Abstract
Structural centrosome aberrations are frequently observed in early stage carcinomas, but their role in malignant transformation is poorly understood. Here, we examined the impact of overexpression of Ninein-like protein (Nlp) on the architecture of polarized epithelia in three-dimensional mammospheres. When Nlp was overexpressed to levels resembling those seen in human tumors, it formed striking centrosome-related bodies (CRBs), which sequestered Ninein and affected the kinetics of microtubule (MT) nucleation and release. In turn, the profound reorganization of the MT cytoskeleton resulted in mislocalization of several adhesion and junction proteins as well as the tumor suppressor Scribble, resulting in the disruption of epithelial polarity, cell-cell interactions and mammosphere architecture. Remarkably, cells harboring Nlp-CRBs displayed an enhanced proliferative response to epidermal growth factor. These results demonstrate that structural centrosome aberrations cause not only the disruption of epithelial polarity but also favor overproliferation, two phenotypes typically associated with human carcinomas.
Introduction
Epithelia are important barriers that line the cavities and external surfaces of the human body.1 In addition to protective functions, they operate as gatekeepers to facilitate absorption or secretion. Epithelial functions depend on apical basal polarity and tight cell-cell interactions mediated by specialized junctions. These properties constitute prerequisites for tissue integrity and the formation of polarized layers of tissue.1 Most human cancers derive from epithelia2 and loss of tissue organization is a prominent feature of cancer (reviewed in Lee and Vasioukhin3). What mechanisms trigger the loss of epithelial integrity during carcinogenesis remains to be fully understood.
Critical for the polarization of epithelial cells are microtubules (MTs), which sustain apical basal polarity through their intrinsic polarity and inherent dynamics.4 MT minus ends are anchored to adherens junctions and structures near the apical membrane adjacent to the lumen,4, 5 while MT plus ends extend to the basal compartment adjacent to the basement membrane. MT plus ends contribute to the positioning of basal adhesion proteins and hence promote the tight connection of epithelial cells to the basement membrane.6 Most MTs are originally nucleated at centrosomes, the main MT-organizing centers of animal cells.7, 8 Centrosomes consist of a pair of centrioles, surrounded by pericentriolar matrix.8, 9, 10, 11 The pericentriolar matrix comprises more than 100 proteins,12, 13 including γ-tubulin ring complexes that are essential for MT nucleation.14 Importantly, in epithelial cells, many MTs are released from centrosomes and anchored at the cell periphery.15, 16 Release of MTs and their transport towards peripheral anchoring sites is mediated by Ninein.16, 17 Thus, the centrosomal abundance of Ninein must be tightly regulated, as even a moderate surplus of centrosomal Ninein abrogates MT release and, in consequence, interferes with MT-dependent functions such as cell migration.18
A large proportion of human carcinomas exhibit centrosome aberrations already during early stages of carcinogenesis.19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Centrosome aberrations have traditionally been subdivided into numerical and structural aberrations.24, 29 Most numerical aberrations are characterized by centrosome amplification, most commonly caused by supernumerary centrioles that result from deregulated centriole duplication or division failure, whereas structural centrosome aberrations refer to altered shapes and compositions of the pericentriolar matrix with normal centriole numbers. Numerical aberrations and their consequences have been studied extensively.30, 31, 32, 33, 34, 35, 36 In contrast, the origin and role of structural centrosome aberrations has received comparatively little attention.37, 38, 39
Here, we have explored the consequences of structural centrosome aberrations for the architecture and function of MCF10A breast epithelial cells grown in two-dimensional (2D) and three-dimensional (3D) tissue culture. We focused on structural centrosome aberrations induced by overexpression of Ninein-like protein (Nlp).40 This distant relative of Ninein localizes preferentially to mother centrioles and contributes to MT organization in interphase cells.40 Cell cycle regulation of Nlp expression and localization is required for mitotic spindle formation40 and accurate progression through mitosis.41, 42, 43, 44, 45 Intriguingly, Nlp is strongly overexpressed in various types of human cancer (including breast cancer),46, 47, 48, 49 and Nlp overexpression is associated with enhanced proliferation as well as reduced sensitivity to anticancer treatment, which has been proposed to contribute to an adverse clinical course of disease.46, 48, 49 Supporting these findings, transgenic mice overexpressing Nlp spontaneously develop invasive breast cancer.47 Moreover, in 2D culture EGFP-Nlp was previously shown to form striking centrosome-related bodies (CRBs)40, 50 that closely resemble the structurally aberrant centrosomes seen in human cancers.33, 36, 37 Here, we demonstrate that Nlp overexpression profoundly impacts on the MT organization of epithelial cells. In 3D culture mammospheres, this results in a drastic disruption of epithelial architecture and enhanced responsiveness to growth factor stimulation.
Results
Overexpression of EGFP-Nlp prevents epithelium formation in 3D tissue culture
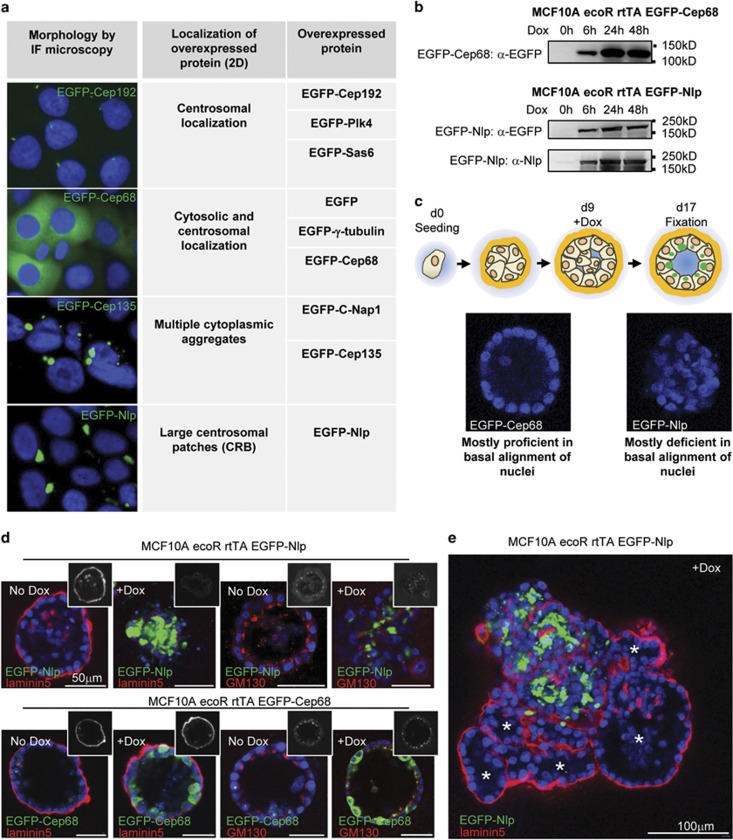
Structural centrosome aberrations in cancer cells are presumed to result from deregulated expression of centrosomal proteins.23, 24, 51 We therefore applied a retroviral system to study the effects of overexpression of selected centrosomal proteins in MCF10A breast epithelial cells (Supplementary Figure S1). Importantly, this experimental set-up made it possible to induce the overexpression of EGFP-tagged proteins at defined time points, allowing the initial morphogenesis of 3D mammospheres to remain unperturbed (Supplementary Figure S1). Analysis of a number of centrosomal proteins revealed distinct consequences of protein overexpression in 2D culture (Figure 1a). While some proteins localized primarily to centrosomes, others additionally filled up the cytoplasm or formed multiple cytoplasmic aggregates, as illustrated by the fates of Cep192, Cep68 or Cep135, respectively (Figure 1a). Most strikingly, the overexpression of Nlp caused large centrosome-associated patches, confirming and extending previous results.40, 50 In the following, we refer to these structural centrosome aberrations as CRBs (Figure 1a).
