Abstract
Objectives:
Existing prognostic models for patients with hepatocellular carcinoma (HCC) have limitations. Analytic morphomics, a novel process to measure body composition using computational image-processing algorithms, may offer further prognostic information. The aim of this study was to develop and validate a prognostic model for HCC patients using body composition features and objective clinical information.
Methods:
Using computed tomography scans from a cohort of HCC patients at the VA Ann Arbor Healthcare System between January 2006 and December 2013, we developed a prognostic model using analytic morphomics and routine clinical data based on multivariate Cox regression and regularization methods. We assessed model performance using C-statistics and validated predicted survival probabilities. We validated model performance in an external cohort of HCC patients from Parkland Hospital, a safety-net health system in Dallas County.
Results:
The derivation cohort consisted of 204 HCC patients (20.1% Barcelona Clinic Liver Cancer classification (BCLC) 0/A), and the validation cohort had 225 patients (22.2% BCLC 0/A). The analytic morphomics model had good prognostic accuracy in the derivation cohort (C-statistic 0.80, 95% confidence interval (CI) 0.71–0.89) and external validation cohort (C-statistic 0.75, 95% CI 0.68–0.82). The accuracy of the analytic morphomics model was significantly higher than that of TNM and BCLC staging systems in derivation (P<0.001 for both) and validation (P<0.001 for both) cohorts. For calibration, mean absolute errors in predicted 1-year survival probabilities were 5.3% (90% quantile of 7.5%) and 7.6% (90% quantile of 12.5%) in the derivation and validation cohorts, respectively.
Conclusion:
Body composition features, combined with readily available clinical data, can provide valuable prognostic information for patients with newly diagnosed HCC.
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide and one of the leading causes of death in patients with cirrhosis.1 The incidence of HCC is rapidly rising in the United States, related to large numbers of patients with advanced hepatitis C virus infection and nonalcoholic steatohepatitis.2 Prognosis for patients with HCC remains poor with 3-year survival rates below 30%, largely driven by advanced tumor burden at the time of diagnosis. Despite improvement over time, the majority of HCCs in the United States are still diagnosed beyond an early stage.3
Accurate tumor staging is not only important for prognostication but also for determining appropriate treatment options. HCC prognosis and treatment decisions are often determined by a combination of tumor burden, degree of hepatic dysfunction, and patient performance status. Several staging systems have been proposed, without any one system being universally accepted.4 The Barcelona Clinic Liver Cancer (BCLC) classification may offer the most prognostic information, has been validated in several populations, and is endorsed by the American Association for the Study of Liver Diseases (AASLD).4, 5 However, there are potential shortcomings with the BCLC, including the subjective nature and low inter-observer reliability of assessing functional status.6 Prior studies have suggested additional prognostic information from a 5-gene score; however, the widespread applicability of this marker has been limited by low rates of biopsy among patients with HCC.7
In contrast, the wide availability of computed tomography (CT) imaging—abdominal or chest CT—in patients with HCC makes this an ideal platform for biomarker discovery. Analytic morphomics is a novel approach that uses high throughput semi-automated image-processing techniques to assess body composition, such as body dimensions, visceral fat, and muscle mass, and link it to clinical outcomes.8, 9 We have previously demonstrated that morphomics offers improved prognostic accuracy over standard clinical assessment in patients after liver transplantation as well as those in motor vehicle accidents.9, 10, 11, 12 We hypothesize that analytic morphomics can identify body factor biomarkers that may improve our ability to prognosticate in patients with HCC. The aims of our study were (i) to develop and validate a prognostic model for patients with HCC using analytic morphomics and pre-treatment objective clinical/tumor information and (ii) to compare the performance of this model to more widely accepted prognostic models, including the BCLC.
Methods
Study populations
Our derivation cohort (Ann Arbor Cohort) consisted of all male patients with a new diagnosis of HCC at the VA Ann Arbor Healthcare System between January 2006 and December 2013 (n=306). Female patients were excluded from the cohort because only two patients were female. Patients were identified through liver tumor conference lists and administrative databases were searched using ICD-9 codes (155.0 and 155.2) as previously described.13 More than 90% of all HCC patients at the VA Ann Arbor Healthcare system were presented at the liver tumor conference.
