Abstract
The stromal cell derived factor-1 (SDF-1) rs1801157 gene polymorphism has been implicated in susceptibility to cancer, but the results were inconclusive. The current study was to precisely investigate the association between SDF-1 rs1801157 polymorphism and cancer risk using meta-analysis and the false positive report probability (FPRP) test. All 17,876 participants were included in the study. The meta-analysis results indicated a significant association between the SDF-1 rs1801157 polymorphism and cancer risk. By subgroup analyses, the results detected that the SDF-1 rs1801157 polymorphism was associated with cancer susceptibility among Asians and Caucasians. Additionally, we also found significant associations between the SDF-1 rs1801157 polymorphism and susceptibility to different types of cancer. However, to avoid a “false positive report”, we further investigated the significant associations observed in the present meta-analysis using the FPRP test. Interestingly, the results of the FPRP test indicated that only 4 gene models were truly associated with cancer risk, especially in Asians. Moreover, we confirmed that the SDF-1 rs1801157 gene polymorphism was only associated with lung and urologic cancer risk. In summary, this study suggested that the SDF-1 rs1801157 polymorphism may serve as a risk factor for cancer development among Asians, especially an increased risk of urologic and lung cancers.
Cancer remains a major cause of mortality worldwide. It is estimated that the cancer death rate will be increased by 5-fold in developing countries, and the global cancer mortality is expected to increase by 104% in 20201. Cancer is a multi-factorial disease that results from complex interactions between numerous genetic and environmental factors2,3. In recent years, an increasing number of studies has focused on the association between chemokine gene variants and malignancy susceptibility. Among them, the stromal cell derived factor-1 (SDF-1) gene, also known as CXC chemokine ligand 12 (CXCL12), has been widely studied4,5,6. The SDF-1 gene is located on chromosome 10q11.1 and spans 10 kb7. The SDF-1 gene mainly encodes a CXC angiogenic chemokine8. One important SDF-1 gene polymorphism named rs1801157 (G801A) involves a guanine to adenine (G→C) substitution at base pair 801 of the 3′-untranslated region of SDF-1 gene9.
Previous studies have indicated that SDF-1 participates in several biological activities, including neuronal and cardiogenesis development, haematopoiesis and lymphocyte trafficking10. Furthermore, growing evidence suggests that the SDF-1 rs1801157 polymorphism plays an important role in the pathogenesis of cancer. Razmkhah et al. reported that the SDF-1 rs1801157 polymorphism increased the risk of breast and lung cancer11,12. Kucukgergin and co-workers showed that the SDF-1 rs1801157 polymorphism was associated with bladder cancer susceptibility13. Hirata et al. reported that the SDF-1 rs1801157 polymorphism is potentially associated with an increased risk of prostate cancer among Japanese individuals14. However, Petersen et al. did not identify an association between the SDF-1 rs1801157 polymorphism and prostate cancer susceptibility15, and similar negative results were also detected by Tee and colleagues16.
The results of those genetic association studies were inconclusive. Moreover, a single study may be insufficient to detect a small effect of the SDF-1 rs1801157 gene polymorphism on cancer susceptibility, especially when the sample size is relatively small. Considering the crucial role of the SDF-1 rs1801157 gene polymorphism in the pathogenesis of cancer, we performed a meta-analysis to accurately investigate the association of the SDF-1 rs1801157 gene polymorphism with cancer risk. Furthermore, to avoid a “false positive report”, we further assessed the significant associations observed in the present meta-analysis using the false positive report probability (FPRP) test. To our knowledge, this study is most recent and accurate meta-analysis exploring the SDF-1 rs1801157 gene polymorphism in cancer susceptibility.
Results
Study characteristics
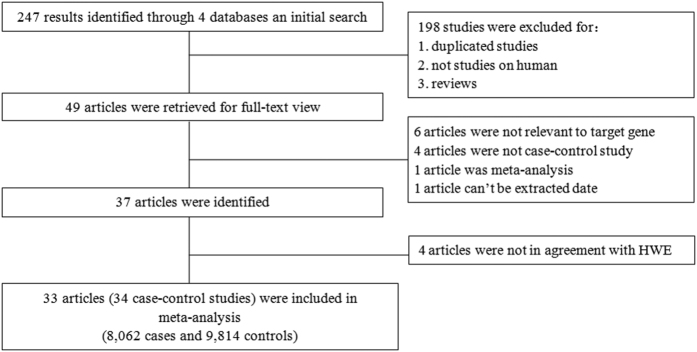
As indicated in the flow diagram (Fig. 1), 247 articles were identified after an initial search. After reading the titles and abstracts, we excluded 198 articles. The remaining 49 articles were further screened by a full-text review. Six articles were excluded because they investigated other SDF-1 gene polymorphisms (e.g., rs2839693 and rs1065297) rather than the rs1801157 polymorphism. Four articles were excluded because they were not case-control studies. One article was not included because it was a meta-analysis, and one article was excluded because the available data could not be extracted to further assess the effect size. Therefore, all 37 articles were identified4,5,6,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44. However, according to the results of the HWE test, 4 articles18,38,39,44 were excluded because they did not achieve HWE in the control group. Finally, a total of 34 eligible case-control studies from 33 articles4,5,6,11,12,13,14,15,16,17,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,40,41,42,43 were included in the study. Among them, 18 articles were performed on Caucasians4,6,15,17,19,20,21,22,23,24,27,28,29,31,33,34,40,43, 13 articles on Asians5,11,12,13,14,16,25,26,30,32,35,36,37, and two studies on individuals of mixed ethnicity41,42. In addition, based on the quality score, eleven studies were considered low quality. The characteristics of included studies are summarized in Tables 1 and 2.
Figure 1.