Figure 1.
Nlp CRBs interfere with apical basal polarity in 3D tissue culture. (a) Localization of overexpressed EGFP-tagged centrosome proteins in 2D culture, as visualized by fluorescence microscopy. (b) Western blots indicate levels of overexpression of EGFP-tagged centrosome proteins after induction (Dox) over time. (c) Top panel: schematic illustrating experimental design for protein induction in 3D culture; bottom panel: DAPI staining of mammospheres, illustrating proficiency or deficiency in basal alignment of nuclei. See also Supplementary Figure S1. (d) Analysis of mammospheres expressing EGFP-Nlp or EGFP-Cep68 by immunofluorescence microscopy. EGFP-Nlp and EGFP-Cep68 are shown in green, Laminin5 and GM130 in red, and DNA in blue (DAPI). See also Supplementary Figure S2. (e) Chimeric mammosphere cluster, illustrating the impact of Nlp CRBs on 3D architecture. Staining as in b. Asterisks indicate parts of the cluster showing glandular morphology and basal laminin5 staining.
As Nlp is frequently overexpressed in human cancers47 and considering that Nlp-induced CRBs closely resemble the structural centrosome aberrations seen in cancer tissue sections,37, 39 we asked how overexpression of Nlp would affect the morphology of mammospheres. Generation of inducible MCF10A cell lines allowed maximal expression of EGFP-Nlp within 24–48 h of doxycycline induction, and similar kinetics of induction were observed for EGFP-Cep68 analyzed for control (Figure 1b). Importantly, the levels of Nlp achieved in these experiments were in a comparable range with those reported for human cancers.47 Mammospheres were allowed to grow until they had reached maximal sizes with polarized epithelia, before overexpression of centrosomal proteins was induced at day 9 and the consequences studied 8 days later (Figure 1c and Supplementary Figure S1b). The proficiency of mammospheres to align their nuclei in a spherical layer provided a first convenient readout to distinguish aberrant from normal tissue architecture (Figure 1c). This analysis revealed that the overexpression of EGFP-Nlp abolished the proper basal alignment of nuclei in most mammospheres, whereas the overexpression of EGFP-Cep68 did not detectably disrupt mammosphere architecture (Figure 1c). As expected, the penetrance of the Nlp-induced phenotype correlated with the extent of overexpression (Supplementary Figures S1c and d).
To more directly address the impact of Nlp CRBs on epithelial polarity, we also probed mammospheres for the basement membrane component laminin5 and the Golgi protein GM130, markers for basal and apical (luminal) compartments, respectively. As shown in Figure 1d, Nlp CRBs caused a major disruption of epithelial polarity, whereas overexpression of Cep68 did not. The consequences of Nlp CRBs on tissue architecture could also be seen in chimeric clusters of mammospheres where parts expressing little or no transgene were located immediately adjacent to cells expressing high levels of EGFP-Nlp (Figure 1e). While gland-like structures were preserved in the former parts of the chimeras, the latter parts exhibited a striking disruption of tissue architecture, reminiscent of tumors (Figure 1e).
To determine the specificity of the phenotype observed upon overexpression of Nlp in 3D mammospheres, we next studied the consequences of overexpression of other centrosomal proteins. When examined in the exact same assay as used for Nlp, none of the other proteins listed in Figure 1a produced major effects on mammosphere architecture, although minor effects were seen sporadically (Supplementary Figure S2). These results not only attest to the specificity of the Nlp phenotype, but also support the hypothesis that Nlp overexpression in human tumors may causally contribute to carcinogenesis.47 In light of a recent study reporting the induction of an invasive phenotype in response to centriole amplification,35 we were surprised that overexpression of the centriole duplication factors Plk4 or Sas-6 did not detectably induce invasive growth or major perturbations of epithelial cell polarity, although both proteins clearly induced centriole amplification (Supplementary Figure S2).
Nlp CRBs alter the organization of the MT cytoskeleton
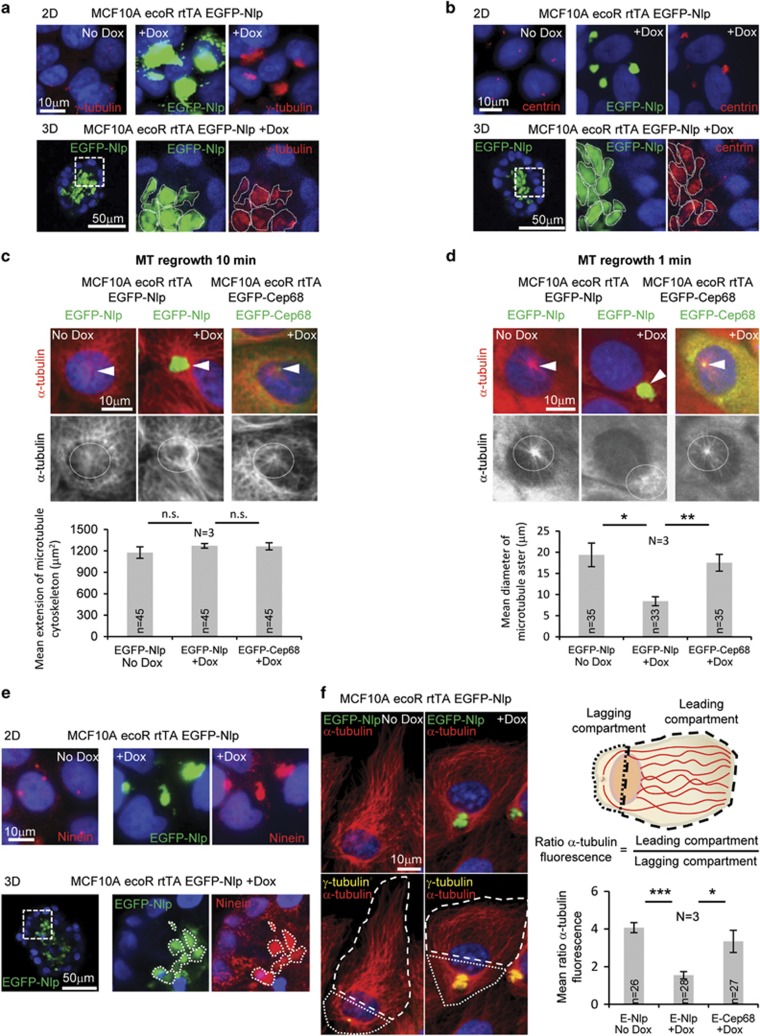
To determine the mechanisms by which excess Nlp interferes with epithelial cell organization, we examined the impact of Nlp CRBs on centriole numbers, MT organization and MT-dependent processes. No evidence was obtained for aberrations in centriole numbers upon overexpression of Nlp (data not shown), in line with previous reports.50 However, both γ-tubulin and centrin-2/3 were recruited to Nlp CRBs in both 2D and 3D tissue culture (Figures 2a and b), confirming and extending previous results.40 In these respects, Nlp CRBs are highly reminiscent of the large γ-tubulin and centrin staining structures observed in sections of breast adenocarcinoma.37, 38 To explore the impact of γ-tubulin recruitment on MT organization, we carried out MT regrowth assays in MCF10A cells (Figures 2c and d). When MTs were allowed to regrow for 10 min, Nlp CRB-containing cells generally displayed denser MT asters than controls (Figure 2c), seemingly in line with the increased MT nucleation activity of structurally aberrant centrosomes in permeabilized tumor sections.37, 39 However, during early phases of MT regrowth, notably after 1 min, we observed that Nlp CRBs caused a reduction in MT nucleation kinetics when compared with Cep68-expressing or control cells (Figure 2d). This surprising result raised the question of how to reconcile initial suppression of MT nucleation activity by excess Nlp with higher density of MT asters at steady state.
