi SR, Williams JH. Methods Mol. Biol. 2008;433:141. doi: 10.1007/978-1-59745-237-3_9. [DOI] [PubMed] [Google Scholar]
- 39.Patil ML, Zhang M, Minko T. ACS Nano. 2011;5:1877. doi: 10.1021/nn102711d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sirsi SR, Schray RC, Guan X, Lykens NM, Williams JH, Erney ML, Lutz GJ. Hum. Gene Ther. 2008;19:795. doi: 10.1089/hum.2007.129. [DOI] [PubMed] [Google Scholar]
- 41.Tian HY, Deng C, Lin H, Sun J, Deng M, Chen X, Jing X. Biomaterials. 2005;26:4209. doi: 10.1016/j.biomaterials.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Kawai F. Appl. Microbiol. Biotechnol. 2002;58:30. doi: 10.1007/s00253-001-0850-2. [DOI] [PubMed] [Google Scholar]
- 43.Vandegriff KD, Malavalli A, Minn C, Jiang E, Lohman J, Young MA, Samaja M, Winslow RM. Biochem. J. 2006;399:463. doi: 10.1042/BJ20060809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veronese FM. Biomaterials. 2001;22:405. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 45.Webster R, Didier E, Harris P, Siegel N, Stadler J, Tilbury L, Smith D. Drug Metab. Dispos. 2007;35:9. doi: 10.1124/dmd.106.012419. [DOI] [PubMed] [Google Scholar]
- 46.Conover C, Lejeune L, Linberg R, Shum K, Shorr RG. Artif. Cells Blood Substit. Immobil. Biotechnol. 1996;24:599. doi: 10.3109/10731199609118885. [DOI] [PubMed] [Google Scholar]
- 47.Bendele A, Seely J, Richey C, Sennello G, Shopp G. Toxicol. Sci. 1998;42:152. doi: 10.1006/toxs.1997.2396. [DOI] [PubMed] [Google Scholar]
- 48.Rudmann DG, Alston JT, Hanson JC, Heidel S. Toxicol. Pathol. 2013;41:970. doi: 10.1177/0192623312474726. [DOI] [PubMed] [Google Scholar]
- 49.Armstrong JK, Hempel G, Koling S, Chan LS, Fisher T, Meiselman HJ, Garratty G. Cancer. 2007;110:103. doi: 10.1002/cncr.22739. [DOI] [PubMed] [Google Scholar]
- 50.Garay RP, El-Gewely R, Armstrong JK, Garratty G, Richette P. Expert. Opin. Drug Deliv. 2012;9:1319. doi: 10.1517/17425247.2012.720969. [DOI] [PubMed] [Google Scholar]
- 51.Luxenhofer R, Han Y, Schulz A, Tong J, He Z, Kabanov AV, Jordan R. Macromol. Rapid Comm. 2012;33:1613. doi: 10.1002/marc.201200354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedlacek O, Monnery BD, Filippov SK, Hoogenboom R, Hruby M. Macromol. Rapid Comm. 2012;33:1648. doi: 10.1002/marc.201200453. [DOI] [PubMed] [Google Scholar]
- 53.Ulbricht J, Jordan R, Luxenhofer R. Biomaterials. 2014;35:4848. doi: 10.1016/j.biomaterials.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 54.Guillerm B, Monge S, Lapinte V, Robin JJ. Macromol. Rapid Comm. 2012;33:1600. doi: 10.1002/marc.201200266. [DOI] [PubMed] [Google Scholar]
- 55.Rehfeldt F, Tanaka M, Pagnoni L, Jordan R. Langmuir. 2002;18:4908. [Google Scholar]
- 56.Woodle MC, Engbers CM, Zalipsky S. Bioconjugate Chem. 1994;5:493. doi: 10.1021/bc00030a001. [DOI] [PubMed] [Google Scholar]
- 57.Zalipsky S, Hansen CB, Oaks JM, Allen TM. J. Pharm. Sci. 1996;85:133. doi: 10.1021/js9504043. [DOI] [PubMed] [Google Scholar]
- 58.Hutter NA, Reitinger A, Zhang N, Steenackers M, Williams OA, Garrido JA, Jordan R. Phys. Chem. Chem. Phys. 2010;12:4360. doi: 10.1039/b923789p. [DOI] [PubMed] [Google Scholar]
- 59.Zhang N, Pompe T, Amin I, Luxenhofer R, Werner C, Jordan R. Macromol. Biosci. 2012;12:926. doi: 10.1002/mabi.201200026. [DOI] [PubMed] [Google Scholar]