Table 1. Characteristics of case-control studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Type | Sample size | Genotyping method | HWE | Score |
|---|---|---|---|---|---|---|---|---|
| Case/Control | ||||||||
| Bodelon C | 2013 | USA | Caucasian | BC | 840/801 | IGGMP | Yes | 6 |
| Bracci PM | 2010 | USA + UK | Caucasian | NHL | 255/511 | IGGMP | Yes | 8 |
| Cacina C | 2012 | Turkey | Asian | EC | 113/139 | PCR-RFLP | Yes | 6 |
| Cai C | 2013 | China | Asian | RC | 322/402 | PCR-RFLP | Yes | 7 |
| de Lourdes Perim A | 2013 | Brazil | Caucasian | ALL | 54/58 | PCR-RFLP | Yes | 6 |
| de Oliveira CE | 2007 | Brazil | Caucasian | CML | 25/60 | PCR-RFLP | Yes | 5 |
| de Oliveira KB | 2009 | Brazil | Caucasian | BC | 103/97 | PCR-RFLP | Yes | 5 |
| de Oliveira KB | 2010 | Brazil | Caucasian | NHL + HL | 106/90 | PCR-RFLP | Yes | 5 |
| Dimberg J | 2007 | Sweden | Caucasian | CRC | 151/141 | PCR-RFLP | Yes | 5 |
| Gawron AJ | 2011 | Poland | Caucasian | GC | 292/414 | TaqMan | Yes | 7 |
| Hidalgo-Pascual M | 2007 | Spain | Caucasian | CRC | 349/516 | Real-time PCR | Yes | 6 |
| Hirata H | 2007 | Japan | Asian | PC | 167/167 | PCR-RFLP | Yes | 5 |
| Isman FK | 2012 | Turkey | Asian | PC | 152/149 | PCR-RFLP | Yes | 7 |
| Khademi B | 2008 | Iran | Asian | HC + NC | 156/262 | PCR-RFLP | Yes | 6 |
| Kontogianni P | 2013 | Greece | Caucasian | BC | 261/480 | PCR-RFLP | Yes | 6 |
| Kruszyna L | 2010 | Poland | Caucasian | BC | 193/199 | PCR-RFLP | Yes | 6 |
| Kruszyna L | 2010 | Poland | Caucasian | LAC | 118/250 | PCR-RFLP | Yes | 6 |
| Kucukgergin C | 2012 | Turkey | Asian | BLC | 142/197 | PCR-RFLP | Yes | 7 |
| Lee YL | 2011 | China | Asian | LC | 247/328 | PCR-RFLP | Yes | 7 |
| Liarmakopoulos E | 2013 | Greece | Caucasian | GC | 88/480 | PCR-RFLP | Yes | 6 |
| Lin GT | 2009 | China | Asian | BC | 220/334 | PCR-RFLP | Yes | 4 |
| Maley SN | 2009 | USA | Caucasian | CC | 899/820 | TaqMan | Yes | 7 |
| Mazur G | 2013 | Poland | Caucasian | MM | 54/75 | PCR-RFLP | Yes | 4 |
| Pemberton NC | 2006 | UK | Caucasian | CLL | 323/108 | PCR-RFLP | Yes | 5 |
| Petersen DC | 2008 | Australia | Caucasian | PC | 815/727 | TaqMan | Yes | 6 |
| Razmkhah M | 2013 | Iran | Asian | GC + CC | 233/262 | PCR-RFLP | Yes | 4 |
| Razmkhah M | 2005 | Iran | Asian | BC | 278/181 | PCR-RFLP | Yes | 6 |
| Razmkhah M | 2005 | Iran | Asian | LC | 72/262 | PCR-RFLP | Yes | 4 |
| Shi MD | 2013 | China | Asian | CRC | 258/300 | DHPLC | Yes | 5 |
| Singh V | 2014 | India | Asian | BLC | 200/200 | PCR-RFLP | Yes | 7 |
| Tee YT | 2012 | China | Asian | CC | 137/337 | PCR-RFLP | Yes | 7 |
| Vairaktaris E | 2008 | Europe | Caucasian | OC | 159/101 | PCR-RFLP | Yes | 6 |
| Vázquez-Lavista LG | 2009 | Mexico | Mixed | BLC | 47/126 | PCR-RFLP | Yes | 7 |
| Wong HL | 2010 | USA | Mixed | NHL | 233/240 | TaqMan | Yes | 7 |
BC = breast cancer; EC = endometrial cancer; RC = renal cancer; ALL = acute lymphatic leukaemia; CML = chronic myeloid leukaemia; NHL non-Hodgkin’s lymphoma; HL = Hodgkin’s lymphoma; CRC = colorectal cancer; GC = gastric cancer; PC = prostate cancer; HC = head cancer; NC = neck cancer; LAC = laryngeal cancer; BLC = bladder cancer; LC = lung cancer; CC = cervical cancer; MM = multiple myeloma; CLL = chronic lymphocytic leukaemia; OC = ovarian cancer; IGGMP = Illumina Golden Gate multiplex platform; PCR-RFLP = polymerase chain reaction-restricted fragment length polymorphism; DHPLC = denaturing high performance liquid chromatography.
Table 2. Distributions of the SDF-1 rs1801157 allele and genotypes in cases and controls.