Figure 2.
Nlp CRBs anchor a surplus of MTs. (a, b) Sequestration of γ-tubulin (a) and centrin (b) to Nlp CRBs in 2D (upper panels; plus/minus Dox) and 3D cultures (lower panels; right hand panels show magnification of boxed area). γ-Tubulin and centrin in red, EGFP-Nlp in green and DNA in blue (DAPI). (c, d) MT regrowth in cells expressing Nlp or Cep68, analyzed for control, before and after protein induction (Dox). Upper panels show merged data in pseudo-color (α-tubulin in red, EGFP-Nlp and EGFP-Cep68 in green, and DNA in blue (DAPI)); arrowheads indicate the location of the centrosome/CRB. Lower panels show α-tubulin staining in grey; dashed ring illustrates MT density in proximity to centrosome/CRB. Histogram to the left shows quantification of mean extension of the MT cytoskeleton. Histogram to the right shows quantification of mean diameter of MT asters; n indicates sample size; data are from three independent experiments (N=3); error bars indicate standard deviation and P-values were derived from unpaired, two-tailed Student's t-test. (*) indicates P-value <0.05, (**) <0.01 and (***) <0.001. (e) Sequestration of Ninein to Nlp CRBs in 2D (upper panels; plus/minus Dox) and 3D cultures (lower panels; right hand panels show magnification of boxed area). Ninein in red, EGFP-Nlp in green and DNA in blue (DAPI). (f) Impact of Nlp CRB on MT reorganization in MCF10A cells in response to monolayer wounding-induced cell polarization by immunofluorescence microscopy. Pseudo-colors show α-tubulin in red, γ-tubulin in yellow, EGFP-Nlp in green and DNA in blue (DAPI) (left panel); cells were compartmentalized (white dashed and dotted lines) to monitor MT reorganization, as illustrated schematically in the upper right panel (dashed lines delineate the leading compartment, dotted lines the lagging compartment). Histogram in the lower right panel shows quantifications of the ratios of MT fluorescence intensities between the leading and lagging compartments; n indicates sample size; data are from three independent experiments (N=3); error bars indicate standard deviation and P-values were derived from unpaired, two-tailed Student's t-test. (*) indicates P-value <0.05, (**) <0.01 and (***) <0.001.
The density of centrosomal MT asters at steady state is determined not only by the rate of MT nucleation, but also by the rate of MT release. Thus, we explored the possibility that excess Nlp might conceivably suppress both MT nucleation and release. To this end, we examined the localization of Ninein, one of the key components regulating MT release from centrosomes.16, 17 Interestingly, Ninein was clearly recruited to Nlp CRBs in both 2D and 3D cultures (Figure 2e). As the centrosomal accumulation of Ninein was previously shown to abrogate MT release from centrosomes,18 these results indicate that the concomitant accumulation of Nlp and Ninein reduces the rates of both MT nucleation and release. Assuming that the latter effect prevails over time, this readily explains the high MT density observed around Nlp CRBs (see also Casenghi et al.40).
To corroborate the notion that accumulation of Ninein has a major role in tethering MTs to Nlp-induced CRBs, we sought a way to determine the impact of Ninein mislocalization on CRB structure and MT organization. We took advantage of a previous study demonstrating that the overexpression of a C-terminal Ninein fragment (Supplementary Figure S3a) acts in a dominant-negative manner and causes the mislocalization of endogenous Ninein.15 High levels of C-terminal Ninein fragment indeed caused a strong reduction in centrosomal Ninein signals in control cells, confirming the functionality of the expression construct (Supplementary Figure S3b). Moreover, when the same C-terminal Ninein fragment was expressed in cells overexpressing EGFP-Nlp, we observed a marked loss of Ninein from Nlp-induced CRBs (Supplementary Figure S3c). Concomitantly, CRBs underwent fragmentation (Supplementary Figures S3c and d) and MT networks lost their focus on CRBs (Supplementary Figure S3d). Together, these data support the view that Ninein accumulation contributes to the characteristic shape and properties of Nlp-induced CRBs.
To investigate the functional consequences of the altered dynamics in MT organization induced by Nlp CRBs, we next examined the ability of the MT cytoskeleton to reorganize in response to monolayer wounding. This assay induces rapid cell polarization towards the wound edge, providing an opportunity to monitor MT re-organization between leading and lagging cell compartments (for definition, see Figure 2f). In line with the observation that centrosomes of epithelial cells do not generally re-orient towards the wound edge,52 108 out of 120 control MCF10A cells at the wound retained their centrosomes in the lagging compartment (data not shown). Yet, in both control and Cep68-expressing cells, the bulk of MTs redistributed to the leading compartment (Figure 2f). In striking contrast, this reorganization of the MT cytoskeleton in response to wounding was markedly suppressed in cells harboring Nlp CRBs (Figure 2f). This strongly supports our conclusion that excess Nlp suppresses the release of MTs from centrosomes and demonstrates that Nlp CRBs prevent the reorganization of the MT network in response to cell polarization.
