Abstract
Background
Angiogenesis is an important biological process involved in the proliferation of endothelial cells, tumor growth and metastasis. Vascular endothelial growth factor (VEGF) is considered as a prominent regulator of angiogenesis which exerts the aforementioned effect(s) through its respective receptors (VEGFR1 and VEGFR2). VEGF receptors are targeted as a therapeutic candidate for cancer growth inhibition. RNAi as a new and promising strategy has provided a useful means to specifically suppress gene expression in cancer cells.
Objectives
The current study aimed to down-regulate expression of the VEGFR1 using siRNA.
Materials and Methods
This experimental study designed specific siRNAs against VEGFR1. Total RNA was extracted from human umbilical vain endothelial cell (HUVEC) and subsequently cDNA was synthetized. PCR was performed using specific primers to amplify the target gene. After double digestion and purification, the gene was cloned into pEFGP-N1 expression vector. Then, AGS cells were transfected with recombinant pEGFP-N1 using lipofectamin. The gene expression and down-regulation were evaluated by fluorescence scanning, reverse transcription PCR (RT-PCR) and Western blot techniques.
Results
Fluorescent scanning, RT-PCR (27.68%) and western blot analysis (31.06%) showed that the expression of VEGFR1 was suppressed effectively.
Conclusions
The results of the current study showed that specifically designed siRNA can be considered as an appropriate strategy to suppress gene expression and might be a promising tool to prevent angiogenesis.
Keywords: Angiogenesis, Gastric Cancer, Small Interfering RNA, Vascular Endothelial Growth Factor Receptor
1. Background
Angiogenesis is an important biological process involved in wound healing, reproduction, organ development and also pathophysiology of different diseases such as cancer (1). Angiogenesis is the formation of new capillaries from blood vessels that plays a prominent role in tumor growth and invasiveness. Angiogenesis is regulated by multiple key effectors, mainly through vascular endothelial cell growth factor A (VEGF-A). VEGF-A is defined to increase micro-vascular growth and permeability via specific receptors such as fms-like tyrosine kinase 1 (FLT-1), VEGFR1 and fetal liver kinase 1 (FLK-1) (2). Moreover, VEGF-A is proposed as a key modulator in revascularization of ischemic diseases (3). VEGF-A plays a central role in the development of several types of angiogenesis and neovascularization (NV), including retinal and choroidal NV (CNV) (4, 5). Furthermore, it is shown that VEGFR1 induces expression of growth factors in liver sinusoidal endothelial cells (6). In addition, it mediates survival of endothelial cells through stimulation of phosphatidylinositol 3-kinase/protein kinase B (AKT) pathway (7). It is reported that expression of VEGFR1 in breast cancer cells is significantly related to high risk for metastasis and relapses. Hence, VEGFR1 is considered as a marker for breast tumor aggressiveness (8). Recent studies revealed that the control of the VEGF and its function is a promising target for either induction or inhibition of angiogenesis in tumor cells (4). High level expression of VEGF stimulates angiogenesis while applying monoclonal antibodies inhibit or degrade VEGF and suppress the angiogenesis process (5). These findings highlight the issue that VEGF pathway might be an important therapeutic target. Clinical trials supported the importance of this growth factor in cancer therapy using an aptamer (9) or an antibody fragment (10) with the ability to bind VEGF-A. Another strategy to antagonize VEGF-A is to block VEGF receptors. A remarkable advantage of this approach is the potency to block multiple VEGF family members at once (11).
RNA interference (RNAi), an absolutely fundamental biological process by which cells regulate gene expression, acts through complementary base-paring with target mRNA and retrieves cellular RNases which in turn degrade mRNA transcripts (12). RNAi strategy rapidly developed from a basic scientific discovery to a powerful research tool and more recently, to a promising therapeutic approach (13). RNAi is now routinely used to evaluate gene function both in vitro and in vivo and many innovative screens reported the use of RNAi to investigate potential drug targets (14). Small interfering RNAs (siRNAs) provide a useful means to selectively reduce the amount of mRNA transcripts and probe the function of gene products (15).
2. Objectives
The present study aimed to down-regulate specifically VEGFR1 expression in human Caucasian gastric adenocarcinoma (AGS) cells using synthetic siRNA.
3. Materials and Methods
3.1. Materials
RPMI-1640, fetal bovine serum (FBS), penicillin-streptomycin, and trypsin enzyme were purchased from GIBCO (Grand Island, NY, USA). RNA extraction, cDNA synthesis and PCR purification kits were from Roche, Germany. Restriction enzymes were obtained from Jena Bioscience.
