Abstract
Chlamydia trachomatis is an important human pathogen that undergoes a characteristic development cycle correlating with stage-specific gene expression profiles. Taking advantage of recent developments in the genetic transformation in C. trachomatis, we constructed a versatile green fluorescent protein (GFP) reporter system to study the development-dependent function of C. trachomatis promoters in an attempt to elucidate the mechanism that controls C. trachomatis adaptability. We validated the use of the GFP reporter system by visualizing the activity of an early euo gene promoter. Additionally, we uncovered a new ompA promoter, which we named P3, utilizing the GFP reporter system combined with 5′ rapid amplification of cDNA ends (RACE), in vitro transcription assays, real-time quantitative RT-PCR (RT-qPCR), and flow cytometry. Mutagenesis of the P3 region verifies that P3 is a new class of C. trachomatis σ66-dependent promoter, which requires an extended −10 TGn motif for transcription. These results corroborate complex developmentally controlled ompA expression in C. trachomatis. The exploitation of genetically labeled C. trachomatis organisms with P3-driven GFP allows for the observation of changes in ompA expression in response to developmental signals. The results of this study could be used to complement previous findings and to advance understanding of C. trachomatis genetic expression.
Chlamydia trachomatis is a Gram-negative obligate intracellular bacterium that is responsible for considerable morbidity and socioeconomic burden worldwide1,2. C. trachomatis serovars A-C produce trachoma, a leading cause of blindness in developing countries. Serovars D–K cause the most common sexually transmitted bacterial infections. Serovars L1–L3 result in lymphogranuloma venereum (LGV), a chronic infection of the lymphatic system. Over 70% of women with C. trachomatis genital tract infections are asymptomatic and, if left untreated, severe sequelae may include pelvic inflammatory disease and infertility. Unfortunately, no vaccine against C. trachomatis is currently available. A better understanding of bacterial adaptation and pathogenesis is imperative for developing effective control strategies against the pathogen.
Bacterial cells of Chlamydia spp. grow solely in membrane-bound vacuoles known as inclusions within the host cell. A hallmark of the chlamydial developmental cycle is the reversible transition between two functionally divergent forms: infectious elementary bodies (EBs) and replicating reticulate bodies (RBs)3,4. During the developmental cycle, the expression of C. trachomatis genes is tightly regulated. Three temporal classes of C. trachomatis genes, early (EB-to-RB germination), middle (RB multiplication), and late (terminal RB-to-EB differentiation) have been revealed5,6,7. Bacterial gene transcription relies on RNA polymerase (RNAP) holoenzyme, which consists of a core enzyme and a σ factor. The σ factor confers the ability to initiate promoter-specific transcription on the enzyme8. C. trachomatis utilizes three σ factors, σ66, σ54, and σ28, in addition to other regulators, to control its transcription9. Many C. trachomatis housekeeping genes are transcribed by σ66 and a set of late-genes are regulated by σ28, whereas the target genes of σ54 remain unclear10,11,12,13,14. Both C. trachomatis σ66 and σ28 belong to the E. coli σ70 family, containing four functionally conserved domains, regions 1 to 415. Despite a significantly condensed genome and limited gene regulation toolbox compared to many other bacteria, C. trachomatis can change its regulatory networks of gene expression rapidly in response to internal metabolic changes and external stimuli for its adaptation and survival7,16,17. A significant hurdle to a detailed understanding of the underlying mechanisms of gene regulation is in part the lack, until recently, of genetic systems to assess the properties of temporal promoters. Whereas the study of C. trachomatis promoters has been often conducted in vitro and in heterogeneous “in vivo” systems, such approaches are unable to elucidate information critical to the understanding of the C. trachomatis developmental cycle.
Recent advances in genetic transformation18 enable the visualization of C. trachomatis growth and the study of its protein localization using shuttle plasmid-encoded fluorescent proteins in situ19,20,21,22. Here, we expanded the repertoire of these powerful tools to investigate the development-dependent action of promoters in C. trachomatis. We constructed a transcription vector, which contains a C. trachomatis promoter linked to green fluorescent protein (GFP) gene. We validated its use by evaluating promoter activity of the C. trachomatis euo (early upstream open reading frame) gene encoding a broad DNA binding protein23. We also uncovered a new ompA promoter, P3, using the reporter assays combined with 5′ rapid amplification of cDNA ends (RACE), real-time RT-qPCR, flow cytometry, and an in vitro transcription assay. C. trachomatis ompA encodes the major outer membrane protein (MOMP), which consists of 60% of total outer membrane proteins24 and serves as a general porin and cytoadhesin25 vital to the infection process. Our results contribute to a deeper understanding of gene regulatory mechanisms by defining transcription signals of ompA promoters during the C. trachomatis development cycle. The use of the GFP reporter system offers an efficient tool to assess development-dependent changes in gene regulation in the intact cells. The knowledge acquired from this study can be used to complement previous findings and to advance studies on regulation of C. trachomatis genes.
Results and Discussion
A shuttle plasmid-based GFP reporter driven by an early C. trachomatis promoter
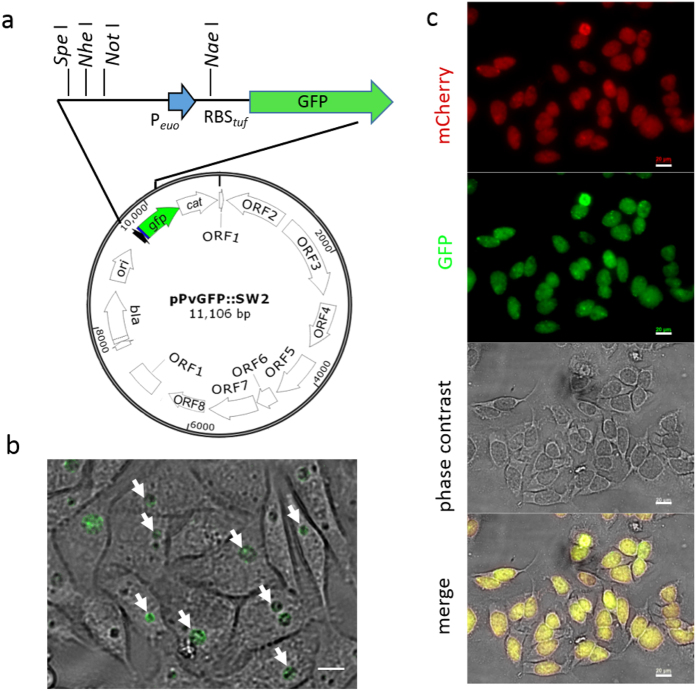
To assess promoter activity in a desirable in situ setting, we constructed a transcriptional reporter vector, pPvGFP::SW2 (Fig. 1a) using the backbone of the Escherichia coli and C. trachomatis shuttle plasmid, pGFP::SW218. Plasmid pPvGFP::SW2 contained (i) a core promoter region (−38 to + 6 relative to the transcription start site (TSS) +1) from the C. trachomatis euo gene26 (Peuo) that is flanked with a multiple cloning site (MCS) to facilitate cloning and to validate the reporter vector, (ii) a 34 bps region containing a ribosome binding site (RBS) from the C. trachomatis tuf gene11, which codes for translation elongation factor EF-Tu at the 5′ end of the gfp gene. Previous studies showed that tuf RBS functioned in both C. trachomatis and in E. coli10,11, ensuring translation of reporter genes. The plasmid also contained the bla gene encoding β-lactamases for positive selection in C. trachomatis. The plasmid pPvGFP::SW2 was transformed into C. trachomatis L2/25667R, a naturally occurring plasmid-free strain. After four rounds of selection with ampicillin, GFP-expressing inclusions were observed by fluorescent microscopy. A clone of the transformed strain, named L2/pPvGFP::SW2, was used to infect HeLa cells. GFP-expressing inclusions appeared at 16 hours post-infection (h pi) (Fig. 1b). The levels of GFP increased proportionately to the expansion of C. trachomatis inclusions and accumulation of organisms. The control, transformed with the promoter-less plasmid, pPLGFP::SW2, did not express GFP. This data indicates that GFP expression is specifically driven by Peuo.
Figure 1. A promoter from the euo gene drives GFP expression in C. trachomatis.