Nlp CRBs affect mammosphere architecture
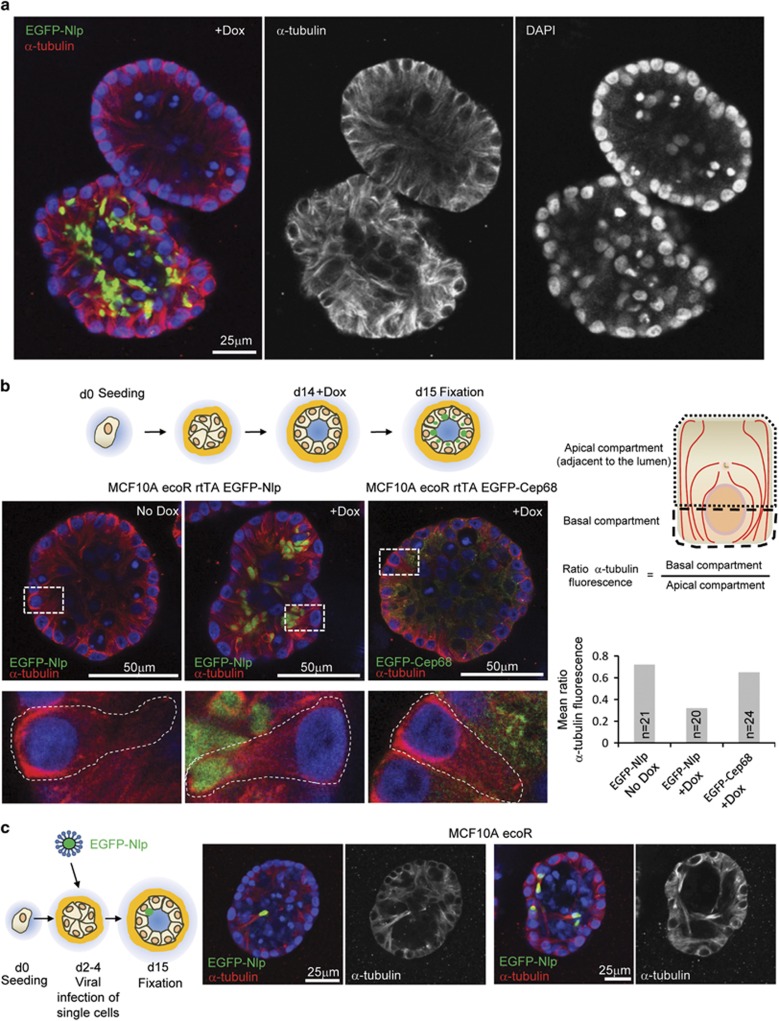
Having established a profound effect of Nlp CRBs on the dynamics of the MT cytoskeleton in 2D culture, we considered it important to extend these results to polarized single cell layers of epithelium in 3D culture. Mammospheres harboring Nlp CRBs lost the prominent enrichment of MTs near the outer confinement and instead showed higher MT densities near the lumen, demonstrating major reorganization of the MT cytoskeleton also in 3D culture (Figure 3a). However, because mammospheres progressively change their architecture in response to Nlp CRBs, it was difficult to analyze the MT phenotype in individual cells. To circumvent this problem, we induced Nlp expression at different times during 3D culture and compared the kinetics of alterations in the MT cytoskeleton relative to the onset of architectural alterations. We indeed found conditions that induced detectable reorganization of the MT cytoskeleton concomitant with CRB formation but prior to major alterations in tissue architecture (Supplementary Figure S4; shaded panel). This made it possible to study the impact of CRBs on the MT cytoskeleton of individual cells (Figure 3b and Supplementary Figure S4). In line with the data obtained in 2D cultures, cells harboring Nlp CRBs typically exhibited a higher density of α-tubulin in the apical, CRB-containing compartments, adjacent to the lumen of the mammosphere (Figure 3b and Supplementary Figure S4).
Figure 3.
Nlp CRBs alter the organization of MTs in polarized epithelia of mammospheres. (a) Impact of Nlp CRBs on MT organization in mammospheres. Micrographs show juxtaposition of two mammospheres, only one of which expresses Nlp in response to Dox. Left panel: merge of pseudo-colors illustrating EGFP-Nlp in green, α-tubulin in red and DNA in blue (DAPI). Middle and right panels: single stainings for α-tubulin highlight disruption of MT organization and concomitant misalignment of nuclei, respectively. (b) Polarization of MT reorganization in mammosphere epithelia at single cell level. Top left panel: schematic illustrating experimental design for protein induction in 3D culture; bottom left panel: micrographs show impact of Nlp CRBs on MT organization (center). Panels show overviews (above) and magnifications of box areas (below). Staining as in a. Right panels: schematic illustrates compartmentalization of polarized mammosphere epithelial cells into apical (luminal) and basal compartments. Histogram shows quantification of ratio of α-tubulin fluorescence intensities in the two compartments (n indicates sample size). Data were obtained from all cells shown in the micrographs of panel (b); virtually identical results were obtained in three independent experiments. See also Supplementary Figure S3. (c) Reorganization of MT cytoskeleton in single cells harboring Nlp CRBs in otherwise unperturbed mammospheres. Viral infections were carried out as indicated in the cartoon to the left. Micrographs to the right illustrate the impact of Nlp CRB on MT organization. Staining in left hand panels as in a, while right hand panels show α-tubulin staining alone (grey).
Tumors are generally thought to originate from individual founder cells within otherwise healthy tissues. To phenocopy this condition in mammospheres and study the behavior of isolated CRB-harboring cells in an otherwise unperturbed epithelial structure, whole mammospheres were incubated with lentiviral particles to stably integrate expression cassettes encoding EGFP-Nlp into the genome of single cells. This approach led to the generation of mammospheres containing isolated cells harboring Nlp CRBs. Importantly, these Nlp CRB-containing cells characteristically showed an intense α-tubulin staining adjacent to the CRB in the apical compartment near the lumen, in stark contrast to normal bystander cells (Figure 3c). This indicates that the localized emergence of structural centrosome aberrations is sufficient to trigger a reorganization of the MT cytoskeleton, which in turn exerts a profound impact on epithelial architecture.
Nlp CRBs interfere with the positioning of basal adhesion and lateral junction proteins
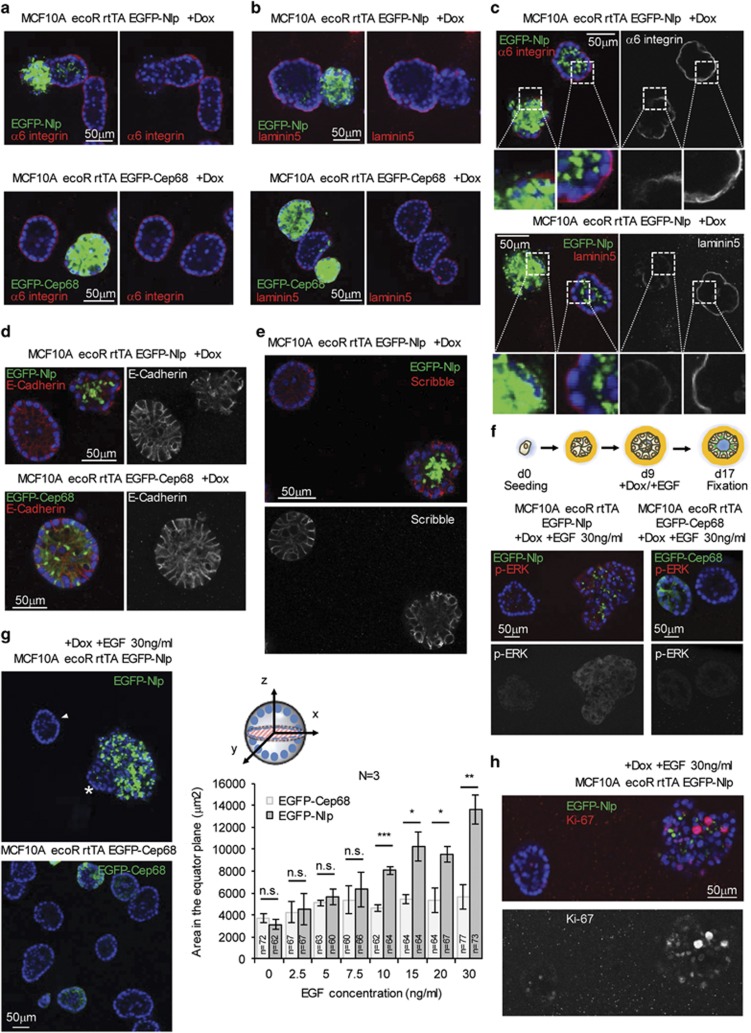
Epithelial cell biology is characterized by two key aspects. First, robust adhesion to the basement membrane is essential for line-up of polarized epithelial cells and maintenance of epithelial architecture.1, 53 Second, tight intercellular connections between neighboring cells are critical for proper lateral adhesion.1 This prompted us to ask to what extent the reduced density of MTs in the basal compartment of cells harboring Nlp CRBs affects the positioning of basal adhesion and lateral junction proteins. While mammospheres formed from cells expressing excess EGFP-Cep68 exhibited proper α6-integrin localization, this basal adhesion component was partially lost from the basal compartment in mammospheres expressing Nlp CRBs (Figures 4a and c), highly reminiscent of early stage human breast cancer.54 Likewise, the ligand of α6-integrin, laminin5, was largely lost from the basal membrane in mammospheres expressing Nlp but normally distributed in Cep68 controls (Figures 4b and c; see also Figures 1d and e). Furthermore, the extent to which these basal adhesion proteins were lost from the basal cortex clearly correlated with the extent to which Nlp CRB formation disrupted tissue architecture (Figure 4c). To determine whether Nlp CRBs also perturb the localization of lateral junction proteins, we probed for E-Cadherin, an important mediator of junction belt initiation.1 While E-Cadherin decorated the lateral boundaries between neighboring cells in control and EGFP-Cep68-overexpressing mammospheres, those harboring Nlp CRBs clearly showed more dispersed and irregular distribution of E-Cadherin (Figure 4d). In addition to being important for junction belt organization, E-Cadherin also recruits the polarity protein and tumor suppressor Scribble to the basolateral membrane.55 As Scribble is frequently lost or mis-localized in breast cancer,55, 56, 57, 58 we next examined the consequences of Nlp CRBs on Scribble localization. Consistent with the results obtained for E-Cadherin, mammospheres harboring Nlp CRBs displayed a profoundly dispersed and irregular Scribble staining pattern when compared with controls (Figure 4e). Taken together these results demonstrate that structural centrosome aberrations, as triggered here by excess Nlp, impact not only the MT cytoskeleton but also profoundly disturb key proteins implicated in epithelial adhesion and cell-cell interactions.