3.2. Methods
This experimental study was conducted in chemical research injury center at Baqiyatallah University of Medical Sciences in 2013.
3.2.1. The siRNA Sequences Designing
Anti-VEGFR1 siRNAs was purchased from Takapuzist Gene Molecular Biotechnology Co. Ltd. (Tehran, Iran). The siRNA was designed to target VEGFR1 at 5‘-CCAGACACTGCATCCAA-3’ sequence. Sense and anti-sense sequences were as follows: siRNA1: Si-Sense1: 5’- GCA,UAA,CUA,AAU,CUG,CCU,G-3’; Si-AntiSense1: 5’- CAG,GCA,GAU,UUA,GUU,AUG,C -3’. Each siRNA was resuspended in double distilled water and the stock solutions (20 μmol/L) were stored at 4°C.
3.2.2. Cell Culture
Human umbilical vein endothelial cells (HUVEC) and AGS cells (from Pasteur institute, Tehran, Iran) were grown in RPMI-1640 media containing 10% fetal bovine serum (FBS). Cells were incubated in a humidified 5% CO2 incubator at 37°C for 48 hours. Viability of cells were examined by trypan blue and then incubated in hypoxic situation for 24 hours to up-regulate the expression of the target genes.
3.2.3. Amplification of VEGFR1 Gene
Total RNA was extracted from HUVEC cells using RNA extraction kit and immediately stored in -80°C. Reverse transcription PCR (RT-PCR) was performed according to the instruction provided by manufacturer (Roche company, Germany). PCR reaction was performed to amplify the target gene by previously designed and synthesized specific primers (Forward: 5’- GAATTCATGGTCAGCTACTGGGACACC-3; Reverse: 5‘- GGATCCGTTAGGTGACGTAACCCGGCAG-3). Primers had restriction sites for EcoRI and BamH I enzymes. PCR product was cleaned up from agarose gel and analyzed by sequencing and digestion using EcoR1 and BamH-1 restriction enzymes.
3.2.4. Cloning of VEGFR1 Gene
Double digested VEGFR1was cloned into N-terminal of GFP gene in PEGFP vector using T4 ligase. The recombinant vectors were transformed into electrocompetent E. coli cells b (DH5α) using electroporation method and then cultured on LB medium containing kanamycin (100 μg/mL) at 37°C and overnight. Plasmids were extracted using plasmid extraction kit (Sinnagen, Tehran, Iran) from grown clones and subjected to sequencing and double digestion by the aforementioned restriction enzymes.
3.2.5. Transfection of Recombinant PEGFP-VEGFR1 Vector and siRNA
AGS cells (institute Pasteur of Iran) were cultured in RPMI-1640 medium. Cells were then transfected with recombinant VEGFr1-pEGFP-N1 vector and pEGFP as a control using lipofectamin 2000 (Invitrogen, USA) and incubated according to the manufacturer’s protocol. In brief, a number of 4 × 105 cells were seeded into six-well plates containing Dulbecco’s modified Eagle medium (DMEM), which was antibiotic-free. Plates were incubated at 37°C overnight. For each well, 2 μL of siRNA was mixed with 50 μL of DMEM. This mixture was then combined with a solution of 1 μL lipofectamine 2000 in 50 μL DMEM followed by 20 minutes incubation at room temperature. Finally, the mixture with the final concentration of 20 pmol/L for each siRNAwas applied to the cells. After incubation for 4 hours at 37°C, media were replaced with fresh RPMI-1640 supplemented with serum and Pen/Strep antibiotic (100 µg/mL). Cells were then cultured for an additional 2 weeks at 37°C before analysis.
3.2.6. Fluorescence Microscopy
Green florescent protein (GFP) expression level was evaluated in different groups (cells transfected with pEGFP-VEGFR1 and untransfected cells as control) by fluorescence microscopy (NikonTS100-F, Japan) 72 hours after transfection of siRNAs in presence and absence of specific siRNAs.