(a) Map of transcription reporter vector pPvGFP::SW2. A core promoter from the C. trachomatis euo gene was cloned upstream of the tuf RBS region and the gfp gene. The unique restriction sites are indicated. Bar = 10 μm. (b) Appearance of living L2/pPvGFP::SW2 infected HeLa cells at 16 h pi. GFP-expressing inclusions (green) are shown with arrows. (c) Appearance of living L2/pBOMB-Peuo infected cells at 40 h pi. Ptet-controlled mCherry (red) was induced by adding αTC (20ng/ml) immediately after infection. Bar = 20 μm.
To facilitate GFP quantitation in C. trachomatis using an internal protein control, a DNA fragment containing MCS-Peuo-tuf RBS from pPvGFP::SW2 was subcloned into pBOMB4-tet-mCherry20 (hereafter called pBOMBm), which contained the fluorescent protein mCherry gene driven by a tetracycline controlled promoter (Ptet). The resultant plasmid, pBOMB-Peuo, was transformed into L2/25667R cells to generate L2/pBOMB-Peuo. Infection of L2/pBOMB-Peuo in HeLa cells formed GFP-expressing inclusions at 16 h pi and beyond, a phenotype similar to that of L2/pPvGFP::SW2. Ptet-driven mCherry expression was induced in the presence of anhydrotetracycline hydrochloride (αTC) (Fig. 1c). Thus, the level of mCherry expressed from the same plasmid provided a control to normalize the levels of GFP expression. Previous studies indicated that the euo gene mRNA was moderately expressed as early as 2 h pi from Chlamydia spp.6,26. The failure to detect Peuo-driven GFP expression prior to 16 h pi by fluorescence microscopy was perhaps due to the small inclusion size and low bacterial accumulation. It is also possible that the core Peuo region used in the construct is intrinsically weak and (an) additional DNA element(s) upstream or downstream may be required for its optimal expression in C. trachomatis. Nevertheless, our results indicate that GFP can be produced under the control of Peuo and the tuf translation machinery ribosome complex.
Prediction of ompA P3
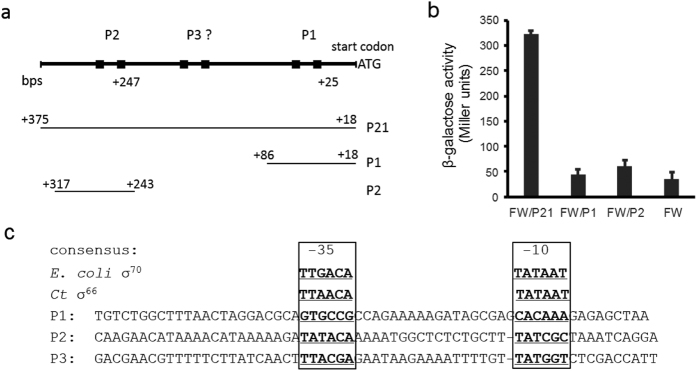
C. trachomatis surface-exposed MOMP, encoded by the ompA gene, is a critical component that directly mediates pathogen-host interactions25. Developmentally regulated ompA transcription has been a topic of extensive study12,14,27,28. Although ompA is expressed as multiple mRNAs, only the P1-derived short transcript and the P2-derived long transcript have been studied in detail12,14,29. To quantify the effects of DNA regions on ompA promoter activity, we created a set of lacZ transcriptional fusions in E. coli based on Whipple’s system30. The regions carrying the promoters (either combined P2 and P1, or P1 or P2 alone) were cloned into the lacZ expression vector, pFW11, and the promoter-lacZ fusion was relocated to an F′ episome in the recipient cells as detailed in the Materials and Methods section. The resultant strains, designated FW/P21, FW/P1, or FW/P2 (Fig. 2a), were subjected to β-galactosidase (β-gal) assays to measure the activity of ompA promoters. We observed a 6-fold increase in β-gal activity in FW/P21 cells compared to the promoter-less control (Fig. 2b). However, no β-gal levels exceeding that of the promoter-less control were produced in FW/P2 or FW/P1. These results are comparable to previous studies using multi-copy plasmid-encoded chloramphenicol acetyltransferase (CAT) as a reporter in E. coli12. Although the activity of P2 has been authenticated in C. trachomatis, the role of P1 is still under debate12,29. It is unsurprising that P2 is not active in E. coli because P2 has a GC-rich −10 hexamer of TATCGC, which is not recognized by E. coli σ70 in vitro29. The high levels of β-gal observed in strain FW/P21 could therefore not be explained by the lack of β-gal activity in strains FW/P1 and FW/P2. We suspected that an additional promoter recognized by E. coli transcription machinery must be present between the P2 and P1 regions.
Figure 2. Studying C. trachomatis ompA promoter activity in E. coli.
(a) Promoter-lacZ constructions. The organization of the regulatory region upstream of the ompA coding region (top) and DNA fragments used to create the lacZ reporter strains are shown. Positions relative to the translation start codon ATG of ompA are indicated. (b) Results of β-gal activity. E. coli strains, FW/P21, FW/P2, and FW/P1, were harvested at the exponential phase and subjected to β-gal assays. Strain FW (vector only) was used as a control. Data are presented as mean ± SD from a representative experiment of three independent experiments. (c) Base sequences of putative ompA promoters used in this work. Putative −35/−10 hexamers are bolded. The E. coli σ70 or C. trachomatis σ66 consensus recognition sequences are listed at the top.
We next searched for the putative promoter (s) from the 607 bp intergenic sequences (IGSs) upstream of the ompA of C. trachomatis strain L2/434/Bu in silico using a dynamic program-based MotifSearch tool10. A two-block-motif, TTAACA (−35)-n16–19-TATAAT (−10), was used for C. trachomatis σ66 recognition sequences31 and motif TAAAGTTT (−35)-n10–14-GTTGACAA (−10) was used for σ28 recognition sequences10. Up to 3 base-pair mismatches (a total 6 bases) were allowed in the −35 hexamer or −10 hexamer. These predictions resulted in the identification of the known ompA P2 region and a region containing the sequence TTACGA-n17-TATGGT, which we named P3 (Fig. 2c). The same P3 region was predicted using a position weight matrix-based program called Footy32. P1 was excluded, as it contained mismatches in five of six positions in the putative −35 hexamer and in three of six positions in the −10 hexamer. No σ28 recognition sequences were found. An alignment of Chlamydia spp. IGSs upstream of ompA available in the NCBI database shows a minor variation in P3 and P2 regions (Table S1). This suggests that these regions may have a conserved role in ompA expression in Chlamydia spp. There are more variations in the ompA P1 region in terms of the length of the “spacer” separating the putative −35/−10 hexamers, ranging from 18 to 26 bps. Chlamydia spp. σ66-dependent promoters typically have a 16–18 bp spacer11,31, resembling those of E. coli σ70, which also senses the promoter spacing and preferentially recognizes a spacer with an average of 17 bps33.
Validation of P3 function in C. trachomatis
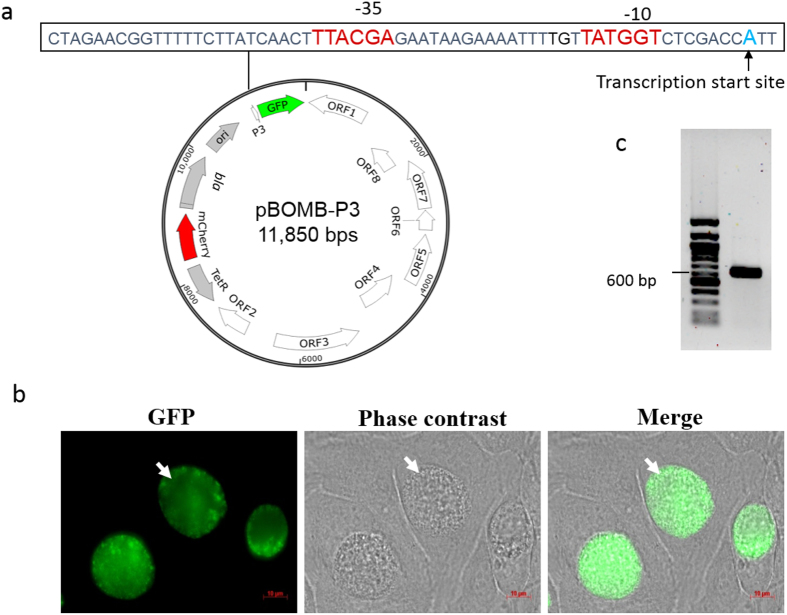
To directly determine whether P3 functions in C. trachomatis, the 63 bp P3 region was cloned into pPvGFP::SW2 and pBOMB-Peuo, in place of Peuo, generating pP3GFP::SW2 and pBOMB-P3 (Fig. 3a), respectively. These plasmids were each transformed into the L2/25667R cells, resulting in L2/pP3GFP::SW2 and L2/pBOMB-P3. After three rounds of ampicillin selection, GFP-expressing inclusions were observed (Fig. 3b). Inclusion morphology in both strains carrying pP3GFP::SW2 and pBOMB-P3 appeared to be normal and not significantly different. Strain L2/pBOMB-P3 was chosen for further characterization.