Figure 4.
Nlp CRBs impair localization of basal and lateral adhesion proteins and confer enhanced responsiveness to EGF stimulation. (a, b) Impact of Nlp CRBs on basal localization of α6-integrin (a) and laminin5 (b) by immunofluorescence (IF) microscopy. Pseudocolors show EGFP-Nlp- and EGFP-Cep68 in green, α6-integrin and laminin5 in red, and DNA in blue (DAPI). (c) Panels to the left show merge of pseudocolors, panels to the right show single stainings in grey for α6-integrin (above) and laminin5 (below). Overviews and blow-ups illustrate correlation between Nlp CRBs and displacement of basal adhesion proteins. (d, e) Impact of Nlp CRBs on lateral localization of E-Cadherin (d) and Scribble (e) by IF microscopy. E-Cadherin and Scribble in red, EGFP-Nlp and EGFP-Cep68 in green, and DNA in blue (DAPI). (f, g, h) Nlp CRBs confer enhanced responsiveness to EGF. EGF treatment of mammospheres, as illustrated schematically in f, enhance p-ERK1/2 staining (f) and, concomitantly, cause mammospheres to grow to larger size (g). Representative micrographs in g show effect of EGF on the proliferative expansion of mammospheres harboring Nlp CRBs; EGFP-Nlp in green, DNA in blue (DAPI); arrowhead points to control mammosphere showing minimal Nlp expression and asterisk marks part of chimeric mammosphere that fails to express EGFP-Nlp. Histogram to the right shows quantification of dose-dependent response to EGF by measurement of equatorial sections (illustrated in graph; dashed plane (red)). Representative micrographs in h show Ki-67-positive cells in mammosphere harboring Nlp CRBs next to a non-expressing mammosphere; upper micrograph shows merge of EGFP-Nlp in green, DNA in blue (DAPI) and Ki-67 in red. Lower micrograph shows Ki-67 in greyscale. n indicates sample size; data are from three independent experiments (N=3); error bars indicate standard deviation and P-values were derived from unpaired, two-tailed Student's t-test. (*) indicates P-value <0.05, (**) <0.01 and (***) <0.001.
Deregulation of epithelial architecture by Nlp CRBs increases responsiveness to growth stimulation
Loss or aberrant localization of Scribble has been reported to enhance cell proliferation in response to epidermal growth factor (EGF).56, 59 This prompted us to speculate that Nlp CRBs might disrupt not only MT networks and epithelial organization, but additionally confer a growth advantage to mammospheres. To explore this hypothesis, we examined the activation state of the MAP kinase pathway, as monitored by phospho-ERK staining (Figure 4f), as well as the proliferative response of mammospheres to EGF (Figure 4g). We found that cells harboring Nlp CRBs displayed a more intense phospho-ERK staining in response to growth factor stimulation than controls expressing Cep68 (Figure 4f). Moreover, the surplus in MAP kinase pathway activity clearly translated into increased proliferation of cells harboring Nlp CRBs. While increasing doses of EGF had only a moderate effect on mammospheres expressing Cep68 (Figure 4g), mammospheres harboring Nlp CRBs showed a clear dose response and grew to progressively larger sizes (Figure 4g, right panel). Remarkably, differential responses to high doses of EGF could be seen even within single mammospheres. Regions with high Nlp expression showed marked expansion, while parts characterized by low expression showed no significant response (Figure 4g). Finally, when grown in the presence of a high dose of EGF, mammospheres harboring Nlp CRBs also showed increased numbers of cells staining positive for the proliferation marker Ki-67 (Figure 4h). This supports our conclusion that structural centrosome aberrations induced by Nlp-CRBs favor proliferation of disrupted epithelia in response to EGF, although additional effects on cell growth (volume) are not excluded.
Discussion
Structural centrosome aberrations are common in human tumors, but their impact on the biological properties of tumor cells is poorly understood. To explore the mechanistic consequences of structural centrosome aberrations on epithelial organization, we have developed a 3D culture model based on overexpression of the centrosomal protein Nlp in mammospheres derived from non-transformed MCF10A cells. We show that overexpression of Nlp results in the formation of CRBs, highly reminiscent of the structural centrosome aberrations described in human cancers.37, 38 Furthermore, we demonstrate that Nlp CRBs trap MT minus ends through sequestration of the MT release factor Ninein. This in turn affects the organization of the MT cytoskeleton as well as the localization of adhesion and junction proteins, resulting in a profound disruption of apical basal polarity. Finally, we show that the Nlp CRB-induced alterations in epithelial architecture favor sustained proliferation in response to growth factor stimulation. Collectively, these results demonstrate that aberrant expression of a single centrosomal protein in otherwise non-transformed epithelial cells induces profound structural centrosome aberrations. These in turn confer properties to 3D mammospheres that closely resemble typical features of human carcinoma cells.
The Nlp CRB model analyzed here shares several important properties with CRBs that are commonly observed in sections of primary human tumor tissue, notably their altered shape and size and the striking accumulation of both γ-tubulin and centrin.37, 38 Moreover, we emphasize that the centrosomal protein Nlp, used here to induce CRBs, is frequently overexpressed in human cancers.47 Although it is difficult to directly compare expression levels between different samples, we emphasize that the extent of Nlp overexpression in MCF10A cells (Figure 1b) appears comparable with that observed in tumor material.47 Previously, CRB formation has been correlated with an increase in the abundance of MTs associated with these structures, both in cultured cells40 and in sections of primary tumor samples.37, 39 However, although our data confirm that CRB formation results in increased MT density, presumably reflecting enhanced MT stability at steady state, MT regrowth assays revealed that excess Nlp exerts a suppressive effect on MT nucleation. This apparent paradox could be resolved by showing that Nlp CRBs also sequester Ninein, which then suppresses MT release,18 resulting in a net increase of MT accumulation at CRBs over time. These observations can explain how structural centrosome aberrations, induced here by Nlp, can trap MT minus ends and thereby impair the ability of the MT network to reorganize in polarizing cells. This in turn has profound consequences for the organization of epithelial cell layers.