Our validation cohort consisted of all patients with newly diagnosed HCC at Parkland Health and Hospital System (Parkland cohort) between January 2005 and March 2012.14 Similar to the VA system, Parkland is an integrated health-care system, so patients often receive their continuity of care through the Parkland health system. However, as the sole safety-net hospital system for Dallas County, Parkland cares for a socioeconomically disadvantaged, racially diverse cohort of patients. Patients in the validation cohort were initially identified using ICD-9 codes for HCC (155.0 and 155.2), tumor conference presentation lists, and prior databases as previously described.15, 16
All HCC cases in both cohorts were adjudicated by two authors (A.S. and G.S.) to confirm they met the diagnostic criteria based on AASLD guidelines.4 For tumors larger than 1 cm, HCC diagnosis required a typical vascular pattern on dynamic imaging (arterial enhancement and delayed washout) or histology. Patients were excluded if they lacked CT imaging prior to HCC-directed treatment, had technical issues with CT imaging precluding analytic morphomics, had incomplete measurements at thoracic vertebral level 11 (T11), or had incomplete clinical data. Of the 306 HCC patients seen at the Ann Arbor VA during the study period, 229 (74.8%) had an abdominal or chest CT scan prior to treatment, with the remainder only having magnetic resonance imaging. All patients in both cohorts were discussed in multidisciplinary Liver Tumor Boards for management decisions, and curative treatments (liver transplantation, resection, and radiofrequency ablation) were recommended for patients with early stage HCC as applicable. Liver transplantation was available to a minority of patients in both cohorts given financial, social, and medical barriers to transplantation in these patient populations.17 This study was approved by the Institutional Review Boards at the Ann Arbor VA Healthcare System and UT Southwestern Medical Center.
Analytic morphomics
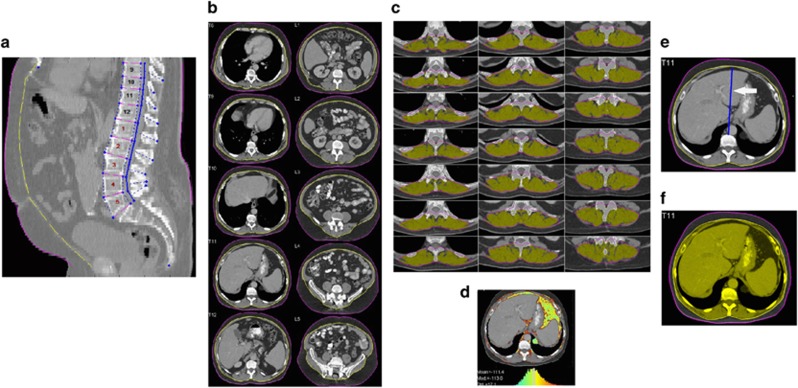
Pretreatment CT studies were analyzed using analytic morphomics as previously described.8, 12, 18, 19 Briefly, all available CT scan DICOM (Digital Imaging and Communications in Medicine) files were loaded into the analytic morphomics server in a de-identified manner. Given the method of processing and required data, both contrast and non-contrast scans could be used. A semi-automated high throughput methodology with algorithms programmed in MATLAB (MathWorks, Natick, MA) enabled image processing and analysis. All algorithms involved a combination of user-defined points, automated image processing, and user editing and verification. All imaging studies were first anatomically indexed using semi-automated identification of spinal vertebral levels to allow for accurate and standardized measurements of the same area in each patient. In this paper, we chose to derive all measurements at the bottom of T11. This anatomic landmark was chosen because this was felt to have the highest likelihood of being available on all abdominal and chest CT scans. A single slice was chosen, as this would include all required body composition features while minimizing processing time and potential radiation exposure for future prospective studies. After anatomic indexing, the fascial envelope and skin outline were automatically defined using key points within the linea alba, dorsal muscles groups, and paraspinus lateral seams at specified vertebra points (Figure 1a–d). In addition to direct measurements of features, we also included measurements standardized to patient body size. We used two surrogates of patient size: (i) the distance between the anterior edge of the vertebra to the anterior edge of the fascial envelope and (ii) the area of the fascial envelope (Figure 1e, f). All geometries were saved in a STL (stereolithography) format in the analytic morphomics database with PostgresSQL and subsequently retrieved to calculate several shape and pixel-based measurements.
Figure 1.
Body composition features as determined by analytic morphomics. (a) Example of identification of spinal vertebral levels that serve as anatomic reference point. (b) Example of fascial envelope (yellow line) and skin outline (red line). (c) Example of the dorsal group muscles (outlined in yellow) defined automatically after delineation of paraspinus lateral seams at specified vertebra points. (d) Example of MATLAB-based 3D image viewer graphical user interface showing the pixel densities which was used to measure the interstitial hounsfeld units at T11 (ITHU). (e) Example of VB2FASCIA or distance between the vertebra to the facial envelope. (f) Example of FASCIAAREA or fascial area.