- 60.Viegas TX, Bentley MD, Harris JM, Fang Z, Yoon K, Dizman B, Weimer R, Mero A, Pasut G, Veronese FM. Bioconjugate Chem. 2011;22:976. doi: 10.1021/bc200049d. [DOI] [PubMed] [Google Scholar]
- 61.Zhang N, Huber S, Schulz A, Luxenhofer R, Jordan R. Macromolecules. 2009;42:6. [Google Scholar]
- 62.Buhler J, Gietzen S, Reuter A, Kappel C, Fischer K, Decker S, Schäffel D, Koynov K, Bros M, Tubbe I, Grabbe S, Schmidt M. Chemistry. 2014;20:12405. doi: 10.1002/chem.201403942. [DOI] [PubMed] [Google Scholar]
- 63.Zhang N, Luxenhofer R, Jordan R. Macromol. Chem. Phys. 2012;213:6. [Google Scholar]
- 64.Zhang N, Luxenhofer R, Jordan R. Macromol. Chem. Phys. 2012;213:8. [Google Scholar]
- 65.Waschinski CJ, Tiller JC. Biomacromolecules. 2005;6:235. doi: 10.1021/bm049553i. [DOI] [PubMed] [Google Scholar]
- 66.Tong J, Zimmerman MC, Li S, Yi X, Luxenhofer R, Jordan R, Kabanov AV. Biomaterials. 2011;32:3654. doi: 10.1016/j.biomaterials.2011.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engelhardt N, Ernst A, Kampmann A, Weberskirch R. Macromol. Chem. Phys. 2013;214:8. [Google Scholar]
- 68.Han YC, He ZJ, Schulz A, Bronich TK, Jordan R, Luxenhofer R, Kabanov AV. Mol. Pharm. 2012;9:2302. doi: 10.1021/mp300159u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luxenhofer R, Schulz A, Roques C, Li S, Bronich TK, Batrakova EV, Jordan R, Kabanov AV. Biomaterials. 2010;31:4972. doi: 10.1016/j.biomaterials.2010.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schulz A, Jaksch S, Schubel R, Wegener E, Di Z, Han Y, Meister A, Kressler J, Kabanov AV, Luxenhofer R, Papadakis CM, Jordan R. ACS nano. 2014;8:2686. doi: 10.1021/nn406388t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eskow Jaunarajs KL, Standaert DG, Viegas TX, Bentley MD, Fang Z, Dizman B, Yoon K, Weimer R, Ravenscroft P, Johnston TH, Hill MP, Brotchie JM, Moreadith RW. Movement Disord. 2013;28:1675. doi: 10.1002/mds.25625. [DOI] [PubMed] [Google Scholar]
- 72.Tong J, Luxenhofer R, Yi X, Jordan R, Kabanov AV. Mol. Pharm. 2010;7:984. doi: 10.1021/mp100102p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konieczny S, Fik CP, Averesch NJ, Tiller JC. J. Biotechnol. 2012;159:195. doi: 10.1016/j.jbiotec.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 74.Tong J, Yi X, Luxenhofer R, Banks WA, Jordan R, Zimmerman MC, Kabanov AV. Mol. Pharm. 2013;10:360. doi: 10.1021/mp300496x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi HJ, Montemagno CD. Nano Lett. 2005;5:2538. doi: 10.1021/nl051896e. [DOI] [PubMed] [Google Scholar]
- 76.Krumm C, Fik CP, Meuris M, Dropalla GJ, Geltenpoth H, Sickmann A, Tiller JC. Macromol. Rapid Comm. 2012;33:1677. doi: 10.1002/marc.201200192. [DOI] [PubMed] [Google Scholar]
- 77.Hsiue GH, Chiang HZ, Wang CH, Juang TM. Bioconjugate Chem. 2006;17:781. doi: 10.1021/bc050317u. [DOI] [PubMed] [Google Scholar]
- 78.Wang CH, Wang WT, Hsiue GH. Biomaterials. 2009;30:3352. doi: 10.1016/j.biomaterials.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 79.Hochleitner G, Hümmer JF, Luxenhofer R, Groll J. Polymer. 2014;55:6. [Google Scholar]
- 80.Thomas M, Lu JJ, Ge Q, Zhang C, Chen J, Klibanov AM. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5679. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cesana S, Auernheimer J, Jordan R, Kessler H, Nuyken O. Macromol. Chem. Physic. 2006;207:183. [Google Scholar]