| First author | Year | Case | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | AA | AG | GG | A | G | ||
| Bodelon C | 2013 | 29 | 288 | 523 | 346 | 1334 | 35 | 251 | 515 | 321 | 1281 |
| Bracci PM | 2010 | 6 | 85 | 164 | 97 | 413 | 14 | 161 | 336 | 189 | 833 |
| Cacina C | 2012 | 12 | 52 | 49 | 76 | 150 | 6 | 64 | 69 | 76 | 202 |
| Cai C | 2013 | 61 | 111 | 150 | 233 | 411 | 29 | 136 | 237 | 194 | 610 |
| de Lourdes Perim A | 2013 | 3 | 18 | 33 | 24 | 84 | 1 | 11 | 46 | 13 | 103 |
| de Oliveira CE | 2007 | 4 | 11 | 10 | 19 | 31 | 3 | 18 | 39 | 24 | 96 |
| de Oliveira KB | 2009 | 3 | 41 | 59 | 47 | 159 | 4 | 32 | 61 | 40 | 154 |
| de Oliveira KB | 2010 | 5 | 43 | 58 | 53 | 159 | 5 | 26 | 59 | 36 | 144 |
| Dimberg J | 2007 | 5 | 62 | 84 | 72 | 230 | 4 | 56 | 81 | 64 | 218 |
| Gawron AJ | 2011 | 99 | 193 | NA | NA | 156 | 258 | NA | NA | ||
| Hidalgo-Pascual M | 2007 | 9 | 128 | 212 | 146 | 552 | 25 | 172 | 319 | 222 | 810 |
| Hirata H | 2007 | 17 | 78 | 72 | 112 | 222 | 13 | 63 | 91 | 89 | 245 |
| Isman FK | 2012 | 17 | 66 | 69 | 100 | 204 | 22 | 57 | 70 | 101 | 197 |
| Khademi B | 2008 | 8 | 84 | 64 | 100 | 212 | 20 | 97 | 145 | 137 | 387 |
| Kontogianni P | 2013 | 29 | 118 | 114 | 176 | 346 | 35 | 198 | 247 | 268 | 692 |
| Kruszyna L | 2010 | 9 | 61 | 123 | 79 | 307 | 5 | 58 | 136 | 68 | 330 |
| Kruszyna L | 2010 | 3 | 46 | 69 | 52 | 184 | 2 | 67 | 181 | 71 | 429 |
| Kucukgergin C | 2012 | 26 | 58 | 58 | 110 | 174 | 23 | 80 | 94 | 126 | 268 |
| Lee YL | 2011 | 36 | 112 | 99 | 184 | 310 | 21 | 136 | 171 | 178 | 478 |
| Liarmakopoulos E | 2013 | 6 | 43 | 39 | 55 | 121 | 46 | 229 | 205 | 321 | 639 |
| Lin GT | 2009 | 16 | 98 | 106 | 130 | 310 | 23 | 136 | 175 | 182 | 486 |
| Maley SN | 2009 | 30 | 276 | 593 | 336 | 1462 | 23 | 274 | 523 | 320 | 1320 |
| Mazur G | 2013 | 1 | 13 | 39 | 15 | 91 | 1 | 21 | 53 | 23 | 127 |
| Pemberton NC | 2006 | 114 | 209 | NA | NA | 43 | 65 | NA | NA | ||
| Petersen DC | 2008 | NA | NA | NA | 326 | 1304 | NA | NA | NA | 276 | 1178 |
| Razmkhah M | 2013 | 18 | 87 | 128 | 123 | 343 | 8 | 39 | 62 | 55 | 163 |
| Razmkhah M | 2005 | 34 | 139 | 105 | 207 | 349 | 13 | 67 | 101 | 93 | 269 |
| Razmkhah M | 2005 | 9 | 38 | 25 | 56 | 88 | 20 | 97 | 145 | 137 | 387 |
| Shi MD | 2013 | 4 | 113 | 141 | 121 | 395 | 0 | 52 | 248 | 52 | 548 |
| Singh V | 2014 | 9 | 132 | 59 | 150 | 250 | 16 | 83 | 101 | 115 | 285 |
| Tee YT | 2012 | 16 | 58 | 63 | 90 | 184 | 33 | 140 | 164 | 206 | 468 |
| Vairaktaris E | 2008 | 4 | 51 | 104 | 59 | 259 | 5 | 41 | 55 | 51 | 151 |
| Vázquez-Lavista LG | 2009 | 3 | 15 | 29 | 21 | 73 | 4 | 39 | 83 | 47 | 205 |
| Wong HL | 2010 | 7 | 78 | 148 | 92 | 374 | 13 | 64 | 163 | 90 | 390 |
NA = not available.
Meta-analysis results
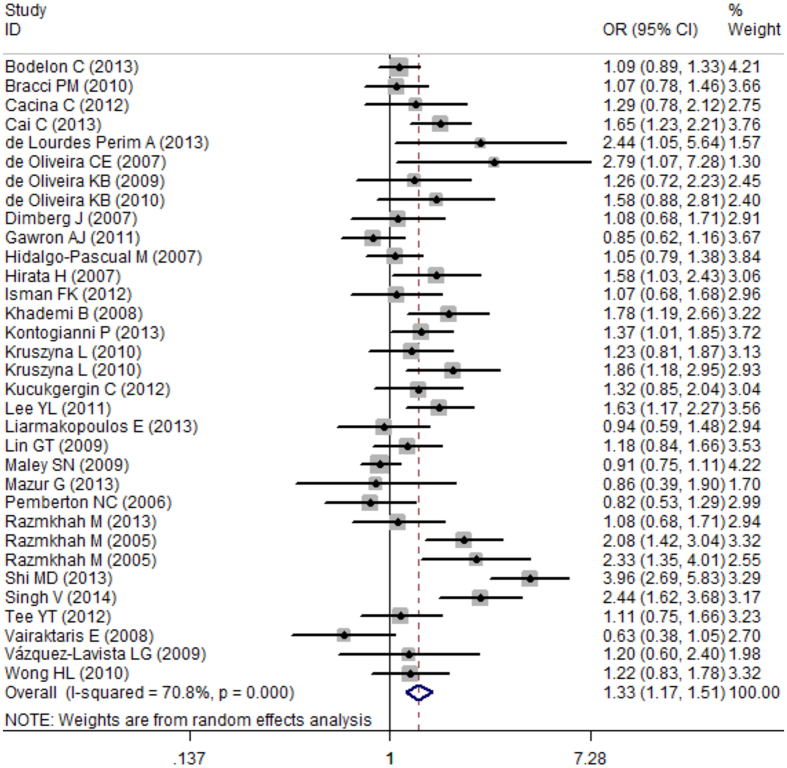
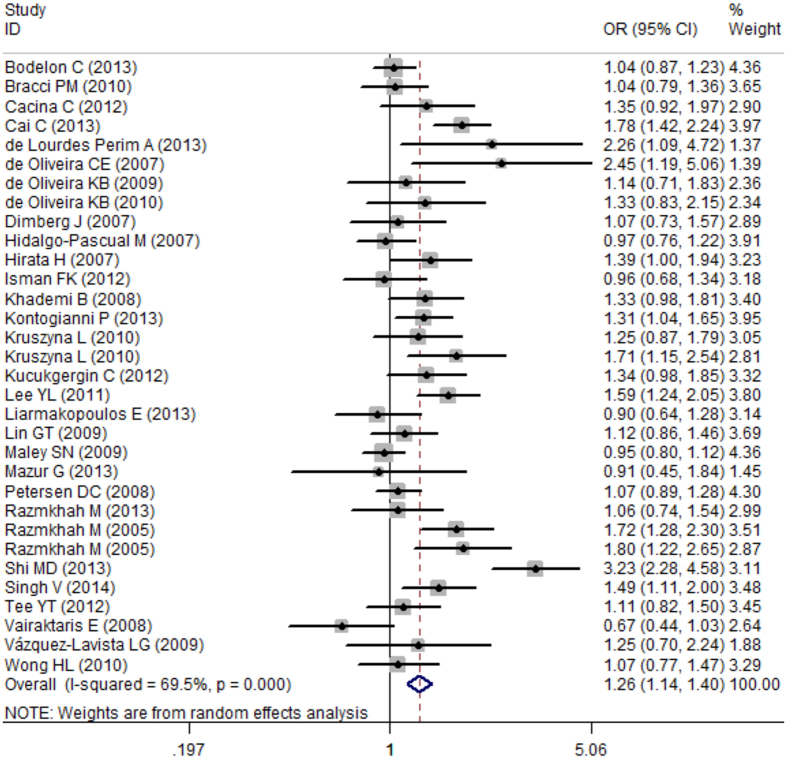
In total, 17,876 participants (8,062 cases and 9,814 controls) from 34 case-control studies were included in the current meta-analysis assessing the relationship between the SDF-1 rs1801157 polymorphism and cancer risk. In the overall meta-analysis, the results suggested a significant association between the SDF-1 rs1801157 polymorphism and cancer susceptibility in the recessive model (AA vs. AG + GG, OR: 1.28, 95% CI: 1.11–1.47, P = 0.001), a co-dominant model (AA vs. GG, OR: 1.43, 95% CI: 1.24–1.65, P < 0.001) as assessed by a fixed-effect model (I2 = 47%, 49.7%, respectively) and other models (AA + AG vs. GG, OR: 1.33, 95% CI: 1.17–1.51, P < 0.001; AG vs. GG, OR: 1.35, 95% CI: 1.19–1.54, P < 0.001; A vs. G, OR: 1.26, 95% CI: 1.14–1.40, P < 0.001) (Figs 2 and 3) as assessed by a random-effect model (I2 = 70.8%, 66.4%, 69.5%, respectively) (Table 3).