Clinical data collection
Patient demographics, clinical history, laboratory data, and imaging results for both cohorts were obtained through review of computerized medical records and extracted using standardized forms. Given our goal was to develop a prognostic model to be used at HCC diagnosis, we only included pre-treatment patient and tumor characteristics. Age, gender, race/ethnicity, liver disease etiology, and presence of hepatic decompensation (ascites or encephalopathy) were recorded for each patient. Laboratory data of interest included platelet count, creatinine, bilirubin, albumin, international normalized ratio, and alpha fetoprotein. Tumor characteristics at diagnosis including number of HCC nodules, presence of portal vein thrombosis, and TNM staging were determined by review of CT imaging.
Statistical analysis
Overall survival distribution was estimated by Kaplan–Meier analysis, with patients' outcomes defined from time of CT scans to death, censored at the time of liver transplantation or date of last follow-up. Prognostic models of survival were developed by the Cox proportional hazard regression models. Because analysis of CT scans with analytic morphomics leads to high dimensional data similar to what occurs with microarray analysis or genome-wide association data, we chose to utilize regularization methods to address the vast number of measurements and multi-collinearity of variables.20 We used elastic net regularization with cross-validation for variable selection to build prognostic models. The elastic net method utilizes a linear combination of L1 (lasso) and L2 (ridge) penalty in minimizing the partial likelihood function. This method addresses model over-fitting and variable co-linearity through the lasso penalty and ridge penalty, respectively. Tuning parameters were optimized through 10-fold cross-validation minimizing the deviance of partial likelihood for the Cox model. With this type of approach for variable selection, all morphomics variables were included in addition to clinical variables. The performance of the model was assessed with C-statistics in the derivation (Ann Arbor VA Healthcare system) cohort and the independent external validation (Parkland Health and Hospital) cohort. C-statistics were also assessed for prognosis based on the TNM and BCLC staging systems, the two most widely used systems in the United States.21 The C-statistics may range from 0 to 1, with 1 indicating perfect prediction and 0.5 indicating prediction by chance alone, with values greater than 0.7 generally being considered a useful model. For prognostic models, values greater than 0.9 are rare.22 To compare the discriminatory validity of the models, we used a modification of the net reclassification improvement methodology that can accommodate survival data to assess discrimination, comparing the fraction of concordant pairs for which one model is more impressive than the other.23, 24 To validate predicted probabilities, we calculated the mean absolute error in predicted probabilities for 1-year survival, and its upper 90% quantile.25, 26, 27, 28 The mean absolute error in predicted probabilities is the average difference in survival between actual survival probabilities and those predicted by the model, and the 90% quantile is the 90% upper quantile for these absolute errors. A lower value suggests a smaller difference in predicted survival probabilities vs. actual survival probabilities, and thereby better calibration. All statistical analyses were performed using R 3.1.0 with packages glmnet, Hmisc, rms.
Results
Patient characteristics
The Ann Arbor VA derivation cohort consisted of 204 patients with HCC, with baseline characteristics shown in Table 1. The median age of the patients was 61 (interquartile range 58–66) years. More than 44% of patients were Caucasian and 100% were male. The most common etiologies of liver disease were hepatitis C (73%), alcohol-induced (7%), and nonalcoholic steatohepatitis/cryptogenic (5%). A total of 57% had Child Pugh class A cirrhosis, 31% Child Pugh B cirrhosis, and 12% Child Pugh C cirrhosis. HCC was multifocal in 50% of patients, and nearly 10% had lymph node involvement and/or distant metastases.
Table 1. Patient demographics.
| Variable | Ann Arbor cohort (n=204) | Parkland cohort (n=225) |
|---|---|---|
| Age at diagnosis (years) | 61 (58–66) | 57 (52–62) |
| Gender (% male) | 204 (100%) | 178 (79%) |
| Race (% Caucasian) | 89 (44%) | 60 (27%) |
| Etiology of liver disease | ||
| Hepatitis C | 148 (73%) | 145 (64%) |
| Alcohol-induced | 14 (7%) | 32 (14%) |
| NASH/cryptogenic | 11 (5%) | 17 (8%) |
| Multifocal HCC | 102 (50%) | 123 (55%) |
| Portal vein thrombosis | 43 (21%) | 76 (34%) |
| Child pugh class | ||
| Child pugh A | 117 (57%) | 83 (37%) |
| Child pugh B | 63 (31%) | 86 (38%) |
| Child pugh C | 24 (12%) | 56 (25%) |
| MELD score | 9 (8–12) | 11 (8–15) |
| Alpha fetoprotein (ng/ml) | 34.9 (9.6–283.2) | 116.0 (13.8–4260.0) |
| ECOG performance status | 1 (0–2) | 2 (1–2) |
| TNM stage (I/II/III/IV) | 86/47/51/20 | 7/109/51/58 |
| BCLC stage (0/A/B/C/D) | 5/36/26/101/36 | 3/47/51/49/75 |
| Treatment | ||
| Resection | 23 | 12 |
| RFA | 23 | 8 |
| TACE | 76 | 70 |
| Sorafenib | 22 | 17 |
| Supportive | 60 | 118 |
BCLC, Barcelona Clinic Liver Cancer; ECOG, Eastern Cooperative Oncology Group; HCC, hepatocellular carcinoma; MELD, Model for End Stage Liver Disease; NASH, nonalcoholic steatohepatitis; RFA, radiofrequency ablation; TACE, transarterial chemoembolization; TNM, tumor node metastases.