Figure 3. Determining P3 activity in C. trachomatis.
(a) Map of pBOMB-P3. Sequences of P3 region are indicated. (b) Appearance of living L2/pBOMB-P3 infected HeLa cells at 30 h pi. GFP-expressing inclusions are marked with white arrows. (c) The ompA P3 transcriptional start site (TSS) was identified by 5′-RACE assays. The PCR products next to the DNA ladder on the agarose gel were subjected to sequencing, and the TSS of P3 appears to be “A” (in blue font), marked with an arrow. The putative −35 and −10 hexamers are indicated in red font.
Next, 5′ RACE assays were performed to determine the transcription start site (TSS) of P3-derived mRNA. Initially, total RNA isolated from the strain L2/434/Bu infected HeLa cells harvested at 18 h pi were used. Our attempt to detect the 5′ end of P3 mRNA was unsuccessful, although a site corresponding to the reported P1 mRNA14,27 was noted. We hypothesized that if the undetectable 5′ end of the P3 transcript is the result of low steady-state levels of P3 mRNA, the use of the C. trachomatis tuf RBS region should improve the stability of P3 mRNA in the reporter system. To test this hypothesis, P3 transcript was determined using a 5′ RACE assay with total RNA isolated from L2/pBomB-P3 infected HeLa cells harvested at 18 h pi. The DNA sequencing data showed that P3 transcript originated with an adenine located at 39 bp upstream from the gfp start codon (Fig. 3c). Based on the TSS at position +1, the sequences of TTACGA and TATGGT are putative −35 and −10 hexamers of P3. Yuan et al.28. have previously reported a similar site in C. psiticci Mn Cal 10 and C. psiticci GPIC (now C. caviae according to current chlamydiae taxonomy)34. We conclude that P3, lying between the previous reported P2 and P1, indeed functions actively in C. trachomatis.
Defining the core P3 promoter
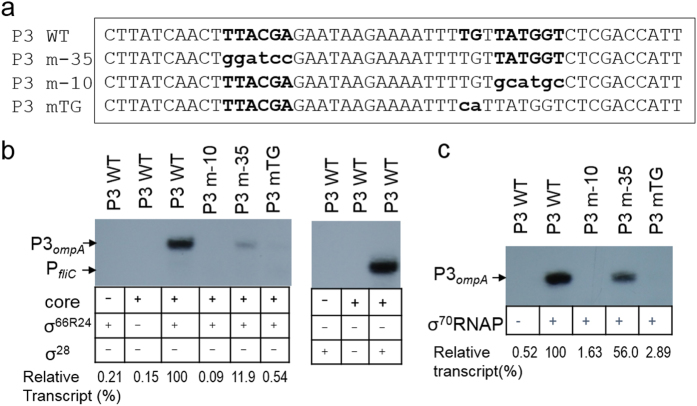
To precisely define the relevant sequence features needed for the recognition of P3, we investigated how specific mutations of P3 could affect transcription in vitro. Plasmids containing wild-type (WT) or mutated P3 regions (Fig. 4a) and a control promoter of the E. coli fliC (PfliC)35 were used as templates. A functional RNAP holoenzyme was reconstituted with core enzyme from E. coli and a hybrid C. trachomatis σ factor, σ66R24, in which σ66 regions 2 to 4 (amino acid residues 315–571) were translationally fused to the region 1 of E. coli σ70 (amino acid residues 1–372). The use of this holoenzyme allows for the study of σ66-dependent promoter activity that relies on the function of σ66 regions 2 to 4. E. coli σ70 or C. trachomatis σ28 were used as controls. In the presence of σ66R24- or σ70-RNAP holoenzyme, transcripts from WT P3 were evident (Fig. 4b,c). In contrast, no P3–derived transcript was observed in the presence of σ28. These data indicate that P3 is specifically recognized by C. trachomatis σ66 and its homolog E coli σ70, consistent with our β-gal reporter assay data in E. coli (Fig. 2). Substitution of the TATGGT (−10 hexamer) with GCATGC abolished P3 transcription activity when either σ66R24RNAP or σ70RNAP was used, indicating the importance of the TATGGT hexamer for P3 activity. Interestingly, substitutions of the TTACGA (−35 hexamer) to GGATCC reduced 80% of P3 transcript in comparison to the WT P3 using σ66R24RNAP, while the use of σ70RNAP reduced P3 transcript only by 50%, suggesting that the TTACGA hexamer is more recognizable by C. trachomatis σ66. These results indicate that the TTACGA sequence of P3 functions as the −35 promoter element and confirm that this sequence is required for the full activity of P3. We noted that the −16TGt−14 sequences in P3 matched the consensus extended -10 TG motif recognized by σ70 from E. coli36,37. Substitution of TG with CA resulted in a significant decrease in P3 transcript levels, suggesting that the TG motif is important for P3 activity (Fig. 4). C. trachomatis σ66-dependent promoters have typically been characterized as −10/−35 promoters. Our data, for the first time, shows that P3 activity requires the −35 hexamer and an extended −10 TGn motif, presenting a new promoter class in C. trachomatis.
Figure 4. Identifying the determinants of ompA P3 region.
(a) Sequences of P3 and its derivatives tested in this study. (b) Autoradiogram of a denaturing acrylamide gel showing the transcript products of in vitro transcription assays with P3 or its derivatives. σ66R24 RNAP holoenzyme (left panel) or σ28 RNAP holoenzyme (right panel) were used. (c) An autoradiogram of a denaturing acrylamide gel showing ompA P3 transcripts produced in the transcription assay. E. coli σ70 RNAP holoenzyme was used. The amounts of transcripts were determined by densitometry using ImageJ49 and are shown relative to WT P3, which is set at 100%, as indicated at the bottom of (b,c). Note: P3 is transcribed by both σ66R24 RNAP and σ70 RNAP, but not σ28 RNAP.
Promoters harboring the TGn motif have been identified from several microbial systems, including E. coli37, Streptococcus pneumoniae38, Bacillus subtilis36, and Mycobacterium tuberculosis39. It has been shown that the recognition of the TGn motif by the principle σ factor plays a role in the formation of an open complex during transcription initiation. Moreover, the TGn motif may compensate for non-canonical −10 hexamers or suboptimal spacer length between −10 and −35 hexamers. The TGn-promoters appear to more often have significant deviations in the –35 hexamer than typical –10/–35 promoters in E. coli. It seems that ompA P3 deviates substantially from both the consensus −10 hexamer and −35 hexamer, because of its GC-rich sequences in these regions. Like E. coli σ70, C. trachomatis σ66 region 2 binds to the promoter −10 hexamer, region 4 of σ66 recognizes the −35 hexamer40. It is likely that the TGn motif allows for extra contact points with σ66 regions 2.5 and 3.0, as has been observed in its E. coli counterpart33,37,41.
Quantifying the contribution of P3 to ompA transcription
To define the role of P3 in ompA transcription and to understand the potential relationship between P3 expression and the action of the well-studied P214,27,29, we quantified the levels of ompA P2 and/or P3 mRNA in the context of chromosomal loci using real-time RT-qPCR. Total RNA from L2/434/Bu infected cells, which were sampled at 0, 2, 4, 12, 24, and 36 h pi, were analyzed (Fig. 5a). P3 transcript was detected early at a low level and continued to accumulate in abundance, exceeding that of P2 at both 2 and 4 h pi (Fig. 5b). P2 transcript rapidly increased with bacteria multiplication and peaked at 12 h pi. However, at 12 h pi, the use of P3 decreased and that of P2 drastically increased. Expression of both ompA P2 and P3 was significantly eliminated at 36 h pi. These data support the notion that P2 is the primary transcript during RB replication and RB-to-EB differentiation, as previously reported27, whereas P3 plays a role in the early stage of the development. Therefore, temporally expressed P3 and P2 coordinate ompA transcription during the C. trachomatis developmental cycle.