Figure 2.
Figure 3.
Table 3. Summary the results of the total and subgroup analyses in different genetic models.
| Variables | AA + AG vs. GG |
AA vs. AG + GG |
AA vs. GG |
AG vs. GG |
A vs. G |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| Total | 1.33 | 1.17–1.51 | <0.001 | 1.28 | 1.11–1.47 | 0.001 | 1.43 | 1.24–1.65 | <0.001 | 1.35 | 1.19–1.54 | <0.001 | 1.26 | 1.14–1.40 | <0.001 |
| Ethnicities | |||||||||||||||
| Caucasian | 1.07 | 0.98–1.16 | 0.15 | 1.03 | 0.82–1.29 | 0.81 | 1.07 | 0.85–1.35 | 0.55 | 1.10 | 1.00–1.21 | 0.05 | 1.07 | 1.00–1.15 | 0.06 |
| Asian | 1.63 | 1.34–1.98 | <0.001 | 1.42 | 1.05–1.93 | 0.02 | 1.71 | 1.28–2.30 | <0.001 | 1.58 | 1.28–1.95 | <0.001 | 1.45 | 1.25–1.68 | <0.001 |
| Mixed | 1.21 | 0.87–1.69 | 0.26 | 0.92 | 0.25–3.34 | 0.90 | 0.82 | 0.37–1.81 | 0.62 | 1.28 | 0.90–1.82 | 0.16 | 1.11 | 0.83–1.47 | 0.48 |
| Types | |||||||||||||||
| Breast cancer | 1.26 | 1.11–1.44 | <0.001 | 1.21 | 0.93–1.59 | 0.16 | 1.36 | 1.03–1.79 | 0.03 | 1.26 | 1.10–1.44 | <0.001 | 1.20 | 1.08–1.33 | <0.001 |
| Gynaecological cancer | 0.98 | 0.83–1.16 | 0.81 | 1.36 | 0.93–1.99 | 0.12 | 1.36 | 0.92–2.01 | 0.13 | 0.94 | 0.79–1.12 | 0.52 | 1.03 | 0.89–1.18 | 0.71 |
| Urologic cancer | 1.56 | 1.32–1.85 | <0.001 | 1.34 | 0.75–2.38 | 0.32 | 1.62 | 0.97–2.69 | 0.06 | 1.46 | 1.10–1.93 | <0.001 | 1.31 | 1.09–1.57 | <0.001 |
| Digestive system cancer | 1.25 | 0.79–1.98 | 0.34 | 0.83 | 0.55–1.27 | 0.40 | 0.88 | 0.57–1.35 | 0.55 | 1.38 | 0.82–2.33 | 0.23 | 1.26 | 0.80–1.97 | 0.32 |
| Hematologic malignancy | 1.17 | 0.97–1.41 | 0.09 | 0.95 | 0.57–1.59 | 0.98 | 1.06 | 0.63–1.78 | 0.83 | 1.29 | 1.04–1.59 | 0.02 | 1.18 | 0.99–1.40 | 0.07 |
| Lung cancer | 1.80 | 1.36–2.39 | <0.001 | 2.24 | 1.41–3.57 | <0.001 | 2.86 | 1.75–4.69 | <0.001 | 1.62 | 1.20–2.18 | <0.001 | 1.65 | 1.34–2.04 | <0.001 |
| Head and neck cancer | 1.3 | 0.68–2.48 | 0.43 | 0.76 | 0.40–1.46 | 0.41 | 0.92 | 0.47–1.80 | 0.81 | 1.35 | 0.70–1.61 | 0.37 | 1.16 | 0.71–1.91 | 0.55 |
OR = odds ratio; CI = confidence interval.
In the subgroup analyses of ethnicity, the meta-analysis results indicated a strong association between the SDF-1 rs1801157 polymorphism and cancer susceptibility among Asians (AA + AG vs. GG, OR: 1.63, 95% CI: 1.34–1.98, P < 0.001) and a weak association in Caucasians (AG vs. GG, OR: 1.10, 95% CI: 1.00–1.21, P = 0.05), but no association in mixed populations using different genetic models (Table 3). Additionally, we also conducted subgroup analyses of cancer types. As shown in Table 3, significant associations between the SDF-1 rs1801157 polymorphism and breast cancer, urologic cancer, hematologic malignancy and lung cancer susceptibility were found in different genetic models. However, no association was observed between the SDF-1 rs1801157 polymorphism and risk of head and neck cancer, gynaecological cancer and digestive system cancer (Table 3).