3.2.7. RNA Extraction and RT-PCR
Total RNA was extracted from the cells using RNA extraction kit according to the manufacturer’s instruction. In brief, 2 µg of total RNA was reversely transcribed and cDNA was synthesized using cDNA synthesis kit (Roche Co., Germany). The cDNA was amplified using the following primers for VEGFR gene: forward 5‘- CAGACACTGCATCTCCAATGCAGG-3’`, reverse 5‘-GTAGAAGCCAGTGTGGTTTG-3’. The following protocol was used for PCR: initial denaturation at 95°C for 3 minutes, 30 cycles of denaturation at 95°C for 50 seconds, annealing at 58°C for 1 minute, extension at 72°C for 1 minute, followed by final extension at 72°C for 5 minutes. The PCR product was electrophoresed on 2% agarose gel containing ethidium bromide. To analyze the gene expression, β-actin was used as reference gene.
3.2.8. Western Blot Analysis
The cells were collected and lysed by RIPA lysis buffer (Sigma-Aldrich Corp., St. Louis, MO, USA). Total protein was extracted and stored in -80°C. The extracts were mixed with 6 × sodium dodecyl sulfate (SDS) buffer and boiled for 4 minutes. Samples were separated by 10% SDS-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride (PVDF) membrane. Membranes were blocked with 5% (w/v) skim milk in phosphate buffered saline (PBS) containing 0.1% Tween-20 for 1 hour at room temperature and then washed with PBST, and probed with primary antibodies overnight at 4°C. Membranes were washed again with PBST and incubated at room temperature with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibodies (Abcam Co., UAS) for 1 hour. The proteins were visualized with enhanced chemiluminescence detection kit (Amersham Bioscience, Buckinghamshire, England. Actin (a goat polyclonal antibody) was used as an internal control.
3.3. Statistical Analysis
Variables in the current study were the amount of mRNA, green fluorescent and receptor protein as the VEGFR1 expression. All tests were done triplicate and then result were explained as mean ± standard deviation (SD). The results were analyzed using Graphpad prism 5.0 program and SPSS (SPSS, Chicago, IL, USA). Because of the large sample size, which was about 5,000,000 cells in each group, based on the central limit theorem the mean has normal dist. Therefore, comparisons were made by one-way ANOVA and t-test; significant P value was considered less than 0.05 and data were shown as mean ± standard deviation (SD).
4. Results
4.1. Construction and Identification of Recombinant Plasmid pEGFP-VEGFR1
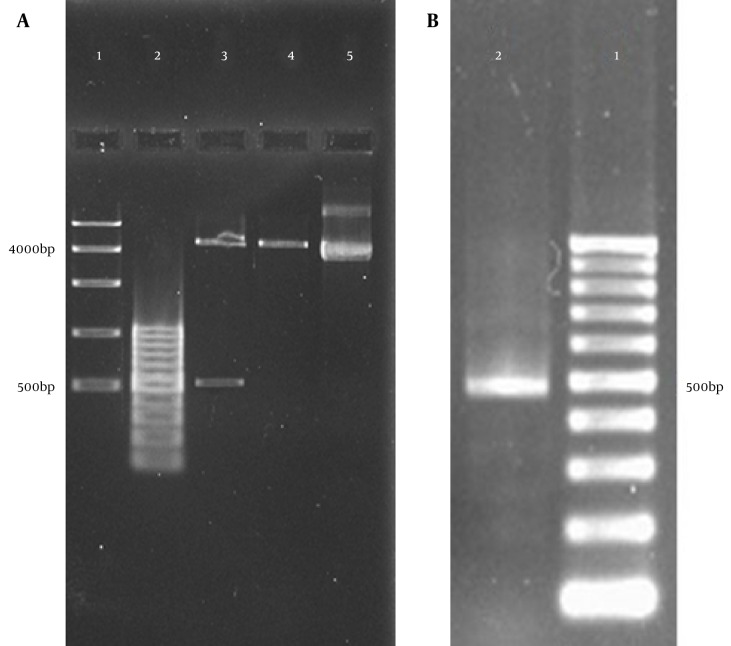
The expected band of VEGFR1 gene, using specific primers, was 504 bp amplified by PCR reaction (Figure 1).The PCR product and pEGFP vectors were both digested by EcoR I and BamH I, and the DNA fragments were ligated into plasmid pEGFP using T4 ligase. The recombinant plasmids (pEGFP-VEGFR1) were transfected into DH5α cells, recombinant plasmids were extracted from cultured DH5α cells, and confirmed by digestion with both EcoR I and BamH I. Figure 1 demonstrates the double-digestion of pEGFP-VEGFR1 vector that shows a 4700 bp and a 504 bp (A; well N.3) band that indicate proper cloning and a 504 bp PCR product shows amplification of VEGFR1 (B; well N.2).