In a recent study, the overexpression of the centrosome duplication factor Plk460, 61 was reported to induce invasive growth of epithelial cells.35 As this work was performed using a 3D culture assay similar to the one used here, we were initially surprised by the apparent lack of phenotype associated with overexpression of either Plk4 or Sas-6 in our mammospheres. Although both of these proteins produced the expected centriole amplification, we saw no evidence for mammosphere deformation indicative of invasive growth. When interpreting these negative results, it is important to consider that mammosphere formation involves different stages.62 While we generally allowed mammospheres to reach maximal size before inducing centrosomal proteins at day 9, Godinho and coworkers35 induced Plk4 overexpression before seeding of cells into matrigel, resulting in cells harboring supernumerary centrioles prior to 3D expansion and initial polarization.62 Furthermore, we note that Godinho and coworkers cultured mammospheres in the presence of type I collagen, which is known to enhance the stiffness of matrigel preparations and sensitize the assay for detection of invasive growth.63 Thus, we consider it likely that our inability to detect invasive growth in response to centrosome amplification reflects the above differences in experimental design and/or assay conditions.
Interestingly, the proficiency of epithelial cells to generate non-centrosomal MTs correlates with differentiation state.64 While cells residing in stem cell compartments mostly exhibit MT asters emanating from the centrosome, differentiated epithelial cells as well as neurons display an abundance of non-centrosomal MTs.64, 65, 66 Release from centrosomes and vectorial transport of MT minus ends depends on Ninein16 and, as we show here, this process is suppressed by Nlp CRBs. We emphasize that structural centrosome aberrations are not the only cause for aberrant MT organization in cancer cells and several important tumor suppressors, including adenomatous polyposis coli, RASSF1A, VHL, E-Cadherin and merlin, were reported to perturb MT networks during early carcinogenesis.67 However, whereas mutational inactivation of a tumor suppressor generally requires two genetic hits, upregulation of Nlp (or other centrosomal proteins conferring similar structural centrosome aberrations) is expected to exert a dominant effect in response to a single genetic event.
The functionality of epithelia depends on their ability to form uniformly polarized layers of differentiated cells.1 From the perspective of carcinogenesis, it is important that the formation of epithelial cell-cell interactions have been recognized as an important barrier against the outgrowth of transformed single cells68, 69, 70 (reviewed in Fujita,71 Hogan,72 and Yamauchi and Fujita73). Conversely, detachment from the epithelial layer and intraluminal proliferation of cells constitutes a hallmark of early breast cancer.68, 74 Concomitantly, the loss of polarity that accompanies the evasion from the epithelial sheet abolishes the segregation of growth factor receptors and their activators into distinct membrane domains, allowing promiscuous interactions.75, 76 Our present study shows that perturbation of the MT network through Nlp CRBs triggers a profound disruption of epithelial integrity, as illustrated by mislocalization of α6-integrin, laminin5 and E-Cadherin. Moreover, the tumor suppressor Scribble, normally associated with basolateral membranes, was found to be dispersed in cells harboring Nlp CRBs. In consideration of data showing mislocalization and downregulation of Scribble in early breast cancer55, 56, 57, 58 and reports linking Scribble depletion to activation of the MAP kinase pathway,57, 59, 77 this observation prompted us to explore the response of mammospheres to growth factor stimulation. Strikingly, when mammospheres were treated with EGF, we observed that structures harboring Nlp CRBs grew to larger sizes than controls. Moreover, the extent of proliferative response was proportional to EGF concentration. These results support the view that structural centrosome aberrations disrupt not only epithelial architecture but also favor cell expansion in response to proliferative stimuli.
In conclusion, our results demonstrate that structural centrosome aberrations exert a profound impact on the biological properties of developing tumor cells, indicating that they deserve as much attention in future studies as numerical centrosome aberrations.
Materials and methods
Cell culture
MCF10A ecoR cells, authenticated by Short Tandem Repeat profiling, were a kind gift from Tilman Brummer (University of Freiburg) and grown as described previously.62 Phoenix ampho cells and HEK293T cells (provided by Stefan Zimmermann and Ralph Wäsch, respectively; University Medical Center Freiburg) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, sodium pyruvate and Pen/Strep. Tissue cultures were routinely tested for mycoplasma contamination by PCR using growth medium from high density cultures as a template. Doxycycline-inducible MCF10A ecoR cell lines allowing expression of selected EGFP-tagged centrosomal proteins were generated as illustrated in Supplementary Figure S1a. For antibiotic selection, hygromycin and puromycin were used at concentrations of 100 μg/ml and of 2 μg/ml, respectively. Doxycycline was used at a final concentration of 2.5 μg/ml. For generation of mammospheres in 3D culture, MCF10A ecoR cells were plated and grown in matrigel using the overlay method as described previously.62
Generation of retroviral and lentiviral expression constructs
The retroviral vector pBabe rtTA Hygro was kindly provided by Cheuk Leung (University of Minnesota).68 Coding sequences of EGFP-Nlp, EGFP-Cep68, EGFP, EGFP-γ-tubulin, EGFP-Cep135, EGFP-C-Nap1, EGFP-Cep192, EGFP-Plk4 and EGFP-Sas-6 were PCR-amplified and ligated into pRetroX tightPur (Clontech, Mountain View, CA, USA). The lentiviral vector pLeGO-iC2 was obtained from Boris Fehse via Addgene (Addgene plasmid #27345; Cambridge, MA, USA).78 For the generation of pLeGO EGFP-Nlp-iC2, the sequence encoding EGFP-Nlp was PCR-amplified and ligated into LeGO-iC2. For the generation of pLeGO Ninein1885-2134-iC2, the sequence encoding Ninein1885-2134 was PCR-amplified and ligated into LeGO-iC2. The template plasmid encoding full-length human Ninein was a kind gift from Yi-Ren Hong (Kaohsiung Medical University, Taiwan). The packaging plasmids psPAX2 (Addgene plasmid #12260) and pMD2.G (Addgene plasmid #12259) were obtained from Didier Trono via Addgene.79 Retroviral particles were generated by transfection of 10 μg of calcium phosphate-precipitated retroviral plasmid DNA onto Phoenix ampho cells according to the Nolan lab protocol (Stanford University Medical Center). Lentiviral particles were generated by cotransfecting 8 μg of lentiviral plasmid DNA with packaging plasmids (8 μg of psPAX2 and 4 μg of pMD2.G) onto HEK293T cells. Viral transductions were carried out by incubating MCF10A ecoR cells in virus particle-containing supernatant in the presence of 8 μg/ml polybrene.