Publication bias and sensitivity analysis
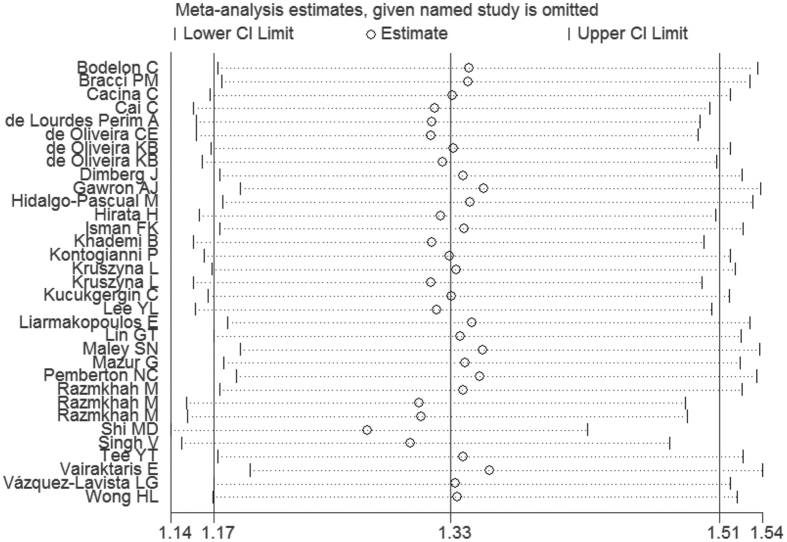
The funnel plot is a symmetrical inverted funnel (Fig. 4). No publication biases were noted using Begg’s (P = 0.329) and Egger’s tests (P = 0.082). Additionally, to further investigate the possible source of heterogeneity, we executed a sensitivity analysis by sequentially excluding studies from the meta-analysis to investigate the influence of each study on the pooled results. The sensitivity analysis results reported that the pooled ORs were not materially altered, suggesting the stability of our meta-analysis (Fig. 5).
Figure 4.
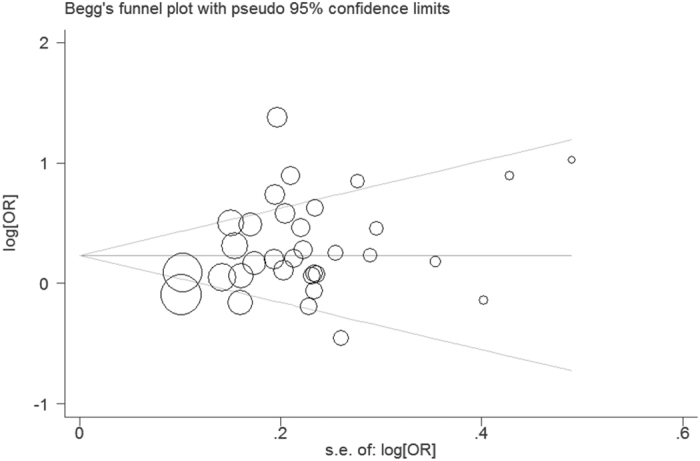
Figure 5.
FPRP test results
Moreover, we further investigated the significant associations (P < 0.05) observed in the present meta-analysis using the FPRP test. As listed in Table 4, the FPRP test results revealed that all 4 gene models (AA + AG vs. GG, AA vs. GG, AG vs. GG, A vs. G) of the SDF-1 rs1801157 gene polymorphism were truly associated with cancer risk (FPRP = 0.011, 0.001, 0.008, 0.017, respectively) at an a priori probability level of 0.001 with an OR of 1.5, especially in Asians. In addition, according to the results of the FPRP test, we confirmed that the SDF-1 rs1801157 gene polymorphism was only associated with lung cancer risk in the allele model (A vs. G, FPRP = 0.019), whereas the polymorphism was associated with susceptibility to urologic cancer in the dominant gene model (AA + AG vs. GG, FPRP = 0.001) at an a priori probability level of 0.001 and an OR of 1.5.
Table 4. The results of the FPRP tests in each gene models.
| Gene models | OR (95% CI) | Power | Prior Probability = 0.001 |
|---|---|---|---|
| AA + AG vs. GG | OR = 1.50 | FPRP | |
| Total | 1.33 (1.17–1.51) | 0.968 | 0.011 |
| Asians | 1.63 (1.34–1.98) | 0.201 | 0.004 |
| Breast cancer | 1.26 (1.11–1.44) | 0.995 | 0.41 |
| Urologic cancer | 1.56 (1.32–1.85) | 0.326 | 0.001 |
| Lung cancer | 1.80 (1.36–2.39) | 0.104 | 0.318 |
| AA vs. AG + GG | |||
| Total | 1.28 (1.11–1.47) | 0.988 | 0.323 |
| Asian | 1.42 (1.05–1.93) | 0.637 | 0.975 |
| Lung cancer | 2.24 (1.41–3.57) | 0.046 | 0.938 |
| AA vs. GG | |||
| Total | 1.43 (1.24–1.65) | 0.744 | 0.001 |
| Asians | 1.71 (1.28–2.30) | 0.193 | 0.668 |
| Breast cancer | 1.36 (1.03–1.79) | 0.758 | 0.974 |
| Lung cancer | 2.86 (1.75–4.69) | 0.005 | 0.856 |
| AG vs. GG | |||
| Total | 1.35 (1.19–1.54) | 0.942 | 0.008 |
| Caucasians | 1.10 (1.00–1.21) | 1 | 0.980 |
| Asians | 1.58 (1.28–1.95) | 0.314 | 0.061 |
| Breast cancer | 1.26 (1.10–1.44) | 0.995 | 0.410 |
| Urologic cancer | 1.46 (1.10–1.93) | 0.575 | 0.932 |
| Hematologic malignancy | 1.29 (1.04–1.59) | 0.921 | 0.949 |
| Lung cancer | 1.62 (1.20–2.18) | 0.306 | 0.826 |
| A vs. G | |||
| Total | 1.26 (1.14–1.40) | 0.999 | 0.017 |
| Asians | 1.45 (1.25–1.68) | 0.674 | 0.001 |
| Breast cancer | 1.20 (1.08–1.33) | 1 | 0.339 |
| Urologic cancer | 1.31 (1.09–1.57) | 0.929 | 0.788 |
| Lung cancer | 1.65 (1.34–2.04) | 0.189 | 0.019 |
OR = odds ratio; FPRP = false positive report probability.