All values are expressed are n (%) or median (interquartile range).
The Parkland Health and Hospital System validation cohort consisted of 225 patients with HCC, with baseline characteristics shown in Table 1. The median age of the patients was 57 (interquartile range 52–62) years. More than one-fourth of patients were Caucasian and 79% were male. The most common etiologies of liver disease were hepatitis C (64%), alcohol-induced (14%), and nonalcoholic steatohepatitis/cryptogenic (8%). A total of 37% had Child Pugh class A cirrhosis, 38% Child Pugh B cirrhosis, and 25% Child Pugh C cirrhosis. HCC was multifocal in 55% of patients, and nearly 26% had lymph node involvement and/or distant metastases.
Despite both centers being in the United States and serving similar socioeconomic levels, there were significant differences between the two populations. The Parkland cohort had higher rates of liver dysfunction, with a lower proportion of patients with Child Pugh A cirrhosis (37 vs. 57%, P<0.001) and higher proportion of Child Pugh C patients (25 vs. 12%, P<0.001). The Parkland cohort also had a higher proportion of patients with lymph node involvement and/or distant metastases (26 vs. 10%, P<0.001). Although the proportions of BCLC stage A patients were similar between the two cohorts (20.9% vs. 17.6%, P=0.47), there were significantly more BCLC stage D patients (33.3 vs. 17.6%, P<0.001) compared with the Ann Arbor cohort. Accordingly, there were lower rates of curative treatment and a higher rate of best supportive care in the Parkland cohort, leading to worse overall survival (median 5.4 vs. 16.8 months).
Model description
To assess the ability of morphomics variables to predict survival in HCC, we built three models. First, we built a model using only morphomics variables (Model 1). Recognizing that tumor factors and liver function are important in determining outcome in HCC, we examined the benefit of adding these factors to morphomics variables for model performance. Model 2 included morphomics variables and pre-treatment clinical factors known to affect survival, whereas Model 3 included morphomics variables, pre-treatment clinical factors, and TNM tumor stage. Only clinical and tumor variables readily available at the time of treatment decisions were included in the models. Variables included in the models and the descriptions of their measurements are detailed in Table 2.
Table 2. Variables in final models.
| Variable name | Description | Type of measurment |
|---|---|---|
| Model 1: Morphomics alone | ||
| FASCIACIRCUMFERENCE BY VB2FASCIA | Circumference of the facial envelope divided by the distance between the vertebra to the facial envelope | Body dimension |
| VISCERALFATAREA BY VB2FASCIA | Area of the visceral fat divided by the distance between the vertebra to the facial envelope | Fat |
| PSPXSECAREA BY VB2FASCIA | Area of the dorsal muscle group divided by distance between the vertebra to the facial envelope | Muscle |
| PSPVOLOFVB BY VB2FASCIA | Volume of the dorsal muscle group divided by the distance between the vertebra to the facial envelope | Muscle |
| TOTALBODYAREA BY VB2FASCIA | Total body area divided by the distance between the vertebra to the facial envelope | Body dimension |
| TOTALBODYCIRCUMFERENCE BY VB2FASCIA | Circumference of the body divided by the distance between the vertebra to the facial envelope | Body dimension |
| VB2FASCIA BY FASCIAAREA | The distance between the vertebra to the facial envelope divided by the fascial area | Body dimension |
| VISCERALFATAREA BY FASCIAAREA | Visceral fat area divided by the fascial area | Fat |
| SUBCUTFATAREA BY FASCIAAREA | Subcutaneous fat area divided by the fascial area | Fat |
| PSPXSECAREA BY FASCIAAREA | Dorsal muscle group area divided by the fascial area | Muscle |
| PSPVOLOFVB BY FASCIAAREA | Dorsal muscle group volume divided by fascial area | Muscle |
| TOTALBODYAREA BY FASCIAAREA | Total body area divided by fascial area | Body dimension |
| TOTALBODYCIRCUMFERENCE BY FASCIAAREA | Body circumference divided by the fascial area | Body dimension |
| ITHU NORMALIZED | Density of the interstitial area normalized | Relative interstitial density |
| Model 2: Morphomics and TNM stage | ||
| FASCIACIRCUMFERENCE BY VB2FASCIA | Circumference of the facial envelope divided by the distance between the vertebra to the facial envelope | Body dimension |