- 82.Hartlieb M, Pretzel D, Englert C, Hentschel M, Kempe K, Gottschaldt M, Schubert US. Biomacromolecules. 2014;15:1970. doi: 10.1021/bm500236y. [DOI] [PubMed] [Google Scholar]
- 83.Rinkenauer AC, Tauhardt L, Wendler F, Kempe K, Gottschaldt M, Traeger A, Schubert US. Macromol. Biosci. 2015;15:414. doi: 10.1002/mabi.201400334. [DOI] [PubMed] [Google Scholar]
- 84.Levy A, Litt M. J. Polym. Sci. Pol. Chem. 1968;6:57. [Google Scholar]
- 85.Zarka MT, Nuyken O, Weberskirch RF. Macromol. Rapid Comm. 2004;25:858. [Google Scholar]
- 86.Liu XM, Quan LD, Tian J, Laquer FC, Ciborowski P, Wang D. Biomacromolecules. 2010;11:2621. doi: 10.1021/bm100578c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhong Z, Feijen J, Lok MC, Hennink WE, Christensen LV, Yockman JW, Kim YH, Kim SW. Biomacromolecules. 2005;6:3440. doi: 10.1021/bm050505n. [DOI] [PubMed] [Google Scholar]
- 88.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR., Jr. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4897. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reul R, Nguyen J, Kissel T. Biomaterials. 2009;30:5815. doi: 10.1016/j.biomaterials.2009.06.057. [DOI] [PubMed] [Google Scholar]
- 90.von Erlach T, Zwicker S, Pidhatika B, Konradi R, Textor M, Hall H, Lühmann T. Biomaterials. 2011;32:5291. doi: 10.1016/j.biomaterials.2011.03.080. [DOI] [PubMed] [Google Scholar]
- 91.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu D, Xu W, Pan R, Ding Y, Sui W, Chen P. Nanomedicine. 2015;11:435. doi: 10.1016/j.nano.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Lee WS, Kim YK, Zhang Q, Park TE, Kang SK, Kim DW, Cho CS, Choi YJ. Nanomedicine. 2014;10:525. doi: 10.1016/j.nano.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 94.Adolph EJ, Nelson CE, Werfel TA, Guo R, Davidson JM, Guelcher SA, Duvall CL. J. Mater. Chem. B, Mater. Biol. Med. 2014;2:8154. doi: 10.1039/C4TB00352G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han S, Wan H, Lin D, Guo S, Dong H, Zhang J, Deng L, Liu R, Tang H, Dong A. Acta Biomater. 2014;10:670. doi: 10.1016/j.actbio.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 96.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. ACS nano. 2013;7:8870. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Biomacromolecules. 2008;9:3294. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nuhn L, Gietzen S, Mohr K, Fischer K, Toh K, Miyata K, Matsumoto Y, Kataoka K, Schmidt M, Zentel R. Biomacromolecules. 2014;15:1526. doi: 10.1021/bm500199h. [DOI] [PubMed] [Google Scholar]
- 99.Ozcan I, Segura-Sánchez F, Bouchemal K, Sezak M, Ozer O, Guneri T, Ponchel G. Int. J. Nanomedicine. 2010;5:1103. doi: 10.2147/IJN.S15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rausch K, Reuter A, Fischer K, Schmidt M. Biomacromolecules. 2010;11:2836. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- 101.Gaertner FC, Luxenhofer R, Blechert B, Jordan R, Essler M. J. Control. Release. 2007;119:291. doi: 10.1016/j.jconrel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 102.Bauer M, Schroeder S, Tauhardt L, Kempe K, Schubert US, Fischer D. J. Polym. Sci. Pol. Chem. 2013;51:1816. [Google Scholar]
- 103.Mack KD, Wei R, Elbagarri A, Abbey N, McGrath MS. J. Immunol. Methods. 1998;211:79. doi: 10.1016/s0022-1759(97)00194-4. [DOI] [PubMed] [Google Scholar]
- 104.Thompson CD, Frazier-Jessen MR, Rawat R, Nordan RP, Brown RT. BioTechniques. 1999;27:824. doi: 10.2144/99274rr05. [DOI] [PubMed] [Google Scholar]