Figure 5. The profile of ompA P3 and P2 expression in chromosomal loci.
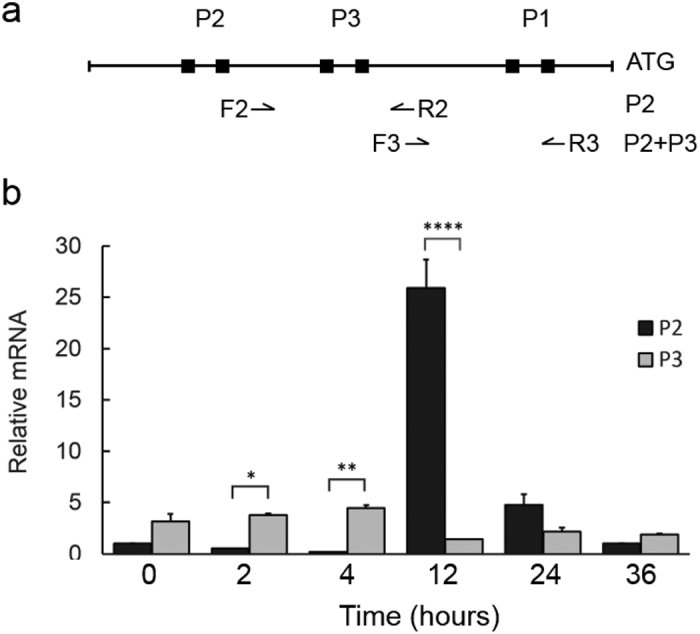
(a) Diagram showing the location of primers designed to assess ompA transcripts by RT-qPCR. Amplification with the primers, F2/R2, produces P2 transcript, whereas amplification with primers, F3/R3, generates the total transcripts of P2 and P3. The individual P3 transcript is obtained by subtracting P2 transcript from both P3 and P2 transcripts. (b) Results of RT-qPCR with total RNA from L2/434/Bu infected HeLa cells harvested at 0, 2, 4, 12, 24, and 36 h pi. Relative amounts of transcript were obtained by normalizing the levels of P2 or P3 transcript to C. trachomatis genomic DNA levels, which were assessed using primers specific to the 16S rRNA gene (Table S3). The values are presented as mean ± SD from an experiment with duplicates. *P < 0.05, **P < 0.005, and ****P < 0.0001. Three independent experiments were performed.
Evaluating the use of P3-GFP as a reporter in C. trachomatis infected epithelial cells
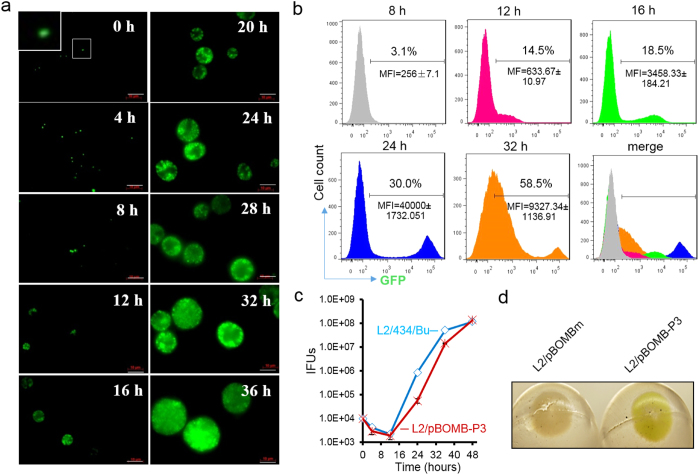
C. trachomatis growth is accompanied by quantitative and qualitative changes in ompA expression (Fig. 5). To investigate the relationship between P3-GFP level and C. trachomatis growth, GFP in L2/pBOMB-P3 infected HeLa cells was monitored throughout the C. trachomatis developmental cycle using fluorescence microscopy and flow cytometry. In parallel, a one-step growth curve was constructed to enumerate infectious EB progeny yields. We visualized GFP-expressing chlamydial organisms associated with the host cells instantly post-infection, and the levels of GFP expression increased through 36 h pi as C. trachomatis inclusions expanded (Fig. 6a). When quantitatively measured using flow cytometry, the GFP signal was detected instantly after infection but the signal-to-noise ratio was low, preventing meaningful quantitative measurements of cells. The distinct cell populations with bona fide P3-GFP expression were separate from the GFP negative cells until 8 h pi (Fig. 6b). The population of GFP-expressing cells increased from ~3.1% at 8 h pi to 30% at 24 h pi; concurrently, the mean florescence intensity (MFI) significantly increased (Fig. 6b). These increases were correlated with the rapid growth of C. trachomatis (Fig. 6c). Interestingly, while GFP-expressing cells increased to 58.5% at 32 h pi, there were more heterogeneous cell populations which displayed a lower average MFI with a larger deviation compared to that at 24 h pi. This could be explained by the asynchronous growth of C. trachomatis, which occurs when decreasing numbers of RBs continue replicating and increasing progeny EBs accumulate and exit from the host cells to initiate a new cycle of infection. The overall P3-driven GFP levels seemed to be quite stable. Thus, it is likely that the GFP signal detected at the late stage is skewed by GFP accumulation along with bacteria accretion and inclusion expansion. We observed that L2/pBOMB-P3 exhibited delayed growth pattern relative to L2/434/Bu (Fig. 6c), possibly due to the stress induced by GFP overexpression. Isolated L2/pBOMB-P3 organisms from infected cells harvested at 40 h pi appeared green-tinged, a change detectable by eye (Fig. 6d), indicating high levels of GFP associated with bacteria. Long-lived P3-GFP used in this study may be applicable for long-term bacteria cell-labeling, which allows for the imagining to track the C. trachomatis infection process in intact cells, e.g. invasion, multiplication, and dissemination.
Figure 6. Inspecting P3-GFP levels during the course of the C. trachomatis infection.
(a) Time course of GFP expression in HeLa cells visualized by fluorescent microscopy. L2/pBOMB-P3 infected HeLa cells were imaged at the times indicated. The inset highlights GFP-expressing C. trachomatis organisms. Bar = 10 μM. (b) Flow cytometric data corroborate microscopic images in (a), showing changes in GFP expression in C. trachomatis infected HeLa cells. GFP-expressing cell population is indicated as percentage. The average mean fluorescence intensity (MFI) and the standard deviation at each time is shown. (c) One-step growth curve of C. trachomatis strains L2/434/Bu and L2/pBOMB-P3. (d) Isolated L2/pBOMB-P3 organisms, but not L2/pBOMBm, appear to be green-colored by direct observation.