Fluorescence microscopy
Immunolabeling of 2D cultures was performed using standard fixation and labeling protocols, using ice-cold methanol.40 Procedures for staining of 3D mammospheres were adapted from previously published protocols.62 For detection of γ-tubulin, centrin and phospho-p44/42 MAPK mammospheres were fixed using ice-cold methanol (15 min). Primary antibodies used in this study were anti-α-tubulin (T9026, Sigma, St Louis, MO, USA), anti-γ-tubulin (for 2D: R60;80 for 3D: T-6557, Sigma), anti-γ-tubulin-Dy647 (ab27076, Abcam, Cambridge, MA, USA), anti-Sas6-Cy5,81 anti-CP110-Cy5,82 anti-Ninein (R154),83 anti-Centrin2/3,84 anti-Laminin V (MAB19562) and anti-α6 integrin (MAB1378) (both from Merck Millipore, Darmstadt, Germany), anti-GM130 (610822) and anti-E-Cadherin (610181) (both from BD, Franklin Lakes, NJ, USA), anti-Scribble (sc-11049, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Ki-67 (18-0191Z, Invitrogen Corporation, Camarillo, CA, USA) and anti-phospho-p44/42 MAPK (9101, Cell Signaling, Danvers, MA, USA). Secondary antibodies (all from Life Technologies, Carlsbad, CA, USA) were AlexaFluor647 goat anti-rabbit (A21245), AlexaFluor568 goat anti-mouse (A11004), AlexaFluor647 goat anti-mouse (A21236), AlexaFluor647 donkey anti-goat (A21447) and AlexaFluor647 goat anti-rat (A21247). Immunofluorescence microscopy on 2D cultures was performed using an Applied Precision DeltaVision system (GE Healthcare, Little Chalfont, UK) equipped with a × 20/0.75 U Plan S Apo air objective as well as × 60/1.2 Plan Apo N and × 100/1.4 UPLS Apo oil immersion objectives. Alternatively, analyses were performed using a FEI MORE wide-field system (FEI Munich, Graefelfing, Germany) equipped with a × 40/0.95 U Plan S Apo air objective and a × 60/1.49 TIRF APON oil immersion objective. Confocal images were acquired using a Leica SP5-II-MATRIX point scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) equipped with × 10/0.30 HC PL Fluotar and × 20/0.70 HCX Plan Apo CS air objectives and a × 63/1.40-1.60 HCX Plan Apo lambda blue oil immersion objective. A 405-nm diode laser light was applied for DAPI staining, 488 nm Argon laser light for visualization of EGFP and 633 nm HeNe laser light for visualization of AlexaFluor647 stainings. Image analyses and final adjustments were carried out in Omero 5.0.2.
Biochemical and cell biological techniques
For western blotting, cell pellets were lysed in Tris lysis buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% IGEPAL CA-630) with protease and phosphatase inhibitors, before 20 or 50 μg of total protein were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. These were incubated with primary antibodies diluted in phosphate-buffered saline/5% skim milk powder (w/v). Primary antibodies were anti-human Nlp (R110),40 anti-GFP (ab290, Abcam) and anti-α-tubulin (DM1A (T9026), Sigma). Horse radish peroxidase-conjugated secondary antibodies were obtained from Bio-Rad (170-6515 and 170-6516, Hercules, CA, USA).
MT regrowth assays were performed essentially as described previously.80 In brief, MCF10A ecoR cells allowing inducible overexpression of centrosomal proteins were seeded on Ibidi eight-well microscopy slides (ibidi GmbH, Planegg-Martinsried, Germany) and grown in the absence or presence of doxycycline. MTs were depolymerized by cold treatment (30 min), before MT regrowth was triggered by addition of warm (37 °C) medium for 1 or 10 min, and cells were fixed using ice-cold methanol.
Monolayer wounding assays were performed essentially as described.85 In brief, 100 000 MCF10A cells were cultured to confluence on Ibidi eight-well microscopy slides, before wounding using a plastic pipette tip. After 24 h, cells were fixed with ice-cold methanol and examined by immunofluorescence microscopy.
Statistical analyses
Statistical analyses were carried out using Microsoft Excel 2010. Sample sizes (n) were chosen to allow the detection of statistically significant differences between subgroups within biological replicates (N). Inclusion and exclusion of samples was carried out based on viability and typical appearance of the negative control sample, which was run side by side with the test sample. Standard deviation and variance were calculated and served as estimates of variation within each group of data. For statistical comparison of groups with similar variance, a homoscedastic t-test was performed. For statistical comparison of groups with diverging variance, a heteroscedastic t-test was applied. A statistically significant effect was assumed for P-values<0.05.
Acknowledgments
We thank Tilman Brummer (University of Freiburg), Stefan Zimmermann and Ralph Wäsch (University Medical Center Freiburg) as well as Heike Brinkhaus and Mohamed Bentires-Alj (FMI Basel) for cell lines and technical advice, and Cheuk Leung (University of Minnesota) as well as Yi-Ren Hong (Kaohsiung Medical University, Taiwan) for viral vectors. We also thank Alexia Loynton-Ferrand and other members of the Imaging Core Facility (IMCF) for help and Elena Nigg-Martignoni for technical assistance. Finally, we are grateful to Felix B. Engel and the 'Emerging fields Initiative' (EFI) for 'Cell Cycle in Disease and Regeneration (CYDER)' from the Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU) for constructive comments and support, as well as to all members of the Nigg lab for helpful discussions. This work was supported by the University of Basel and the Swiss National Science Foundation (310030B_149641).
Author contributions
DS performed all experiments. DS and EAN conceived experiments, analyzed the data and wrote the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 2014; 15: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T et al. Cancer statistics, 2008. CA Cancer J Clin 2008; 58: 71–96. [DOI] [PubMed] [Google Scholar]
- Lee M, Vasioukhin V. Cell polarity and cancer—cell and tissue polarity as a non-canonical tumor suppressor. J Cell Sci 2008; 121: 1141–1150. [DOI] [PubMed] [Google Scholar]
- Musch A. Microtubule organization and function in epithelial cells. Traffic 2004; 5: 1–9. [DOI] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 2008; 135: 948–959. [DOI] [PubMed] [Google Scholar]
- Hotta A, Kawakatsu T, Nakatani T, Sato T, Matsui C, Sukezane T et al. Laminin-based cell adhesion anchors microtubule plus ends to the epithelial cell basal cortex through LL5alpha/beta. J Cell Biol 2010; 189: 901–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci 2007; 120: 2139–2142. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. Deconstructing the centriole: structure and number control. Curr Opin Cell Biol 2012; 24: 4–13. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 2012; 13: 425–435. [DOI] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 2007; 8: 161–167. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003; 426: 570–574. [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 2011; 30: 1520–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol 2011; 12: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 2005; 118: 1565–1575. [DOI] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 2000; 113: 3013–3023. [DOI] [PubMed] [Google Scholar]
- Moss DK, Bellett G, Carter JM, Liovic M, Keynton J, Prescott AR et al. Ninein is released from the centrosome and moves bi-directionally along microtubules. J Cell Sci 2007; 120: 3064–3074. [DOI] [PubMed] [Google Scholar]
- Abal M, Piel M, Bouckson-Castaing V, Mogensen M, Sibarita JB, Bornens M. Microtubule release from the centrosome in migrating cells. J Cell Biol 2002; 159: 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HQ, Gao M, Ma J, Xiao T, Zhao LL, Gao Y et al. Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res 2007; 9: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Caraway NP, Sabichi AL, Zhang HZ, Ruitrok A, Grossman HB et al. Centrosomal abnormality is common in and a potential biomarker for bladder cancer. Int J Cancer 2003; 106: 661–665. [DOI] [PubMed] [Google Scholar]
- Kayser G, Gerlach U, Walch A, Nitschke R, Haxelmans S, Kayser K et al. Numerical and structural centrosome aberrations are an early and stable event in the adenoma-carcinoma sequence of colorectal carcinomas. Virchows Arch 2005; 447: 61–65. [DOI] [PubMed] [Google Scholar]
- Kronenwett U, Huwendiek S, Castro J, Ried T, Auer G. Characterisation of breast fine-needle aspiration biopsies by centrosome aberrations and genomic instability. Br J Cancer 2005; 92: 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2002; 2: 815–825. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer 2006; 119: 2717–2723. [DOI] [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res 2003; 63: 1398–1404. [PubMed] [Google Scholar]