Discussion
Cancer has high morbidity and mortality worldwide. Previous studies have suggested that cancer risk factors include an unhealthy life style, environmental pollution, radiation, infection, and immunity dysfunction45,46,47. Additionally, studies have increasingly focused on genetic factors48,49,50. In recent years, numerous studies reported an association between the SDF-1 rs1801157 polymorphism and cancer susceptibility, but the results were inconsistent or inconclusive. In addition, a single study may lack sufficient power to detect the potentially small effect of the SDF-1 rs1801157 polymorphism on cancer, especially when the sample size is relatively small. Meta-analysis is a useful method for investigating cancer associations with genetic factors because this method uses a quantitative approach by combining the results of different studies on the same topic, potentially providing more reliable conclusions. Thus, we conducted a pooled analysis to investigate the association of the SDF-1 rs1801157 polymorphism and risk of cancer.
In fact, two recently published meta-analyses51,52 investigated the association between the SDF-1 rs1801157 gene polymorphism and cancer risk. One study51 included 29 papers involving 4932 cases and 7917 controls, and another article52 only included 4,435 cancer cases and 6,898 controls from 25 studies. Additionally, the two meta-analyses did not assess the quality of the included studies. Furthermore, the previous two studies did not use the FPRP test to explore truly significant associations, and the results of those studies were potentially unable to reflect the true association between the SDF-1 rs1801157 gene polymorphism and cancer risk. Therefore, we performed a meta-analysis and FPRP test to assess the association between the SDF-1 rs1801157 gene polymorphism and cancer risk.
In total, 8,062 cases and 9,814 controls were included in the current meta-analysis. Based on our overall meta-analysis and FPRP tests, we found that the SDF-1 rs1801157 polymorphism obviously increased the risk of cancer. Additionally, we identified significant heterogeneity between studies in the present meta-analysis. Although heterogeneity can be considered using the random-effect model, heterogeneity increases the probability of type-I errors. The following factors may have contributed to the significant heterogeneity: 1) different demographic and genetic characteristics of Caucasian, Asian and mixed populations; 2) different types of cancer may be caused by different mechanisms; 3) different stages of cancer in each study among the studied cancer patients; 4) different genotyping methods of each study; and 5) different qualities among included studies.
Hence, to identify the cause of heterogeneity, we conducted subgroup analyses based on ethnicity and cancer type. The results suggested that the SDF-1 rs1801157 gene polymorphism strongly increased the cancer risk among Asians, whereas the heterozygote genotype (AG) of the SDF-1 rs1801157 gene polymorphism only weakly increased cancer susceptibility in Caucasians. Unfortunately, the SDF-1 rs1801157 gene polymorphism did not increase or decrease the cancer risk among mixed populations. Furthermore, the results of sub-group analyses after stratification based on cancer type indicated that the SDF-1 rs1801157 polymorphism increased the risk of breast cancer, urologic cancer, lung cancer, head and neck cancer, and haematological malignancies. However, the SDF-1 rs1801157 polymorphism did not influence gynaecological cancer and digestive system cancer susceptibility. Despite the fact that the heterogeneity was still detected when we performed stratified analyses based on ethnicity and cancer type, the subgroup analyses may provide more precise results compared with the overall analysis.
Several previous studies indicated that the published “statistically significant” results for genetic variants were false-positive findings, even in large and well-designed studies53,54. Fortunately, we could use the FPRP test to estimate true significant associations in the current meta-analysis. The FPRP is calculated from the statistical power of the test, the observed p-value, and a given a priori probability for the association.
Therefore, based on the positive results of the current meta-analysis, we further investigated whether a significant association between the SDF-1 rs1801157 gene polymorphism and cancer risk is “noteworthy”. Interestingly, the FPRP test results revealed that the SDF-1 rs1801157 gene polymorphism actually increased cancer susceptibility. Additionally, the FPRP test also demonstrated that the SDF-1 rs1801157 gene polymorphism could increase cancer risk among Asians, but it did not increase cancer risk in Caucasians. Moreover, the results of the FPRP test confirmed that the SDF-1 rs1801157 gene polymorphism increased the risk of lung cancer and urologic cancer. Surprisingly, the significant associations with breast cancer, head and neck cancer, and haematological malignancies in the present meta-analysis were false positive at the a priori probability level of 0.001 and an OR of 1.5. Some of the discoveries may be false-positive findings due to the limited sample size in each stratum, so it is important to perform FPRP analysis to avoid these findings, especially when the sample size is not of sufficient size.
The mechanism by which the SDF-1 rs1801157 polymorphism affects cancer risk is unclear. Previous studies indicated that SDF-1 is a chemokine that plays a pivotal role in different stages of cancer development and metastasis55,56. Combined with previous results57 and our present meta-analysis results, we put forward a simple hypothesis that the SDF-1 rs1801157 polymorphism, which has a G > A transition in the 3′-UTR, may have an important regulatory function of up-regulating SDF-1 production and the high serum concentrations of SDF-1 might contribute to cancer susceptibility. Finally, people who carried the variant A allele are more likely to develop cancer, especially urologic cancer and lung cancer.
Several limitations in the present meta-analysis should be noted. First, the published studies include small sample sizes, and only published articles were included in a few databases. Thus, these limitations might lead to additional biases or overall overestimated associations. Moreover, a language bias potentially also occurred because the including articles were only published in English and Chinese. Second, given the lack sufficient data for each included study, we failed to perform further subgroup analysis to investigate the risk factors of cancer, such as gender, gene-environment/gene-gene interactions, life style and age. Third, a small number of studies were included in the subgroup analysis to investigate the association between the SDF-1 rs1801157 polymorphism and head and neck cancer risk, so we must be cautious when referring to the pooled results. A study with larger samples is needed to confirm the results in the future. Despite these limitations, we minimized the likelihood of bias throughout the entire process by creating a detailed protocol and carefully performing study identification, statistical analysis and data selection. Thus, the reliability of the results is guaranteed.
In conclusion, the current study suggested that the SDF-1 rs1801157 polymorphism may contribute to the risk of cancer in Asians. In particular, the polymorphism could increase urologic and lung cancer susceptibility. More well-designed studies with larger sample sizes focusing on ethnicities or cancer types should be conducted to confirm the results in the future.