Determining the strength of P3 in C. trachomatis
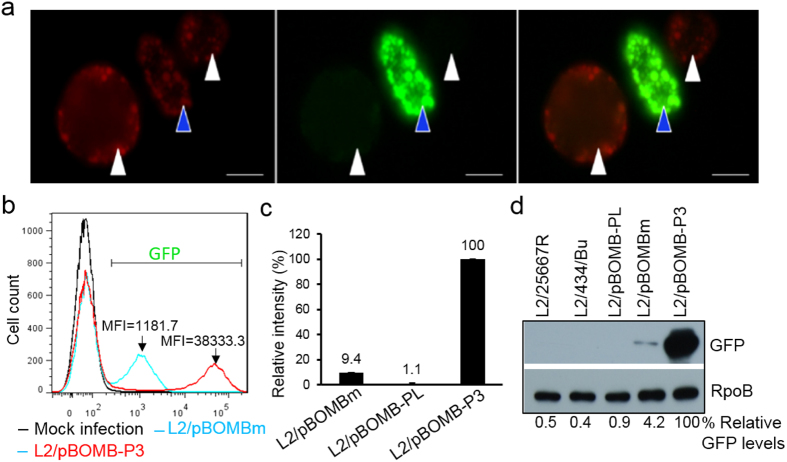
To evaluate the strength of P3, we assessed the P3-driven GFP levels in L2/pBOMB-P3 compared to those driven by the Neisseria meningitidis promoter, Pnm, in L2/pBOMBm18,20. Four different quantitative methods (microscopy, flow cytometry, microspectrometry, and immunoblotting) were used. First, to facilitate a direct comparison, an equivalent mixture of L2/pBOMB-P3 and L2/pBOMBm was used to infect HeLa cells, followed by fluorescence microscopy analysis. In the same field of vision, L2/pBOMB-P3 exhibited a brighter green fluorescence than L2/pBOMBm (Fig. 7a). In the presence of equivalent mCherry levels, which were induced by the addition of αTC, L2/pBOMB-P3 displayed higher levels of GFP signal than L2/pBOMBm. Next, cells infected with either L2/pBOMB-P3 or L2/pBOMBm with similar infectivity (~30%) were harvested at 24 h pi and subjected to flow cytometry. As determined by assessment of the MFI, the average level of P3–driven GFP expression was ~32 times stronger than Pnm-driven GFP at 24 h pi, (Fig. 7b). Further, isolated C. trachomatis organisms from cells harvested at 24 h pi were measured for their relative fluorescence intensities (RFI) using microspectrometry. Figure 7c shows that the RFI of L2/pBOMB-P3 was ~10-fold higher than that of L2/pBOMBm. Lastly, the level of GFP protein in isolated C. trachomatis organisms was determined by immunoblotting. The levels of GFP in lysates of L2/pBOMB-P3 were ~24-fold higher than those of L2/pBOMBm (Fig. 7d). With the use of similar plasmid constructs, these results demonstrate that, relative to Pnm, P3 is highly active in C. trachomatis. Furthermore, bacterial organisms expressing P3-GFP are more noticeable and easier to measure using fluorescence microscopy, flow cytometry and immunoblotting, rather than microspectrometry.
Figure 7. Quantitative analysis of the ompA P3 strength in C. trachomatis.
(a) Appearance of GFP-expressing inclusions (green) upon induction of mCherry (red) expression by the addition of αTC. HeLa cells co-infected with strains L2/pBOMB-P3 (blue arrow) and L2/pBOMB-m (white arrow) were photographed at 24 h pi. Bar = 5 μm. (b) Flow cytometry measures of P3-GFP intensity compared to that of Pnm–GFP. C. trachomatis infected HeLa cells were harvested at 24 h pi and subjected to flow cytometry. (c) Assessing GFP levels of cell-free C. trachomatis organisms using microspectrometry as detailed in Materials and Methods. (d) Immunoblotting of GFP protein. Bacteria with the same OD600 values (OD600 = 0.4) were used in the experiments. The amount of protein were determined by densitometry using ImageJ. The relative amount of GFP was obtained by normalizing the GFP intensity to the corresponding RpoB intensity and value is reported as a percentage relative to L2/pBOMB-P3, as indicated at the bottom.
OmpA P3 displays weak transcription activity in E. coli
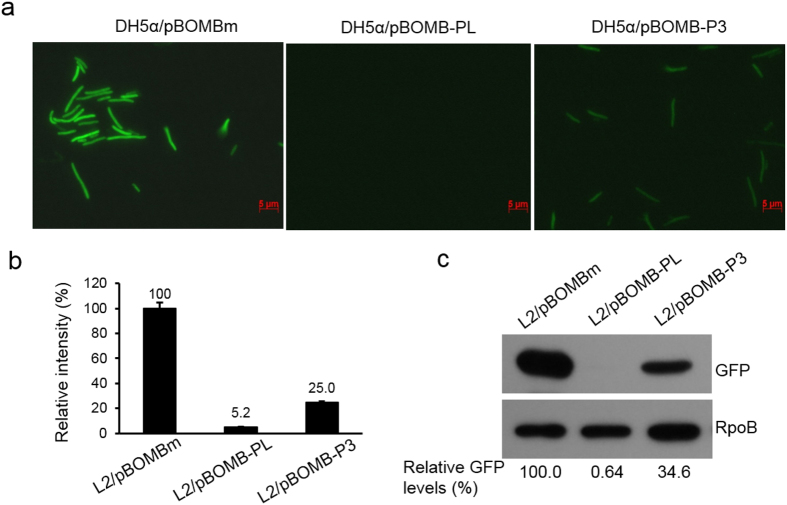
Our next step was to determine whether P3 activity varies with the E. coli σ70 paradigm. This information may provide new insight into the diversity of bacterial gene transcription and also may provide an explanation for the previous detection of P3 in E. coli systems. To this end, E. coli strains carrying pBOMB-P3, pBOMBm, and promoter-less pBOMB-PL were harvested at the exponential phase and their GFP expression was measured using fluorescence microscopy, microspectrometry, and immunoblotting. A significant increase in GFP expression in E. coli cells was observed as the bacterial culture grew (Fig. 8a), confirming that P3 was recognized by E. coli RNAP. This data is consistent with the earlier observations as shown in Figs 2 and 4b. Unlike in C. trachomatis, where P3 is highly expressed, P3-driven GFP expression was much lower than Pnm-driven GFP in E. coli (Fig. 8b,c). The weak activity of P3 in E. coli is not surprising, as perfect P3 function may be achieved only in the context of a regulatory network unique to C. trachomatis; different regulatory mechanisms exist in E. coli.
Figure 8. The OmpA P3 is weakly active in E. coli.
(a) Visualizing GFP expression in E. coli DH5α cells harboring pBOMBm, promoter-less pBOMB-PL, and pBOMB-P3 by fluorescence microscopy. Fixed E. coli cells harvested from LB cultures grown for 3 hours were used. Bar = 5 μm. (b) Assessment of GFP intensity of E. coli cells using microspectrometry as detailed in the Materials and Methods section. (c) Quantification of GFP by immunoblotting. Bacterial lysates were used for immunoblotting with antibodies to GFP or RpoB which was used as a loading control for protein amounts. The relative amount of GFP was obtained by normalizing the GFP intensity to the corresponding RpoB intensity and value is reported as a percentage relative to L2/pBOMB-P3, as indicated at the bottom.
Given the high similarity between C. trachomatis σ66 and E. coli σ70 in their regions 1 to 4, the use of heterogeneous E. coli systems has provided useful information to understand some questions regarding chlamydial promoter recognition as described both previously and in this study. However, these heterogeneous systems do not allow for the expression of all C. trachomatis genes, such as ompA P229. P3 functions weakly in E. coli and appears to rely on the availability of endogenous conditions for optimal expression in C. trachomatis. Such variety might be attributed to the non-conserved regions between C. trachomatis σ66 and E. coli σ70, including the area between regions 1 and 2, as well as the regions at the N- and C-termini of σ factors42,43. Differences between C. trachomatis σ28 and E. coli σ28 have been studied previously10,44. Additionally, physiological signals within the host cells (such as immunity and metabolic capacity) that C. trachomatis encounters affect bacterial gene expression. Therefore, it is necessary to validate the promoters that were predicted or identified by the heterologous systems in C. trachomatis. A major advantage of the transcription reporter shuttle plasmid with an MCS and tuf RBS upstream of GFP is to allow a preferred promoter to be inserted conveniently so that promoter functioning can be studied by assessing the levels of GFP expression. Promoters detected in C. trachomatis could be directly compared to those in E. coli, thus providing an opportunity to examine the extent of the differences in their activity.
Conclusion
This study resulted in three significant conclusions. First, the ompA regulatory region is complex and exhibits novel features. In particular, ompA transcription is exactly derived from both mid-cycle expressed P2 and the newly identified, early-expressed P3. Second, P3 represents a new class of C. trachomatis promoter, which requires a −35 DNA element and an extended −10 TGn motif for its transcription. Third, expression of ompA P3-GFP, but not Peuo-GFP, in living cells produces a unique, robust signal. These results provide a basis for a deeper understanding of novel characteristics of the gene regulation in C. trachomatis.