| VISCERALFATAREA BY VB2FASCIA | Area of the visceral fat divided by the distance between the vertebra to the facial envelope | Fat |
| PSPXSECAREA BY VB2FASCIA | Area of the dorsal muscle group divided by distance between the vertebra to the facial envelope | Muscle |
| PSPVOLOFVB BY VB2FASCIA | Volume of the dorsal muscle group divided by the distance between the vertebra to the facial envelope | Muscle |
| TOTALBODYCIRCUMFERENCE BY VB2FASCIA | Circumference of the body divided by the distance between the vertebra to the facial envelope | Body dimension |
| VISCERALFATAREA BY FASCIAAREA | Visceral fat area divided by the fascial area | Fat |
| ITHU NORMALIZED | Density of the interstitial area normalized | Relative interstitial density |
| TNM stage III | TNM stage III | Tumor factors |
| TNM stage IV | TNM stage IV | Tumor factors |
| Model 3: Morphomics, TNM stage, and clinical factors | ||
| PSPXSECAREA BY VB2FACIA | Area of the dorsal muscle group divided by distance between the vertebra to the facial envelope | Muscle |
| PSPVOLOFVB BY VB2FACIA | Volume of the dorsal muscle group divided by the distance between the vertebra to the facial envelope | Muscle |
| TOTALBODYAREA BY VB2FASCIA | Total body area divided by the distance between the vertebra to the facial envelope | Body dimension |
| TOTALBODYCIRCUMFERENCE BY VB2FASCIA | Circumference of the body divided by the distance between the vertebra to the facial envelope | Body dimension |
| VB2FASCIA BY FASCIAAREA | The distance between the vertebra to the facial envelope divided by the fascial area | Body dimension |
| PSPXSECAREA BY FASCIAAREA | Dorsal muscle group area divided by the fascial area | Muscle |
| PSPVOLOFVB BY FASCIAAREA | Dorsal muscle group volume divided by fascial area | Muscle |
| TOTALBODYAREA BY FASCIAAREA | Total body area divided by fascial area | Body dimension |
| ITHU NORMALIZED | Density of the interstitial area normalized | Relative interstitial density |
| Multifocal | Presence of multifocal tumor | Tumor factor |
| PVT Y NY | Presence of portal vein thrombosis | Clinical factor |
| LogTBili | Log of total bilirubin | Clinical factor |
| LogINR | Log of INR | Clinical factor |
| Albumin | Albumin | Clinical factor |
| Hepatic encephalopathy | Presence of hepatic encephalopathy | Clinical factor |
| Ascites | Presence of ascites | Clinical factor |
| TNM stage III | TNM stage III | Tumor factors |
| TNM stage IV | TNM stage IV | Tumor factors |
INR, international normalized ratio; TNM, tumor node metastases.
Model performance
Median transplant-free survival of the derivation cohort was 16.8 months, with 1-year and 3-year survival rates of 59% and 21%, respectively. In the derivation cohort, morphomics variables when used alone had a C-statistic of 0.72 (95% confidence interval (CI) 0.63–0.82) for predicting survival. Adding routine clinical information to the analytic morphomics model increased the C-statistic to 0.76 (95% CI 0.67–0.85). Finally, a model combining analytic morphomics, TNM tumor stage, and routine clinical information achieved the highest C-statistic of 0.80 (95% CI 0.71–0.89) when combined with routine clinical information (P<0.001 compared with both prior models). In a post hoc subgroup analysis, the analytic morphomics model, there appeared to be a trend towards better C index in patients with Child Pugh B or C cirrhosis compared with those with Child Pugh A cirrhosis (C-statistics 0.82, 95% CI 0.73–0.91 vs. 0.73, 95% CI 0.58–0.88). The final analytic morphomics model had a significantly higher C-statistic compared with the TNM (0.71, 95% CI 0.61–0.80) and BCLC (0.66, 95% CI 0.56–0.76) staging systems (P<0.001) (Table 3). For calibration, the mean absolute error in predicted 1-year survival probabilities was 5.3%, with a 90% quantile of 7.5%.
Table 3. Performance of model in derivation and validation cohorts.
|
Derivation cohort |
Validation cohort |
|||
|---|---|---|---|---|
| C-statistic | Mean absolute error in predicted probability of 1-year survival | C-statistic | Mean absolute error in predicted probability of 1-year survival | |
| Model 1a | 0.72 95% CI 0.63–0.82 | |||
| Model 2a | 0.76 95% CI 0.67–0.85 | |||
| Model 3a | 0.80 95% CI 0.71–0.89 | 5.3% 90% quantile 7.5% | 0.75 95% CI 0.68–0.82 | 7.6% 90% quantile 12.5% |
| TNM System | 0.71 95% CI 0.61–0.80 | 0.67 95% CI 0.60–0.74 | ||
| BCLC System | 0.66 95% CI 0.56–0.76 | 0.70 95% CI 0.62–0.78 | ||
BCLC, Barcelona Clinic Liver Cancer; CI, confidence interval; TNM, tumor node metastases.