- Sato N, Mizumoto K, Nakamura M, Nakamura K, Kusumoto M, Niiyama H et al. Centrosome abnormalities in pancreatic ductal carcinoma. Clin Cancer Res 1999; 5: 963–970. [PubMed] [Google Scholar]
- Segat D, Cassaro M, Dazzo E, Cavallini L, Romualdi C, Salvador R et al. Pericentriolar material analyses in normal esophageal mucosa, Barrett's metaplasia and adenocarcinoma. Histol Histopathol 2010; 25: 551–560. [DOI] [PubMed] [Google Scholar]
- Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent? Trends Cell Biol 2009; 19: 334–346. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci 2014; 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A et al. Centrosome amplification can initiate tumorigenesis in flies. Cell 2008; 133: 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet 2011; 27: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 2008; 121: 1–84. [DOI] [PubMed] [Google Scholar]
- D'Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R et al. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat 2002; 75: 25–34. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 2014; 510: 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA 2002; 99: 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA 1998; 95: 2950–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am J Pathol 1999; 155: 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Lingle WL, White RA, Cordes LE, Barrett S. Microtubule nucleating capacity of centrosomes in tissue sections. J Histochem Cytochem 1999; 47: 1265–1274. [DOI] [PubMed] [Google Scholar]
- Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R, Nigg EA. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell 2003; 5: 113–125. [DOI] [PubMed] [Google Scholar]
- Casenghi M, Barr FA, Nigg EA. Phosphorylation of Nlp by Plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J Cell Sci 2005; 118: 5101–5108. [DOI] [PubMed] [Google Scholar]
- Li J, Zhan Q. The role of centrosomal Nlp in the control of mitotic progression and tumourigenesis. Br J Cancer 2011; 104: 1523–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhan Q. Cell cycle-dependent expression of centrosomal ninein-like protein in human cells is regulated by the anaphase-promoting complex. J Biol Chem 2007; 282: 17712–17719. [DOI] [PubMed] [Google Scholar]
- Yan J, Jin S, Li J, Zhan Q. Aurora B interaction of centrosomal Nlp regulates cytokinesis. J Biol Chem 2010; 285: 40230–40239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jin S, Song Y, Zhan Q. Cdc2/cyclin B1 regulates centrosomal Nlp proteolysis and subcellular localization. Cancer Biol Ther 2010; 10: 945–952. [DOI] [PubMed] [Google Scholar]
- Qu D, Qu H, Fu M, Zhao X, Liu R, Sui L et al. Increased expression of Nlp, a potential oncogene in ovarian cancer, and its implication in carcinogenesis. Gynecol Oncol 2008; 110: 230–236. [DOI] [PubMed] [Google Scholar]
- Shao S, Liu R, Wang Y, Song Y, Zuo L, Xue L et al. Centrosomal Nlp is an oncogenic protein that is gene-amplified in human tumors and causes spontaneous tumorigenesis in transgenic mice. J Clin Invest 2010; 120: 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Song Y, Zhang Q, Zhan Q. Ninein-like protein is overexpressed in head and neck squamous cell carcinoma and contributes to cancer growth and resistance to apoptosis. Oncol Rep 2009; 22: 789–798. [DOI] [PubMed] [Google Scholar]
- Zhao W, Song Y, Xu B, Zhan Q. Overexpression of centrosomal protein Nlp confers breast carcinoma resistance to paclitaxel. Cancer Biol Ther 2012; 13: 156–163. [DOI] [PubMed] [Google Scholar]
- Rapley J, Baxter JE, Blot J, Wattam SL, Casenghi M, Meraldi P et al. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol Cell Biol 2005; 25: 1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeron ML, Muller H, Schade S, Dreher F, Lehmann V, Kuhnel A et al. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat Commun 2013; 4: 1531. [DOI] [PubMed] [Google Scholar]
- Yvon AM, Walker JW, Danowski B, Fagerstrom C, Khodjakov A, Wadsworth P. Centrosome reorientation in wound-edge cells is cell type specific. Mol Biol Cell 2002; 13: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA 1992; 89: 9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natali PG, Nicotra MR, Botti C, Mottolese M, Bigotti A, Segatto O. Changes in expression of alpha 6/beta 4 integrin heterodimer in primary and metastatic breast cancer. Br J Cancer 1992; 66: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Nola S, Audebert S, Santoni MJ, Arsanto JP, Ginestier C et al. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene 2005; 24: 4330–4339. [DOI] [PubMed] [Google Scholar]
- Feigin ME, Akshinthala SD, Araki K, Rosenberg AZ, Muthuswamy LB, Martin B et al. Mislocalization of the cell polarity protein scribble promotes mammary tumorigenesis and is associated with basal breast cancer. Cancer Res 2014; 74: 3180–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ 2011; 18: 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 2008; 135: 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene 2008; 27: 5988–6001. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 2005; 15: 2199–2207. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 2005; 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30: 256–268. [DOI] [PubMed] [Google Scholar]
- Xiang B, Muthuswamy SK. Using three-dimensional acinar structures for molecular and cell biological assays. Methods Enzymol 2006; 406: 692–701. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci 2006; 119: 4155–4163. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol 2007; 176: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM. Microtubule organizing centers in polarized epithelial cells. In: Nigg EA (ed). Centrosomes in Development and Disease vol. 1. WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, 2004, pp 299–320. [Google Scholar]
- Hernandez P, Tirnauer JS. Tumor suppressor interactions with microtubules: keeping cell polarity and cell division on track. Dis Model Mech 2010; 3: 304–315. [DOI] [PubMed] [Google Scholar]
- Leung CT, Brugge JS. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature 2012; 482: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen JI, Nieminen AI, Makela TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci USA 2007; 104: 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DR, Yu M, Zheng S, Zhao Z, Muthuswamy SK, Tansey WP. Epithelial cell organization suppresses Myc function by attenuating Myc expression. Cancer Res 2011; 71: 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. Interface between normal and transformed epithelial cells: a road to a novel type of cancer prevention and treatment. Cancer Sci 2011; 102: 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C. Impact of interactions between normal and transformed epithelial cells and the relevance to cancer. Cell Mol Life Sci 2012; 69: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi H, Fujita Y. Epithelial self-defense against cancer. Cell Res 2012; 22: 1527–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol 2001; 3: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway CA, Carraway KL. Sequestration and segregation of receptor kinases in epithelial cells: implications for ErbB2 oncogenesis. Sci STKE 2007; 2007: re3. [DOI] [PubMed] [Google Scholar]
- Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, Inghirami G et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell 2006; 126: 489–502. [DOI] [PubMed] [Google Scholar]
- Nagasaka K, Pim D, Massimi P, Thomas M, Tomaic V, Subbaiah VK et al. The cell polarity regulator hScrib controls ERK activation through a KIM site-dependent interaction. Oncogene 2010; 29: 5311–5321. [DOI] [PubMed] [Google Scholar]
- Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther 2008; 16: 698–706. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 1997; 15: 871–875. [DOI] [PubMed] [Google Scholar]
- Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J 1998; 17: 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell 2007; 13: 190–202. [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD et al. Control of centriole length by CPAP and CP110. Curr Biol 2009; 19: 1005–1011. [DOI] [PubMed] [Google Scholar]
- Yan X, Habedanck R, Nigg EA. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol Biol Cell 2006; 17: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol 2007; 178: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007; 2: 329–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.