- 105.Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Bioconjugate Chem. 2008;19:1987. doi: 10.1021/bc800144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lemieux P, Guerin N, Paradis G, Proulx R, Chistyakova L, Kabanov A, Alakhov V. Gene Ther. 2000;7:986. doi: 10.1038/sj.gt.3301189. [DOI] [PubMed] [Google Scholar]
- 107.Yang Z, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. J. Control. Release. 2005;108:496. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 108.Twigg HL. Semin. Respir. Crit. Care Med. 2004;25:21. doi: 10.1055/s-2004-822302. [DOI] [PubMed] [Google Scholar]
- 109.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Science. 2010;327:656. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Trends Immunol. 2002;23:549. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 111.Sica A, Schioppa T, Mantovani A, Allavena P. Eur. J. Cancer. 2006;42:717. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 112.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. Crit. Rev. Oncol. Hematol. 2008;66:1. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 113.Haney MJ, Zhao Y, Harrison EB, Mahajan V, Ahmed S, He Z, Suresh P, Hingtgen SD, Klyachko NL, Mosley RL, Gendelman HE, Kabanov AV, Batrakova EV. PloS one. 2013;8:e61852. doi: 10.1371/journal.pone.0061852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Zhao Y, Haney MJ, Gupta R, Bohnsack JP, He Z, Kabanov AV, Batrakova EV. PloS one. 2014;9:e106867. doi: 10.1371/journal.pone.0106867. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Walter E, Thiele L, Merkle HP. Stp. Pharma. Sci. 2001;11:45. [Google Scholar]
- 116.Mahor S, Dash BC, O'Connor S, Pandit A. Bioconjugate Chem. 2012;23:1138. doi: 10.1021/bc200599k. [DOI] [PubMed] [Google Scholar]
- 117.Parsa S, Wang Y, Fuller J, Langer R, Pfeifer BA. Pharm. Res. 2008;25:1202. doi: 10.1007/s11095-008-9563-x. [DOI] [PubMed] [Google Scholar]
- 118.Foreman MB, Coffman JP, Murcia MJ, Cesana S, Jordan R, Smith GS, Naumann CA. Langmuir. 2003;19:6. [Google Scholar]
- 119.Naka K, Nakamura T, Ohki A, Maeda S. Macromol. Chem. Phys. 1997;198:15. [Google Scholar]
- 120.Luxenhofer R, Sahay G, Schulz A, Alakhova D, Bronich TK, Jordan R, Kabanov AV. J. Control. Release. 2011;153:73. doi: 10.1016/j.jconrel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han M, Bae Y, Nishiyama N, Miyata K, Oba M, Kataoka K. J. Control. Release. 2007;121:38. doi: 10.1016/j.jconrel.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 122.Miyata K, Fukushima S, Nishiyama N, Yamasaki Y, Kataoka K. J. Control. Release. 2007;122:252. doi: 10.1016/j.jconrel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 123.Gebhart CL, Kabanov AV. J. Control. Release. 2001;73:401. doi: 10.1016/s0168-3659(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 124.Sriadibhatla S, Yang Z, Gebhart C, Alakhov VY, Kabanov A. Mol. Ther. 2006;13:804. doi: 10.1016/j.ymthe.2005.07.701. [DOI] [PubMed] [Google Scholar]
- 125.Astafieva I, Maksimova I, Lukanidin E, Alakhov V, Kabanov A. FEBS Lett. 1996;389:278. doi: 10.1016/0014-5793(96)00607-2. [DOI] [PubMed] [Google Scholar]
- 126.Gaymalov ZZ, Yang Z, Pisarev VM, Alakhov VY, Kabanov AV. Biomaterials. 2009;30:1232. doi: 10.1016/j.biomaterials.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]