Figure 1. Agarose Gel Electrophoresis of VEGFR1 PCR and Double-Digestion Products.
A, double-digestion of pEGFP-VEGFR1 vector shows a 4700 bp and a 504 bp (well N.3) band that indicates proper cloning; B, a 504 bp PCR product shows amplification of VEGFR1.
Electrophoresis analysis of recombinant plasmids digestion showed two bands of 4700 bp and 501 bp, which suggested that the target fragment, had been inserted into plasmid pEGFP correctly.
4.2. Fluorescence Microscopy Analysis
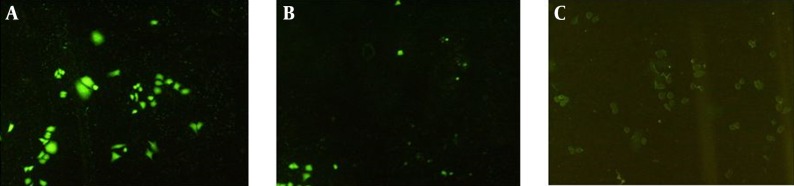
Efficiency of transfection methods and expression level were investigated by fluorescence microscopy. The VEGFR1 fragment in recombinant vector was co-expressed by GFP, therefore, reduction in VEGFR1 expression causes decline in fluorescence. After transfection of both recombinant vector and specific siRNAs into AGS cells, reduction of green fluorescence of GFP expression indicated that VEGFR1 gene expression significantly reduced in the treated cells when compared with those of the control cells 100.88 ± 13.82 (lux/inch) for control and 93.5 ± 9.29 (lux/inch) for the treated group (P < 0.05) (Figure 2).
Figure 2. Analysis of GFP Gene Expression in AGS Cells Using Fluorescent Microscope (20 X) in Presence and Absence of Anti-VEGFR1 siRNA.
A, pEGFP-VEGFR1 transfected cells without siRNA; B, pEGFP-VEGFR1 transfected cells after treating with anti-VEGFR siRNA; C, Cells without any transfection as control. As it is shown, after transfecting cells with anti-VEGFR1, the level of GFP significantly decreased, compared to cells without siRNA transfection.
4.3. RT-PCR of VEGFR1mRNA
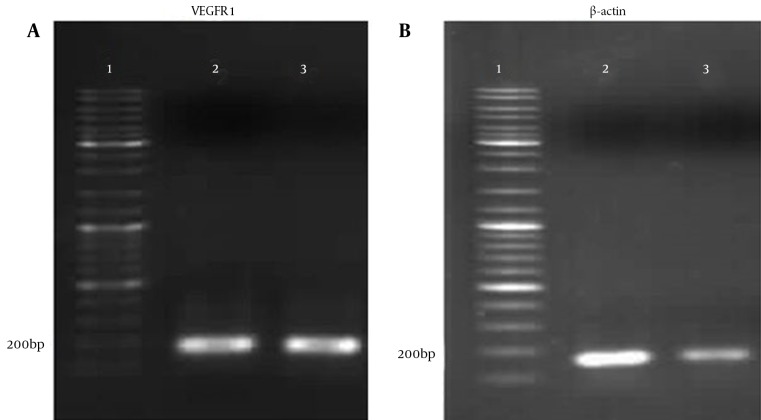
Gene expression was analyzed in cells using RT-PCR. Total RNA was extracted from both pEGFP and pEGFP-VEGFR1 transfected cells after treating with anti-VEGFR1 siRNAs. The intensity of cDNA bands demonstrated the expression of genes (well N.3 in comparison with well N.2). β-actin was used as a reference to compare gene expression in different cells. RT-PCR analysis revealed that the intensity of VEGFR1 bands was different in the two groups and mRNA level of VEGFR1 decreased significantly in cells treated with specific siRNAs compared to pEGFP transfected cells without siRNA; 54.9 ± 3.2 ng/μL mRNA for the control and 15.2 ± 0.93 ng/μL for the treated groups respectively (P < 0.001) (Figure 3).
Figure 3. Comparison of VEGFR1 Gene Expression in AGS Cells Using Semi-Quantitative RT-PCR in Presence and Absence of Anti-VEGFR1 siRNA.