Tandemly arranged promoters have been reported to involve developmentally regulated C. trachomatis gene expression, including ompA27,28 and tuf11. To complement the earlier finding that P2 plays a primary role in ompA synthesis, we have shown that P3 also contributes to ompA transcription (Figs 2, 3, 4, 5). Previous studies have indicated the detection of steady-state levels of ompA transcript mid-cycle6,7. However, the levels of ompA transcripts often do not fully reflect the relative activities of the promoters because the fate of each transcript may be varied and is affected by various processes. The switch in utilization of P3 and/or P2 is evidently a regulatory mechanism of ompA transcription. The manner in which P3 and P2 activity is coordinated or mutually influenced can be affected by multiple factors. A 9-bp inverted repeat adjacent to the −35 hexamer of the P3 region may play a role in this process. Mathews and Stephens12 have proposed that this repeat, resembling an operator in DNA structure, might be a target of a yet not-identified protein repressor. Alternatively, a stable stem-loop structure that resembles a rho-independent terminator capable of stopping P2 transcription could be formed by this region. Additionally, DNA supercoiling has been reported to modify C. trachomatis transcription, including ompA P245. Only a subset of early genes can be affected by DNA supercoiling46. In general, plasmids are supercoiled to a higher degree than chromosomal DNA. We found similar strong activity of P3 regardless of its plasmid and chromosomal locations, suggesting that supercoiling is less likely to be a main factor involved in P3 expression. Moreover, posttranscriptional regulation of ompA mRNA may occur during development. Transcript derived from P3 seems less stable than that of P2 by mRNA structural prediction. Despite its role in early transcription, P3 mRNA might limit its activity through quick turnover when RBs rapidly replicate and convert to EBs. We cannot exclude the possibility that P1 signal detected is the processed product of longer transcripts14, perhaps from P3 and/or P2. It is worth noting that the utility of tuf RBS in mediating the translation of gfp may be mechanistically different from, and possibly more efficient than, the native ompA RBS. Because of the apparent conservation of the P3 and P2 regions, their importance in Chlamydia spp. adaptation to changing environments can be predicted. The proposed P1 remains unproven, but phylogenetic analysis suggest that P1 is less likely to be the conserved regulatory mechanism of ompA in Chlamydia spp.
Our findings underscore the role of new promoter element, the extended −10 TGn, in C. trachomatis transcription. By sequence inspection, early promoters in C. trachomatis, Peuo and the promoter from C. trachomatis groES13, also contain potential extended −10 TGn motifs. It is possible that such a motif represents a signature element for early promoter-RNAP recognition and is affected differently from the typical −35/−10 promoters by a transcription factor. For example, we have found that anti-σ factor, CT663 (or Scc4)40, strongly inhibited transcription from the −35/−10 promoters but was less effective in suppressing transcription from an extended −10 promoter lacking a −35 element. Despite the limited number of transcription factors in C. trachomatis, the arrangements of promoter modules may be largely diverse and greatly contribute to developmentally regulated gene expression profiles.
The consistency of the different quantitative methods employed show that robust P3-GFP signal is present throughout the developmental cycle. These properties make P3-GFP valuable for long-term bacterial cell-labeling to facilitate the visualization and tracking of infectious process in live cells. Coupled with flow cytometry and microscopy, this GFP-reporter assay may also be applicable in probing ompA changes induced by various insults at both population and single cell level. Adverse growth conditions, such as exposure to IFNγ or antibiotics, cause C. trachomatis ompA downregulation and the formation of an “altered persistent form”17,47. With regard to this, low levels of GFP expression driven by a weak promoter, such as Peuo, could be challenging to interpret. At the same time, the use of long-lived GFP may be limited, as it is incapable of fully reflecting the highly dynamic process of gene expression. An improved short-lived GFP reporter may be more suitable for studying the transient mRNA dynamics in C. trachomatis. All together, the results obtained and approaches used in this study will allow for the design of future studies in defining promoter signatures, relationships between promoter structure, RNAP recognition, and transcriptional activity in C. trachomatis.
Materials and Methods
Cell cultures and bacteria growth
HeLa 229 cells (human cervical epithelial carcinoma cells; ATCC CCL-2) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1 mM glutamic acid, 10% fetal bovine serum and 20 μg/ml gentamycin (DMEM-10) at 37 °C in an incubator supplied with 5% CO2. HeLa cells were infected with C. trachomatis as previously described17. To construct a one-step growth curve, cells infected at an MOI of 1 were lysed and cultured on fresh HeLa cells at 0, 4, 12, 24, 32, and 48 h pi. Infectious EB progeny was then evaluated by enumeration of inclusion forming units (IFUs) in 1 ml. E. coli DH5α was used as the host for cloning. A methylation deficient E. coli strain (ER2925, New England Biolabs) was used to prepare the plasmids for C. trachomatis transformation. The E. coli cells were grown in Luria-Bertani (LB) broth or agar plates containing the appropriate antibiotics.
Molecular cloning
Plasmids, primers, and oligonucleotides used in this study are listed in Table S2 and Table S3, respectively. Primers and oligonucleotides were synthesized by Integrated DNA Technologies. To construct pPvGFP::SW2 (Fig. 1a), two separated PCR fragments were obtained with primers, Euo_tuf priF/ORF2 priF or euoPriR/ORF1priF, using pGFP::SW218 (a gift form Dr. Ina Clarke, University of Southampton, UK) as template. These PCR fragments were mixed, annealed, and used as templates for subsequent PCR using primers ORF2 priF/ORF1priF. The Bam HI-cut PCR fragment was then inserted into the sites of Bam HI in pGFP::SW2. To create pP3GFP::SW2, the two annealed complementary oligonucleotides consisting of ompA P3 were cloned into pPvGFP::SW2 at the Spe I/Nae I sites. The Puv I/Nco I fragment of the pP3GFP::SW2 was cloned into the Pvu I/Nco I sites in pBOMB4-tet-mCherry (a gift form Ted Hackstadt, Rocky Mountain Lab, NIH) to yield pBOMB-P3 (Fig. 3a). To create a promoter-free plasmid, pPvGFP::SW2 was digested by Spe I/Nae I to remove Peuo. After Kenow fragment treatment, the DNA was self-ligated to create pPLGFP::SW2. The Puv I/Nco I fragment was subcloned into pBOMB to create pBOMB-PL. The plasmids used for the in vitro transcription assay were derived from pPflic35. Two annealed complementary oligonucleotides consisting of WT or mutated P3 were inserted into pPflic at the Xba I/EcoR V sites to yield pP3WT-PfliC, pP3m10-PfliC, pP3mTG-PfliC or pP3m35-PfliC. The expression plasmid, pLN-σ66R24, containing genes encoding C. trachomatis σ66 regions 2–4 and E. coli σ70 region 1, was derived from pLNH-12. The PCR fragment consisting of region 1 of E. coli σ70 was amplified with primers s70priF/Sig7066Rg24BLo. The PCR fragment containing σ66 regions 2–4 was amplified with primers sig7066Rg24CUp/S66prRBam. These DNA fragments were annealed and used as a template for subsequent PCR using primers EC70priF/S66prRBam. The resultant PCR fragments were digested with Nco I/Bam HI and were then inserted into the Nco I/BamH I sites of pLN-7066 to generate pLN-σ66R24, which encodes a fusion σ66R24 protein. The identities of all constructs were confirmed by PCR and DNA sequencing.
Generating ompA promoter-lac Z fusion in E. coli
A DNA fragment containing the ompA P1 and P2 regions (+375 to +18 relative to the translation start codon ATG of ompA) was amplified by PCR with primers ompApriF/P1priR using the genome of C. trachomatis serovar D as a template. The EcoR I/Sal I digested PCR fragment was then inserted into the Eco RI/Sal I sites of pFW11 to yield pFW/P21. Two annealed oligonucleotides containing P2 (+317 to +243 relative to the ompA start codon) or P1 (+87 to +18 relative to the ompA start codon) were inserted into pFW11 at Eco RI/Sal I sites, resulting in pFW/P2 or pFW/P1 (Fig. 2a). These constructs were introduced into E. coli CSH100 for homologous recombination of the ompA promoter::lacZ onto an F’ episome as previously described30. Further mating with E. coli strain FW102 was performed to finalize the construction of the reporter strains, designated as FW/P21, FW/P2, and FW/P1, respectively. The identities of all constructs and strains were confirmed by PCR and DNA sequencing.