Model variables are detailed in Table 2. Model 1 includes analytic morphomics variables alone. Model 2 includes analytic morphomics variables and TNM tumor stage. Model 3 includes analytic morphomics variables, TNM tumor stage, and pre-treatment clinical variables.
To examine the generalizability of our model, we examined the validity of the model using an external cohort from Parkland Health and Hospital System. Median transplant-free survival of the validation cohort was 5.4 months, with 1-year and 3-year survival rates of 34 and 16%, respectively. The analytic morphomics model performed well with a C-statistic of 0.75 (95% CI 0.68–0.82), which was significantly higher than C-statistics for the TNM (0.67, 95% CI 0.60–0.74) and BCLC (0.70, 95% CI 0.62–0.78) staging systems (P<0.001) (Table 3). As our derivation cohort only had males and the external cohort had males and females, we re-examined C-statistics of the model after excluding females and data were similar to those above (data not shown). For calibration, the mean absolute error in predicted 1-year survival probabilities was 7.6%, with a 90% quantile of 12.5%.
Discussion
We found using analytic morphomics, we can build an accurate prognostic model for patients with HCC. In order to ascertain the relative accuracy of our prognostic model, we sought to compare it with the most prognostic or widely used staging systems, BCLC and TNM, respectively.21 Our model achieved a C-statistic of 0.80 in the derivation cohort, which was significantly better than that of the TNM and BCLC staging systems. We validated our model on an external cohort of patients from a non-VA setting located in a geographically and ethnically distinct region of the United States. Despite these differences in the two cohorts, our model continued to perform well in the validation cohort with a C-statistic of 0.75. The analytic morphomics model demonstrated good calibration in both derivation and validation cohorts, with a mean absolute error in predicted 1-year survival probability of 5.3% and 7.6%, respectively.
Analytic morphomics takes advantage of the wide availability of cross-sectional imaging in patients with HCC and offers a potential source for prognostic information. Our analytic morphomics model includes three components: tumor burden, liver function, and analytic morphomics data. These measures parallel the inclusion of tumor burden, liver function, and performance status included in other staging systems, such as the BCLC. However, our model was able to achieve higher C-statistics for survival in both the derivation and validation cohorts. Prior studies have suggested high inter-observer variability in assessing performance status as well as unclear cutoffs for discriminating ECOG performance status.6, 29 The use of analytic morphomics data may allow more objective, reliable, and continuous measures of patient performance status over a provider's subjective assessment of ECOG status.30
Using analytic morphomics, we are able to quantify features of body composition that may be an important biomarker for prognosticating clinical outcomes. This information can be important for describing prognosis for patients in clinic, risk stratification for clinical trials, and potentially for treatment decisions. For example, it may be possible to define a subgroup of patients who derive less benefit or are at an increased risk of harms from palliative treatment such as TACE and sorafenib. Our previous work showed that analytic morphomics can be used to predict the presence of cirrhosis among patients with liver disease with very high accuracy (Area Under Receiver Operating Characteristics, AUROC>0.90), which was significantly better than other serum-based methods.9 Similarly, we have shown that analytic morphomics can predict mortality after transplantation in patients with chronic liver disease.10 The ability of analytic morphomics to predict outcome in such a diverse population of patients supports the hypothesis that understanding underlying patient features (phenotype) is a very important first step to personalizing care. This is particularly important in a disease such as HCC, in which it is not only the aggressive nature of the tumor but also underlying patient characteristics, such as functional status and liver status, which are critical in determining prognosis. Analytic morphomics can provide an accurate method to quantitate these features.
Our morphomics model incorporates several characteristics, including dorsal muscle area and volume, total body circumference, and interstitial tissue density (Figure 1). It is possible that, if not likely, dorsal muscle area and volume serve as objective surrogates for sarcopenia and/or frailty.8, 10, 30 This hypothesis is further supported by the fact that adding performance status to the analytic morphomics model did not change the predictive accuracy in the derivation cohort (C-statistic 0.793 vs. 0.796). There has been increasing literature on the prognostic importance of these features in patients with cirrhosis and early data suggest exercise programs to reverse frailty and/or sarcopenia may improve prognosis in patients with cirrhosis;31, 32 however, it is unclear whether this would be equally true among patients with HCC. Interstitial tissue density may serve as an early marker for portal hypertension, as it has moderate correlation with the presence of ascites (data not shown).