1, DNA marker; 2, pEGFP-VEGFR1 transfected cells without siRNA treatment; 3, pEGFP-VEGFR1 transfected cells after treatment with siRNA. As shown, VEGFR1mRNA level decreased in presence of specific anti-VEGFR1 siRNA. β-actin was used a reference gene for comparison.
4.4. Western Blot Analysis
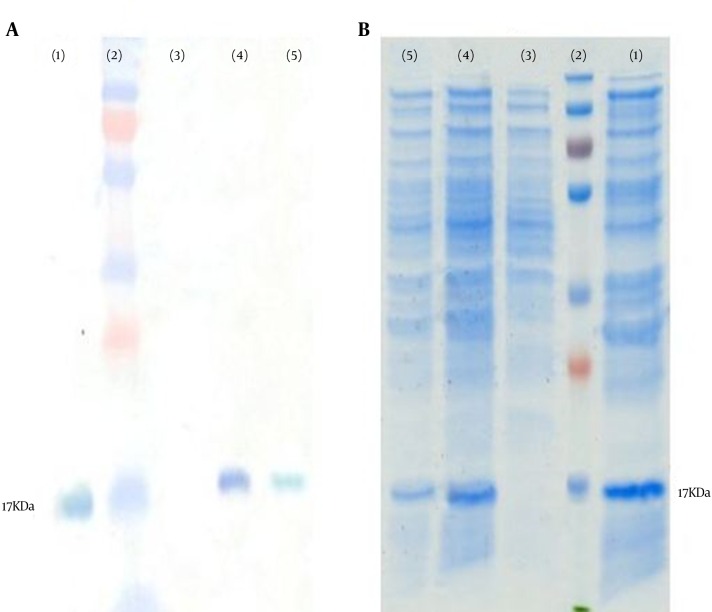
Total protein was extracted from cells and analyzed on SDS-PAGE and then western blot was used to evaluate the VEGFR1 protein expression in AGS cells. As indicated by Figure 4, pEGFP-VEGFR1 transfected cells expressed a 17 kDa VEGFR1 band while cells transfected with pEGFP vector did not express any VEGFR1. Treating transfected cells with anti-VEGFR siRNA causes significant reduction in VEGFR1 protein level compared to the control group. This finding showed that the use of anti-VEGFR1 siRNA, specifically degraded VEGFR1 mRNA and influenced its protein production (well N.5 in compare with well N.4). β-actin was used as a positive control in this experiment. The VEGFR1 concentration was 63.45 ± 4.6 μg/mL for the control and 19.71 ± 2.3 μg/mL for treated group which was a significant decrease (P < 0.003).
Figure 4. Analysis of siRNA Effect on VEGFR1 Protein Expression in AGS Cells Using.
A, western blot and B, SDS-PAGE; 1, β-actin as positive control; 2, Protein marker; 3, AGS cells transfected with pEGFP vector as negative control; 4, pEGFP-VEGFR1 transfected cells without siRNA treatment; 5, pEGFP-VEGFR1 transfected cells after treatment with siRNA. As shown, a 17 kDa protein was expressed in cells transfected with pEGFP-VEGFR1 and transfection with siRNA significantly reduced the level of VEGFR1 protein.
5. Discussion
There are different anti-VEGF agents including chemicals and antibodies that are used to control angiogenesis (16), in addition, inhibition of VEGF receptor1 is a main point in angiogenesis and also is another strategy which can block several VEGF family including VEGF-A at once. VEGF receptors are considered for cancer therapy and production of anti-cancer drugs (17). Several VEGFR inhibitors such as SU6668, ZD6474, PTK787 compounds and mono-clonal antibodies are developed (18).
RNAi is a post translational silencing mechanism of gene expression which targeting mRNA. Small interference RNA (siRNA) has attracted great attention between researchers as a new and powerful approach (19); therefore, the current study used this technology. A powerful point in siRNA usage is the less immune system stimulation. The siRNA is used as a therapeutic strategy to control angiogenesis through inhibition of different angiogenesis factors (12). A research on RPE cells demonstrated that inhibiting the expression of VEGF and HIF-α factors using specific siRNA prevented angiogenesis through down-regulating molecules involved in angiogenesis such as neurophilin, angiogenin, interleukin 6 and 8, MCP-1 and TGF-β (2). Applying siRNAs against VEGF in a mouse model showed significant reduction of the gene expression and prevention of tumor growth (20).