Methods
Study selection
A systematic literature search of the PubMed, Embase, and Wanfang databases and the China National Knowledge Internet (CNKI) was performed to identify studies involving the relationship between the SDF-1 rs1801157 gene polymorphism and cancer susceptibility, with the last updated search being conducted on May 14, 2015. The following key words were used: (stromal cell derived factor-1 OR SDF-1 OR CXC chemokine ligand 12 OR CXCL12) AND (cancer OR tumour OR neoplasm OR malignancy OR leukaemia OR myeloma OR sarcoma OR lymphoma) AND (polymorphism OR mutation OR variant). The language was restricted to English and Chinese.
Inclusion and exclusive criteria
The inclusion criteria were defined as follows: 1) the study should be a case-control study; 2) the association between the SDF-1 rs1801157 polymorphism and cancer risk should be evaluated; 3) the available genotype or allele frequencies for counting the odds ratio (OR) and 95% confidence interval (CI) should be provided in the study; 4) the study should evaluate humans; and 5) the genotype distributions of control cohorts should be in with Hardy-Weinberg equilibrium (HWE). The following exclusive criteria were adopted: 1) the study was not designed as a case-control study; 2) review, abstract or overlapping study; and 3) the study cannot provide available genotype or allele frequencies for determining the effect size.
Quality score evaluation
The qualities of included studies were assessed by the Newcastle-Ottawa Scale (case control study). The scale estimates quality based on three aspects: selection, comparability and exposure in the study. The total score ranged from 0 to 9, and a score greater than 6 was considered to represent high quality. In addition, we assessed the quality of the studies in a consensus meeting with all authors.
Data extraction
The independent authors (Xiang Tong and Huajiang Deng) collected detailed data from each study according to the inclusive criteria. If a disagreement was encountered, the third author (Xixi Wang) evaluated the articles. For each study, the first author, publication year, country, ethnicity, number of case and control groups, type of cancer, genotype and allele distribution, and genotyping method were extracted. The information is listed in Tables 1 and 2.
Statistical methods
The OR and 95% CI were used to estimate the effect strength of association between the SDF-1 rs1801157 polymorphism and cancer susceptibility. We calculated heterogeneity using the χ2-based Q-test and I-squared (I2) statistics tests. The pooled OR was assessed by the random-effect model when the heterogeneity was considered to be statistically significant (I2 > 50% and P < 0.10); alternatively, the fixed-effect model was applied. We explored the association between the SDF-1 rs1801157 polymorphism and cancer risk in different gene models (AA + AG vs. GG, AA vs. AG + GG, AA vs. GG, AG vs. GG and A vs. G). To evaluate the effects of ethnicity and type of cancer, we also performed subgroup analyses based on ethnicity group and type of cancer.
In addition, to evaluate whether significant associations (P < 0.05) detected in the present study are “noteworthy”, we further calculated the FPRP value at a probability level of 0.001 and an OR of 1.5 58. As suggested by the previous study59, we set a FPRP cut-off value of 0.2, and only FPRP results < 0.2 were considered to be “noteworthy”. Publication bias was assessed by several methods. A visual inspection of asymmetry in funnel plots was performed. Furthermore, the Begger’s and Egger’s tests were used to assess the publication bias60. Additionally, HWE was assessed in each study using the Chi-square before the present meta-analysis was performed. All analyses were conducted using STATA 11.0 software.
Additional Information
How to cite this article: Tong, X. et al. The SDF-1 rs1801157 Polymorphism is Associated with Cancer Risk: An Update Pooled Analysis and FPRP Test of 17,876 Participants. Sci. Rep. 6, 27466; doi: 10.1038/srep27466 (2016).
Acknowledgments
We thank Elsevier Language Editing Services for its linguistic assistance during the preparation of this manuscript.
Footnotes
Author Contributions H.F. designed the study, coordinated the study and directed its implementation. X.T., Y.M. and H.D. searched the publications, extracted the data and wrote the manuscript. X.T., Y.M., X.W., S.L. and Z.Y. were responsible for data synthesis and figures creation. Y.M., H.D. and S.P. wrote the abstract and made the tables. All authors read and approved the final manuscript.
References
- Rastogi T., Hildesheim A. & Sinha R. Opportunities for cancer epidemiology in developing countries. Nat. Rev. Cancer 4, 909–917, doi: 10.1038/nrc1475 (2004). [DOI] [PubMed] [Google Scholar]
- Bredberg A. Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer 117, 440–445, doi: 10.1002/cncr.25440 (2011). [DOI] [PubMed] [Google Scholar]
- Pharoah P. D., Dunning A. M., Ponder B. A. & Easton D. F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 4, 850–860, doi: 10.1038/nrc1476 (2004). [DOI] [PubMed] [Google Scholar]
- Cacina C., Bulgurcuoglu-Kuran S., Iyibozkurt A. C., Yaylim-Eraltan I. & Cakmakoglu B. Genetic variants of SDF-1 and CXCR4 genes in endometrial carcinoma. Mol. Biol. Rep. 39, 1225–1229, doi: 10.1007/s11033-011-0852-9 (2012). [DOI] [PubMed] [Google Scholar]
- Cai C. et al. Association of CXCL12 and CXCR4 gene polymorphisms with the susceptibility and prognosis of renal cell carcinoma. Tissue Antigens 82, 165–170, doi: 10.1111/tan.12170 (2013). [DOI] [PubMed] [Google Scholar]