The performance of prognostic models is often lower in validation cohorts than derivation cohorts given possible over-fitting of the model, measurement error related to inter-observer variability of predictors, and/or differences in patient case mix.33, 34 However, the prognostic accuracy of our analytic morphomics was similar in our independent external validation cohort despite large differences in patient characteristics. Our derivation cohort sampled from a VA population and consisted primarily of Caucasian elderly males, whereas our validation cohort sampled from a safety-net population and included a large number of Hispanics and Blacks, younger patients, and both sexes. Furthermore, our derivation cohort was sampled from Dallas County and included a large number of overweight, obese, and even morbidly obese patients.
Some limitations of our study warrant further discussion. Although analytic morphomics is versatile, at the present time, we can only perform image analysis on CT data files, and for patients who only had an magnetic resonance imaging, we were unable to include them in the study. Nevertheless, this is proof of concept to demonstrate image analysis software that quantifies morphologic features in patients may provide valuable prognostic information in patients with newly diagnosed HCC. Furthermore, we increased the versatility of our tool by focusing on the anatomic level that is often present on both abdominal and chest CT scans and included all protocol scans. Although magnetic resonance imaging is becoming more preferred in some centers for diagnosis of HCC, it is not always widely available especially in rural and underserved areas. Even in patients who only received magnetic resonance imaging for diagnosis of HCC, there is often accompanying CT of the chest to rule out metastasis that can be used for analysis. A second concern may be that analysis of the data requires specialized expertise and software that is not readily available. We are currently in the process of developing stand-alone software packages that can be distributed to academic sites as well as developing web portals and other modalities for easy acquisition of the standard DICOM data files available in any CT scan. We have shown the feasibility of this process by acquiring CT scan data from completely different health-care systems such as the Veterans Administration Health Systems and Parkland Health Systems. The analytic software required for image analysis was not available at Parkland Health Systems, demonstrating the capability of performing this analysis remotely. Thus, we feel that this process can be easily generalized for different types of practices given time and development. Because of its retrospective nature, our study was also limited by missing data and the potential for measurement bias. The possibility of measurement bias is particularly applicable to variables, such as performance status, which are subjective and were not consistently reported in clinical notes. Finally, the majority of patients in both cohorts underwent palliative therapies with TACE and/or sorafenib, so larger studies are needed to determine whether our results are equally valid in patients undergoing curative treatments.
In summary, we found proof of principle that analytic morphomics may offer prognostic information in diverse cohorts of patients with HCC. Incorporation of body composition features from CT imaging can likely provide objective data regarding sarcopenia as a potential marker of performance status and interstitial edema as a potential marker of early portal hypertension. Further research is needed to prospectively validate these findings in large cohorts of patients with HCC.
Study Highlights
Guarantor of the article: Grace L. Su, MD.
Specific author contributions: Singal AG: data collection, interpretation of data, drafting of manuscript, and critical revision for intellectual concept; Zhang P: data analysis, interpretation of data, drafting of manuscript, and critical revision for intellectual concept; Waljee AK: data analysis, interpretation of data, and critical revision for intellectual concept; Ananthakrishnan L: data collection and critical revision for intellectual concept; Parikh ND: interpretation of data and critical revision for intellectual concept; Sharma P: interpretation of data and critical revision for intellectual concept; Barman P: data collection and critical revision for intellectual concept; Krishnamurthy V: data collection and critical revision for intellectual concept; Wang L: data analysis and critical revision for intellectual concept; Wang SC: study concept and critical revision for intellectual concept; Su GL: study concept, data collection, data analysis, interpretation of data, drafting of manuscript, and critical revision for intellectual concept.
Financial support: Singal was supported in part by the AHRQ Center for Patient-Centered Outcomes Research (R24 HS022418). Waljee's research is funded by a VA HSR&D CDA-2 Career Development Award 1IK2HX000775. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality or the VA.
Potential competing interests: None.