Transformation of C. trachomatis
C. trachomatis was transformed with the shuttle plasmids according to the method described18 with minor modifications. Briefly, plasmid-free C. trachomatis L2/25667R EBs (1 × 107) were mixed with 7 μg of plasmid DNA in 100 μl of CaCl2 buffer (5 mM Tris HCl, pH7.4, 100 mM CaCl2) and incubated at room temperature for 30 minutes. Freshly trypsinized HeLa cells (6 × 106) resuspended in 200 μl CaCl2 buffer were added to the plasmid/EB mixture and incubated at 37 °C for an additional 20 min. Aliquots of this mixture were then added to a 6-well plate with 1.0 ml of pre-warmed medium in each well. After culturing in DMEM-10 without antibiotics at 37 °C for 24 h, cells were incubated in the presence of ampicillin (5 μg/ml) and cycloheximide (20 μg/ml) for an additional 24 hours. The infected cells were harvested and lysed by vortexing with glass beads. The cell debris was removed by spinning at 233 g for 10 min. The Chlamydia-containing supernatant was collected and added onto a HeLa monolayer in a T175 flask. After incubation at 37 °C for 1 hour with gentle shaking, the infected cells were cultured in DMEM-10 containing ampicillin and cycloheximide for 48 hours. The C. trachomatis organisms were isolated by 30% renografin density gradient purification to enrich infectious EBs. For the first passage, a HeLa monolayer in a 6-well plate was inoculated with EBs, followed by centrifugation to enhance infection. Passages were continued 2–3 times, until inclusions positive for green fluorescence were observed. To isolate a single clone of C. trachomatis transformant, GFP-expressing cells were lysed in sterile dH2O, serially diluted, and inoculated onto a HeLa cell monolayer in a 96-well plate. At 24 h pi, the plate was examined by fluorescence microscopy. The culture wells containing only a single inclusion were marked and continued to be cultured until subsequent harvest for C. trachomatis large scale amplification.
Phase contrast and fluorescence microscopy
HeLa cell monolayers grown in a glass culture chamber (NUNC) or 24-well culture plate were infected with C. trachomatis to achieve ~30% infection. At various times post infection, the culture wells were observed and photographed with an inverted fluorescence microscope (Zeiss Axio Observer D1). Images were processed using AxioVision software version 4.8.
Microspectrometry
Isolated chlamydial organisms from infected cells harvested at 36 h pi were washed with PBS, diluted to OD600 = 0.4, and subjected to fluorescence intensity analysis using a Synergymx microplate reader (BioTek) with excitation wavelengths of 48 nm and 528 nm. A volume of 100 μl of bacterial cell dilutions was added into each well of a 96-well plate. The following formula was used to determine relative fluorescence intensity (RFI): RFI = (bacterial fluorescence intensity–background)/bacterial OD600. The overnight cultures of E. coli DH5α carrying GFP expression plasmids were diluted in fresh LB (1:100) and grown at 37 °C. E. coli cells grown at log phase (4 hours after inoculation) were collected (0.5 ml) by centrifugation and resuspended in the same volume of PBS. The fluorescence intensity of E.coli was obtained in the same manner as described above for C. trachomatis.
Flow cytometry
C. trachomatis infected HeLa 229 cells were detached by trypsinization at different times as indicated in the results and fixed with 2% paraformaldehyde at room temperature for 20 minutes. Cells were then detected in a FACSVantage flow cytometer (BD Biosciences) using the FL-1 (green) channel. Flow cytometry data was recorded for at least 3 × 104 cells per sample. Mock infection cells were used as a blank. Data were analyzed using FlowJo software Version 7.6 (TreeStar Inc) for both the percentage of GFP-expressing cell populations and mean fluorescence intensity.
Immunoblotting analysis
Chlamydial organisms were lysed with 2 × SDS loading buffer, separated on 4–20% SDS-PAGE, and transferred onto a PVDF membrane for immunoblotting. The membrane was incubated with a polyclonal anti-GFP antibody (Pierce) or a monoclonal antibody to bacterial RNAP β subunit (RpoB) (Neoclone), followed by incubation with horseradish peroxidase-conjugated goat anti-mouse IgG. RpoB was used as a protein loading control. The protein bands were visualized by an enhanced chemiluminescence kit (Pierce).
5′ Rapid amplification of cDNA ends (RACE)
C. trachomatis-infected cells with ~90% infectivity were harvested at 18 hours. Total RNA was isolated using TRIzol reagent (Invitrogen). The transcription start site for P3 in C. trachomatis was probed with the rapid amplification of the cDNA ends (5′-RACE) using the FirstChoice ® RLM-RACE Kit version 2.0 (Life Technologies) following the manufacturer’s protocol. Briefly, 10 μg of total RNA were treated with calf intestine alkaline phosphatase to remove free 5′-phosphates. The RNA was then treated with tobacco acid pyrophosphatase to remove the cap structure from full-length mRNA, leaving a 5′-monophosphate. An RNA adapter oligonucleotide was then ligated to the RNA with 5′-phosphate using T4 RNA ligase. The 5′ end of the gfp transcript, which is initiated by P3, was then amplified with random-primer reverse transcription and nested PCR using gfp specific primers with the provided primers in the kit. PCR products were then inserted into pUC19 (Promega, Madison, WI) for DNA sequencing.
Real-time reverse transcription quantitative PCR (RT-qPCR)
C. trachomatis infected HeLa cells were harvested at indicated times post infection. Genomic DNA was isolated from cells using the DNeasy® Blood & Tissue Kit (Qiagen). RNA was isolated from an equivalent number of cells using the Direct-zol™ RNA Kit (Zymo). DNase treatment was performed to remove residual DNA. A total of 1 μg of RNA was reverse transcribed into cDNA and the genes of interests were amplified using the VeriQuest Fast SYBR Green qPCR (USB) with appropriate primers (Table S2). The PCR cycle conditions were as follows: 50 °C for 2 min., 95 °C for 5 min., 95 °C for 3 sec., and 60 °C for 30 sec. The latter two steps were repeated for 35 to 45 cycles, and fluorescence was detected at the end of each cycle. Transcripts were normalized to C. trachomatis genomic DNA levels, which were quantified by qPCR with primers specific to the 16S rRNA gene48 (Table S3). The 2-ddCt method was used to obtain relative transcript levels.
Recombinant protein purification, RNAP and the in vitro transcription assays
Recombinant C. trachomatis σ66R24 was expressed in E. coli RosettaTM (DE3) pLysS cells harboring pLN-σ66R24. Cells were grown in LB broth at 37 °C to A600 = 0.8, and then protein expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 0.5 mM. Protein purification was performed using anion exchange chromatography (Source Q, GE Healthcare), followed by gel filtration purification as previously described40. Recombinant C. trachomatis σ28 was purified as previously described35. To make a functional RNAP holoenzyme, the purified σ66R24 (2.0 μg) was mixed with 1 unit of E. coli RNAP core (Epicentre Technologies) and incubated on ice for 30 min. The σ28-RNAP holoenzyme was reconstituted using E. coli core and purified σ28. E. coli σ70 RNAP holoenzyme was purchased from USB. An in vitro transcription assay was performed as described previously10,40, in a 10 μl reaction containing 1 μl 10 × RNA Pol reaction buffer (New England Biolab), 1 μl RNAP holoenzyme, 1 μg of plasmid template, 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.2 μM [α-32P]CTP, 100 μM 3′-O-methylguanosine 5′-triphosphate (GE HealthCare), 2 mM DTT, and 20 U RNase inhibitor (USB). The reaction was incubated for 15 min at 37 °C and terminated by adding loading buffer. The transcripts were separated by electrophoresis with 6% polyacrylamide/8 M urea gel and visualized by autoradiography.
Statistical analyses
Data analyses were performed using GraphPad PRISM software. Statistical significance was determined by two-way analysis of variance (ANOVA). Values of *P < 0.05 were considered statistically significant.
Additional Information
How to cite this article: Cong, Y. et al. Quantifying promoter activity during the developmental cycle of Chlamydia trachomatis. Sci. Rep. 6, 27244; doi: 10.1038/srep27244 (2016).
Supplementary Material
Acknowledgments
We are grateful to Drs Ina Clarke (University of Southampton, UK), David Hackstadt (Rocky Mountain Laboratories, NIH), and Ann Hochschild (Harvard University Medical School) for their generous gifts of plasmids. This work is supported by NIAID/NIH Grant R56 AI093565 and the Louisiana State University School of Medicine’s Dean’s Research Bridge Funding. This work is also supported by the National Natural Science Foundation of China 81370777.
Footnotes
Author Contributions L.S. designed the study. Y.C, L.S., Y. Z., L.G., Y.Z., Y.X. and H.E. performed experiments. L.S and Y.C wrote the main manuscript and all authors reviewed the results and approved the final version of the manuscript.