Jiang et al. showed that anti-HIF-α and anti-VEGF-165 siRNAs significantly prevented angiogenesis in mouse eyes (21). Jiang et al. by specific siRNA against HIF-α gene in vitro showed significant reduction of angiogenesis rate (22) and Xu et al. showed that anti-HIF-α siRNA leads to inhibition of angiogenesis and tumor growth in vivo (23). Gu et al. concluded that specific and non-specific siRNAs against VEGFR2 and VEGF-A decreased the expression of VEGFR2 and VEGF-A genes about 40% and 55%, respectively (12).
The current study aimed to inhibit antigenic effect of VEGF but not others; therefore, RNAi mechanism was used to down-regulate the expression of VEGFR1 gene involved in angiogenesis. For this purpose, first, VEGFR1 gene was cloned and expressed into AGS cells. Then, two different specific siRNAs were designed against VEGFR1 gene to evaluate their efficiency to inhibit gene expression. RT-PCR results indicated that specific siRNAs designed against a unique sequence of external fraction of VEGFR1 significantly silenced the gene expression at mRNA level. It is probably that siRNA acts specifically on target gene at mRNA level, but, either the silencing effect is not long enough or gene product has big half-life; therefore the protein level does not reduce. The Western blot result showed that the effect of anti-VEGFR1 siRNAs was enough to significantly reduce the protein level of VEGFR1. Results showed that both siRNAs silenced the target gene to an approximately same level. The current study, in accordance with the other studies, showed that siRNAs can specifically and efficiently suppress expression of different genes involved in angiogenesis including VEGFR1 and thus it can be useful to inhibit angiogenesis and tumor growth.
The present study employed the process known as RNAi to down regulate the expression of VEGFR1 gene using specific siRNAs. Results showed that the specific siRNAs can be considered as appropriate molecules to (31.06% decrease in the protein level and 27.68% decreased in mRNA) reduce the expression of specific genes. The designed siRNAs reduced VEGFR1 level and can be used for VEGFR1 signaling studies and in vivo angiogenesis inhibition as a therapeutic approach.
Acknowledgments
The authors wish to thank the chemical injuries research center and nano-biotechnology research center of Baqiyatallah University of Medical Sciences for project support and technical help.
Footnotes
Authors’ Contribution:Study concept and design: Ali Zarei Mahmudabadi and Moslem Jafari Sani; acquisition of data: Majid Rahmati and Foad Yazdi; analysis and interpretation of data: Moslem Jafari Sani; drafting of the manuscript: Moslem Jafari Sani; critical revision of the manuscript for important intellectual content: Javad Alizadeh; statistical analysis: Majid Rahmati; administrative, technical, and material support: Ali Zarei Mahmudabadi, Moslem Jafari Sani; study supervision: Ali Zarei Mahmudabadi.
Financial Disclosure:Ali Zarei Mahmudabadi reported receiving research grants and honoraria.
Funding/Support:This study was supported in part by grant from the Baqiyatallah University of Medical Sciences.
References
- 1.Kim B, Suvas S, Sarangi PP, Lee S, Reisfeld RA, Rouse BT. Vascular endothelial growth factor receptor 2-based DNA immunization delays development of herpetic stromal keratitis by antiangiogenic effects. J Immunol. 2006;177(6):4122–31. doi: 10.4049/jimmunol.177.6.4122. [DOI] [PubMed] [Google Scholar]
- 2.Forooghian F, Das B. Anti-angiogenic effects of ribonucleic acid interference targeting vascular endothelial growth factor and hypoxia-inducible factor-1alpha. Am J Ophthalmol. 2007;144(5):761–8. doi: 10.1016/j.ajo.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Zheng M, Klinman DM, Gierynska M, Rouse BT. DNA containing CpG motifs induces angiogenesis. Proc Natl Acad Sci U S A. 2002;99(13):8944–9. doi: 10.1073/pnas.132605599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenyon BM, Voest EE, Chen CC, Flynn E, Folkman J, D'Amato RJ. A model of angiogenesis in the mouse cornea. Invest Ophthalmol Vis Sci. 1996;37(8):1625–32. [PubMed] [Google Scholar]