References
- Jemal A, Bray F, Center MM et al. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012; 142: 1264–1273 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol 2009; 27: 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2010; 53: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo U, Vitale A, Grigoletto F et al. Prospective validation of the Barcelona Clinic Liver Cancer staging system. J Hepatol 2006; 44: 723–731. [DOI] [PubMed] [Google Scholar]
- Ando M, Ando Y, Hasegawa Y et al. Prognostic value of performance status assessed by patients themselves, nurses, and oncologists in advanced non-small cell lung cancer. Br J Cancer 2001; 85: 1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault JC, De Reynies A, Villanueva A et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology 2013; 145: 176–187. [DOI] [PubMed] [Google Scholar]
- Englesbe MJ, Lee JS, He K et al. Analytic morphomics, core muscle size, and surgical outcomes. Ann Surg 2012; 256: 255–261. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy V, Zhang P, Ethiraj S et al. Use of analytic morphomics of liver, spleen, and body composition to identify patients at risk for cirrhosis. Clin Gastroenterol Hepatol 2014; 13: 360–368 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesbe MJ. Quantifying the eyeball test: sarcopenia, analytic morphomics, and liver transplantation. Liver Transpl 2012; 18: 1136–1137. [DOI] [PubMed] [Google Scholar]
- Parenteau CS, Zhang P, Holcombe S et al. Can anatomical morphomic variables help predict abdominal injury rates in frontal vehicle crashes? Traffic Inj Prev 2014; 15: 619–626. [DOI] [PubMed] [Google Scholar]
- Zhang P, Parenteau C, Wang L et al. Prediction of thoracic injury severity in frontal impacts by selected anatomical morphomic variables through model-averaged logistic regression approach. Accid Anal Prev 2013; 60: 172–180. [DOI] [PubMed] [Google Scholar]
- Barman PM, Sharma P, Krishnamurthy V et al. Predictors of mortality in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Dig Dis Sci 2014; 59: 2821–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yopp AC, Mansour JC, Beg MS et al. Establishment of a multidisciplinary hepatocellular carcinoma clinic is associated with improved clinical outcome. Ann Surg Oncol 2014; 21: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Yopp AC, Singal AG. Diagnostic delays are common among patients wtih hepatocellular carcinoma. J Natl Compr Canc Netw 2014; 13: 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal AG, Yopp AC, Gupta S et al. Failure rates in the hepatocellular carcinoma surveillance process. Cancer Prev Res (Phila) 2012; 5: 1124–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal AG, Chan V, Getachew Y et al. Predictors of liver transplant eligibility for patients with hepatocellular carcinoma in a safety net hospital. Dig Dis Sci 2012; 57: 580–586. [DOI] [PubMed] [Google Scholar]
- Huhdanpaa H, Douville C, Baum K et al. Development of a quantitative method for the diagnosis of cirrhosis. Scand J Gastroenterol 2011; 46: 1468–1477. [DOI] [PubMed] [Google Scholar]
- Harbaugh CM, Terjimanian MN, Lee JS et al. Abdominal aortic calcification and surgical outcomes in patients with no known cardiovascular risk factors. Ann Surg 2013; 257: 774–781. [DOI] [PubMed] [Google Scholar]
- Waldmann P, Meszaros G, Gredler B et al. Evaluation of the lasso and the elastic net in genome-wide association studies. Front Genet 2013; 4: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero JA, Fontana RJ, Barrat A et al. Prognosis of hepatocellular carcinoma: comparison of 7 staging systems in an American cohort. Hepatology 2005; 41: 707–716. [DOI] [PubMed] [Google Scholar]
- Kamath PS, Wiesner RH, Malinchoc M et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001; 33: 464–470. [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 2011; 30: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008; 27: 157–172; discussion 207–212. [DOI] [PubMed] [Google Scholar]
- May M, Royston P, Egger M et al. Development and validation of a prognostic model for survival time data: application to prognosis of HIV positive patients treated with antiretroviral therapy. Stat Med 2004; 23: 2375–2398. [DOI] [PubMed] [Google Scholar]
- Stallard N. Simple tests for the external validation of mortality prediction scores. Stat Med 2009; 28: 377–388. [DOI] [PubMed] [Google Scholar]
- Cox DR, Snell EJ. A general definition of residuals (with discussion). JRSSB 1968; 30: 248–275. [Google Scholar]
- Kooperberg C, Stone CJ, Truong YK. Hazard regression. J Am Stat Assoc 1995;90:78–94. [Google Scholar]
- Hsu CY, Lee YH, Hsia CY et al. Performance status in patients with hepatocellular carcinoma: determinants, prognostic impact, and ability to improve the Barcelona Clinic Liver Cancer system. Hepatology 2013; 57: 112–119. [DOI] [PubMed] [Google Scholar]
- Miller AL, Min LC, Diehl KM et al. Analytic morphomics corresponds to functional status in older patients. J Surg Res 2014; 192: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai T, Shiraki M, Nishimura K et al. Sarcopenia impairs prognosis of patients with liver cirrhosis. Nutrition 2015; 31: 193–199. [DOI] [PubMed] [Google Scholar]
- Montano-Loza AJ. Clinical relevance of sarcopenia in patients with cirrhosis. World J Gastroenterol 2014; 20: 8061–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal AG, Mukherjee A, Joseph Elmunzer B et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol 2013; 108: 1723–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waljee AK, Higgins PD, Singal AG. A primer on predictive models. Clin Transl Gastroenterol 2014; 5: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]