References
- Rekart M. L. et al. Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ectopic pregnancy. J Infect Dis 207, 30–38, doi: 10.1093/infdis/jis644 (2013). [DOI] [PubMed] [Google Scholar]
- Newman L. et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PloS one 10, e0143304, doi: 10.1371/journal.pone.0143304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder J. W. Interaction of chlamydiae and host cells in vitro. Microbiological reviews 55, 143–190 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelrahman Y. & Belland R. The chlamydial developmental cycle. FEMS Microbiology Review 29, 949–959 (2005). [DOI] [PubMed] [Google Scholar]
- Shaw E. I. et al. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Molecular Microbiology 37, 913–925, doi: 10.1046/j.1365-2958.2000.02057.x (2000). [DOI] [PubMed] [Google Scholar]
- Belland R. J. et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 100, 8478–8483, doi: 10.1073/pnas.1331135100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson T. L., Olinger L., Chong K., Schoolnik G. & Stephens R. S. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol 185, 3179–3189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C. A. et al. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol 63, 141–155 (1998). [DOI] [PubMed] [Google Scholar]
- Stephens R. S. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759, doi: 10.1126/science.282.5389.754 (1998). [DOI] [PubMed] [Google Scholar]
- Shen L. et al. Selective promoter recognition by chlamydial sigma28 holoenzyme. J Bacteriol 188, 7364–7377, doi: 10.1128/JB.01014-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. et al. Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Archives of biochemistry and biophysics 379, 46–56, doi: 10.1006/abbi.2000.1854 (2000). [DOI] [PubMed] [Google Scholar]
- Mathews S. A. & Stephens R. S. DNA structure and novel amino and carboxyl termini of the Chlamydia σ70 analogue modulate promoter recognition. Microbiology 145, 1671–1681, doi: doi: 10.1099/13500872-145-7-1671 (1999). [DOI] [PubMed] [Google Scholar]
- Tan M., Wong B. & Engel J. N. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol 178, 6983–6990 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. L. & Hatch T. P. Functional analysis of the major outer membrane protein gene promoters of Chlamydia trachomatis. J Bacteriol 177, 6286–6289 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M. & Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 174, 3843–3849 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R. J. et al. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci USA 100, 15971–15976, doi: 10.1073/pnas.2535394100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Altered protein secretion of Chlamydia trachomatis in persistently infected human endocervical epithelial cells. Microbiology 157, 2759–2771, doi: 10.1099/mic.0.044917-0 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kahane S., Cutcliffe L. T., Skilton R. J., Lambden P. R. & Clarke I. N. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7, e1002258, doi: 10.1371/journal.ppat.1002258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Yang Z., Lei L., Shen L. & Zhong G. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195, 3819–3826, doi: 10.1128/jb.00511-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauler L. D. & Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196, 1325–1334, doi: 10.1128/JB.01290-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H. & Derré I. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PloS one 8, e57090, doi: 10.1371/journal.pone.0057090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vromman F., Laverrière M., Perrinet S., Dufour A. & Subtil A. Quantitative monitoring of the Chlamydia trachomatis developmental cycle using gfp-expressing bacteria, microscopy and flow cytometry. PloS one 9, e99197, doi: 10.1371/journal.pone.0099197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Douglas A. L. & Hatch T. P. Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect Immun 66, 1167–1173 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell H. D., Kromhout J. & Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31, 1161–1176 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Watkins N., Zhang Y.-X. & Caldwell H. D. Chlamydia trachomatis-host cell interactions: Role of the chlamydial major outer membrane protein as an adhesion. Infect. Immun 58, 1017–1025 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichlan D. G. & Hatch T. P. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol 175, 2936–2942 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R. S., Wagar E. A. & Edman U. Developmental regulation of tandem promoters for the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol 170, 744–750 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Zhang Y. X., Manning D. S. & Caldwell H. D. Multiple tandem promoters of the major outer membrane protein gene (omp1) of Chlamydia psittaci. Infect Immun 58, 2850–2855 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. L. & Hatch T. P. Mutagenesis of the P2 promoter of the major outer membrane protein gene of Chlamydia trachomatis. J Bacteriol 178, 5573–5578 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple F. W. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Research 26, 3700–3706, doi: 10.1093/nar/26.16.3700 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallios R. R., Ojcius D. M. & Ardell D. H. An iterative strategy combining biophysical criteria and duration hidden Markov models for structural predictions of Chlamydia trachomatis sigma66 promoters. BMC bioinformatics 10, 271, doi: 1471-2105-10-271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grech B., Maetschke S., Mathews S. & Timms P. Genome-wide analysis of chlamydiae for promoters that phylogenetically footprint. Research in microbiology 158, 685–693 (2007). [DOI] [PubMed] [Google Scholar]
- Dombroski A. J., Johnson B. D., Lonetto M. & Gross C. A. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc Natl Acad Sci USA 93, 8858–8862 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse K. et al. Emendation of the family Chlamydiaceae: Proposal of a single genus, Chlamydia, to include all currently recognized species. Systematic and Applied Microbiology 38, 99–103, doi: 10.1016/j.syapm.2014.12.004 (2015). [DOI] [PubMed] [Google Scholar]
- Shen L., Li M. & Zhang Y. X. Chlamydia trachomatis sigma28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae. Microbiology 150, 205–215 (2004). [DOI] [PubMed] [Google Scholar]
- Voskuil M. I., Voepel K. & Chambliss G. H. The—16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Molecular Microbiology 17, 271–279, doi: 10.1111/j.1365-2958.1995.mmi_17020271.x (1995). [DOI] [PubMed] [Google Scholar]
- Hook-Barnard I., Johnson X. B. & Hinton D. M. Escherichia coli RNA Polymerase Recognition of a σ70-Dependent Promoter Requiring a −35 DNA Element and an Extended −10 TGn Motif. Journal of Bacteriology 188, 8352–8359, doi: 10.1128/jb.00853-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelnikov A. G., Greenberg B. & Lacks S. A. An Extended −10 Promoter Alone Directs Transcription of the DpnII Operon of Streptococcus pneumoniae. J Mol Biol 250, 144–155, doi: doi: 10.1006/jmbi.1995.0366 (1995). [DOI] [PubMed] [Google Scholar]
- Agarwal N. & Tyagi A. K. Role of 5′-TGN-3′ motif in the interaction of mycobacterial RNA polymerase with a promoter of ‘extended −10’ class. FEMS Microbiology Letters 225, 75–83, doi: 10.1016/s0378-1097(03)00483-x (2003). [DOI] [PubMed] [Google Scholar]
- Rao X. et al. A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes & development 23, 1818–1829, doi: 10.1101/gad.1784009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barne K. A., Bown J. A., Busby S. J. W. & Minchin S. D. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended-10” motif at promoters. EMBO J. 16, 4034 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Nickels B. E. & Fan H. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of sigma66. Proc Natl Acad Sci USA 109, 16870–16875, doi: 10.1073/pnas.1207300109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler J. E., Burgess R. R., Thompson N. E. & Stephens R. S. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. The Journal of biological chemistry 265, 13206–13214 (1990). [PubMed] [Google Scholar]
- Hua Z., Rao X., Feng X., Luo X., Liang Y. & Shen L. Mutagenesis of region 4 of sigma 28 from Chlamydia trachomatis defines determinants for protein-protein and protein-DNA interactions. J Bacteriol 191, 651–660, doi: 10.1128/JB.01083-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E., Cheng E. & Tan M. DNA Supercoiling-Dependent Gene Regulation in Chlamydia. Journal of Bacteriology 190, 6419–6427, doi: 10.1128/jb.00431-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E. & Tan M. Differential Effects of DNA Supercoiling on Chlamydia Early Promoters Correlate with Expression Patterns in Midcycle. Journal of Bacteriology 194, 3109–3115, doi: 10.1128/jb.00242-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty W. L., Morrison R. P. & Byrne G. I. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiological reviews 58, 686–699 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Bailey L., Önskog T., Bergström S. & Johansson J. A comparative study of RNA and DNA as internal gene expression controls early in the developmental cycle of Chlamydia pneumoniae. FEMS Immunology & Medical Microbiology 58, 244–253, doi: 10.1111/j.1574-695X.2009.00631.x (2010). [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to Image J: 25 years of image analysis. Nat Meth 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.