- 5.Zheng M, Schwarz MA, Lee S, Kumaraguru U, Rouse BT. Control of Stromal Keratitis by Inhibition of Neovascularization. Am J Pathol. 2001;159(3):1021–9. doi: 10.1016/s0002-9440(10)61777-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber HP, Dixit V, Ferrara N. Vascular endothelial growth factor induces expression of the antiapoptotic proteins Bcl-2 and A1 in vascular endothelial cells. J Biol Chem. 1998;273(21):13313–6. doi: 10.1074/jbc.273.21.13313. [DOI] [PubMed] [Google Scholar]
- 7.Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr Rev. 1997;18(1):4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 8.Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, et al. Vascular Endothelial Growth Factor Regulates Endothelial Cell Survival through the Phosphatidylinositol 3'-Kinase/Akt Signal Transduction Pathway: REQUIREMENT FOR Flk-1/KDR ACTIVATION. J Biol Chem. 1998;273(46):30336–43. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 9.Kim B, Tang Q, Biswas PS, Xu J, Schiffelers RM, Xie FY, et al. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165(6):2177–85. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng M, Deshpande S, Lee S, Ferrara N, Rouse BT. Contribution of vascular endothelial growth factor in the neovascularization process during the pathogenesis of herpetic stromal keratitis. J Virol. 2001;75(20):9828–35. doi: 10.1128/JVI.75.20.9828-9835.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura H, Sasaki Y, Uno M, Yoshikawa T, Asano T, Ban HS, et al. Synthesis and biological evaluation of benzamides and benzamidines as selective inhibitors of VEGFR tyrosine kinases. Bioorg Med Chem Lett. 2006;16(19):5127–31. doi: 10.1016/j.bmcl.2006.07.075. [DOI] [PubMed] [Google Scholar]
- 12.Gu L, Chen H, Tuo J, Gao X, Chen L. Inhibition of experimental choroidal neovascularization in mice by anti-VEGFA/VEGFR2 or non-specific siRNA. Exp Eye Res. 2010;91(3):433–9. doi: 10.1016/j.exer.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Autiero M, Luttun A, Tjwa M, Carmeliet P. Placental growth factor and its receptor, vascular endothelial growth factor receptor-1: novel targets for stimulation of ischemic tissue revascularization and inhibition of angiogenic and inflammatory disorders. J Thromb Haemost. 2003;1(7):1356–70. doi: 10.1046/j.1538-7836.2003.00263.x. [DOI] [PubMed] [Google Scholar]
- 14.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4(6):609–15. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–85. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrara N, Hillan KJ, Novotny W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem Biophys Res Commun. 2005;333(2):328–35. doi: 10.1016/j.bbrc.2005.05.132. [DOI] [PubMed] [Google Scholar]
- 17.Frumovitz M, Sood AK. Vascular endothelial growth factor (VEGF) pathway as a therapeutic target in gynecologic malignancies. Gynecol Oncol. 2007;104(3):768–78. doi: 10.1016/j.ygyno.2006.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glade Bender J, Yamashiro DJ, Fox E. Clinical development of VEGF signaling pathway inhibitors in childhood solid tumors. Oncologist. 2011;16(11):1614–25. doi: 10.1634/theoncologist.2011-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadj-Slimane R, Lepelletier Y, Lopez N, Garbay C, Raynaud F. Short interfering RNA (siRNA), a novel therapeutic tool acting on angiogenesis. Biochimie. 2007;89(10):1234–44. doi: 10.1016/j.biochi.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Kanazawa T, Sugawara K, Tanaka K, Horiuchi S, Takashima Y, Okada H. Suppression of tumor growth by systemic delivery of anti-VEGF siRNA with cell-penetrating peptide-modified MPEG-PCL nanomicelles. Eur J Pharm Biopharm. 2012;81(3):470–7. doi: 10.1016/j.ejpb.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Xia XB, Xu HZ, Xiong Y, Song WT, Xiong SQ, et al. Inhibition of retinal neovascularization by gene transfer of small interfering RNA targeting HIF-1alpha and VEGF. J Cell Physiol. 2009;218(1):66–74. doi: 10.1002/jcp.21566. [DOI] [PubMed] [Google Scholar]
- 22.Jiang M, Wang CQ, Wang BY, Huang DJ. [Inhibitory effect of siRNA targeting HIF-1alpha on differentiation of peripheral blood endothelial progenitor cells]. Ai Zheng. 2005;24(11):1293–300. [PubMed] [Google Scholar]
- 23.Xu HZ, Liu SZ, Xiong SQ, Xia XB. [HIF-1alpha siRNA reduces retinal neovascularization in a mouse model of retinopathy of prematurity]. Zhongguo Dang Dai Er Ke Za Zhi. 2011;13(8):680–3. [PubMed] [Google Scholar]