- Maley S. N. et al. Genetic variation in CXCL12 and risk of cervical carcinoma: a population-based case-control study. Int. J. Immunogenet. 36, 367–375, doi: 10.1111/j.1744-313X.2009.00877.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borish L. C. & Steinke J. W. 2. Cytokines and chemokines. J. Allergy Clin. Immunol. 111, S460–475 (2003). [DOI] [PubMed] [Google Scholar]
- Pillarisetti K. & Gupta S. K. Cloning and relative expression analysis of rat stromal cell derived factor-1 (SDF-1)1: SDF-1 alpha mRNA is selectively induced in rat model of myocardial infarction. Inflammation 25, 293–300 (2001). [DOI] [PubMed] [Google Scholar]
- Dean M., Carrington M. & O’Brien S. J. Balanced polymorphism selected by genetic versus infectious human disease. Annu. Rev. Genomics Hum. Genet. 3, 263–292, doi: 10.1146/annurev.genom.3.022502.103149 (2002). [DOI] [PubMed] [Google Scholar]
- Nagasawa T. et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382, 635–638, doi: 10.1038/382635a0 (1996). [DOI] [PubMed] [Google Scholar]
- Razmkhah M., Doroudchi M., Ghayumi S. M., Erfani N. & Ghaderi A. Stromal cell-derived factor-1 (SDF-1) gene and susceptibility of Iranian patients with lung cancer. Lung Cancer 49, 311–315, doi: 10.1016/j.lungcan.2005.04.014 (2005). [DOI] [PubMed] [Google Scholar]
- Razmkhah M., Talei A. R., Doroudchi M., Khalili-Azad T. & Ghaderi A. Stromal cell-derived factor-1 (SDF-1) alleles and susceptibility to breast carcinoma. Cancer Lett. 225, 261–266, doi: 10.1016/j.canlet.2004.10.039 (2005). [DOI] [PubMed] [Google Scholar]
- Kucukgergin C. et al. The role of chemokine and chemokine receptor gene variants on the susceptibility and clinicopathological characteristics of bladder cancer. Gene 511, 7–11, doi: 10.1016/j.gene.2012.09.011 (2012). [DOI] [PubMed] [Google Scholar]
- Hirata H. et al. CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin. Cancer Res. 13, 5056–5062, doi: 10.1158/1078-0432.ccr-07-0859 (2007). [DOI] [PubMed] [Google Scholar]
- Petersen D. C. et al. No association between common chemokine and chemokine receptor gene variants and prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 17, 3615–3617, doi: 10.1158/1055-9965.epi-08-0896 (2008). [DOI] [PubMed] [Google Scholar]
- Tee Y. T. et al. G801A polymorphism of human stromal cell-derived factor 1 gene raises no susceptibility to neoplastic lesions of uterine cervix. Int. J. Gynecol. Cancer 22, 1297–1302, doi: 10.1097/IGC.0b013e318265d334 (2012). [DOI] [PubMed] [Google Scholar]
- Bodelon C. et al. Common sequence variants in chemokine-related genes and risk of breast cancer in post-menopausal women. Int. J. Mol. Epidemiol. Genet. 4, 218–227 (2013). [PMC free article] [PubMed] [Google Scholar]
- Chang C. C. et al. Stromal cell-derived factor-1 but not its receptor, CXCR4, gene variants increase susceptibility and pathological development of hepatocellular carcinoma. Clin. Chem. Lab. Med. 47, 412–418, doi: 10.1515/cclm.2009.092 (2009). [DOI] [PubMed] [Google Scholar]
- de Lourdes Perim A. et al. CXCL12 and TP53 genetic polymorphisms as markers of susceptibility in a Brazilian children population with acute lymphoblastic leukemia (ALL). Mol. Biol. Rep. 40, 4591–4596, doi: 10.1007/s11033-013-2551-1 (2013). [DOI] [PubMed] [Google Scholar]
- de Oliveira C. E. et al. Stromal cell-derived factor-1 chemokine gene variant in blood donors and chronic myelogenous leukemia patients. J. Clin. Lab. Anal. 21, 49–54, doi: 10.1002/jcla.20142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira K. B. et al. CXCL12 rs1801157 polymorphism in patients with breast cancer, Hodgkin’s lymphoma, and non-Hodgkin’s lymphoma. J. Clin. Lab. Anal. 23, 387–393, doi: 10.1002/jcla.20346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimberg J., Hugander A., Lofgren S. & Wagsater D. Polymorphism and circulating levels of the chemokine CXCL12 in colorectal cancer patients. Int. J. Mol. Med. 19, 11–15 (2007). [PubMed] [Google Scholar]
- Gawron A. J. et al. Polymorphisms in chemokine and receptor genes and gastric cancer risk and survival in a high risk Polish population. Scand. J. Gastroenterol. 46, 333–340, doi: 10.3109/00365521.2010.537679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Pascual M. et al. Analysis of CXCL12 3′ UTR G > A polymorphism in colorectal cancer. Oncol. Rep. 18, 1583–1587 (2007). [DOI] [PubMed] [Google Scholar]
- Isman F. K. et al. Association between SDF1-3′A or CXCR4 gene polymorphisms with predisposition to and clinicopathological characteristics of prostate cancer with or without metastases. Mol. Biol. Rep. 39, 11073–11079, doi: 10.1007/s11033-012-2010-4 (2012). [DOI] [PubMed] [Google Scholar]
- Khademi B., Razmkhah M., Erfani N., Gharagozloo M. & Ghaderi A. SDF-1 and CCR5 genes polymorphism in patients with head and neck cancer. Pathol. Oncol. Res. 14, 45–50, doi: 10.1007/s12253-008-9007-2 (2008). [DOI] [PubMed] [Google Scholar]
- Kontogianni P. et al. The impact of the stromal cell-derived factor-1-3′A and E-selectin S128R polymorphisms on breast cancer. Mol. Biol. Rep. 40, 43–50, doi: 10.1007/s11033-012-1989-x (2013). [DOI] [PubMed] [Google Scholar]
- Kruszyna L. et al. CXCL12-3′ G801A polymorphism is not a risk factor for breast cancer. DNA Cell Biol. 29, 423–427, doi: 10.1089/dna.2010.1030 (2010). [DOI] [PubMed] [Google Scholar]
- Kruszyna L., Lianeri M., Rydzanicz M., Szyfter K. & Jagodzinski P. P. SDF1-3′ a gene polymorphism is associated with laryngeal cancer. Pathol. Oncol. Res. 16, 223–227, doi: 10.1007/s12253-009-9214-5 (2010). [DOI] [PubMed] [Google Scholar]
- Lee Y. L. et al. Association of genetic polymorphisms of CXCL12/SDF1 gene and its receptor, CXCR4, to the susceptibility and prognosis of non-small cell lung cancer. Lung Cancer 73, 147–152, doi: 10.1016/j.lungcan.2010.12.011 (2011). [DOI] [PubMed] [Google Scholar]
- Liarmakopoulos E. et al. Effects of stromal cell-derived factor-1 and survivin gene polymorphisms on gastric cancer risk. Mol. Med. Rep. 7, 887–892, doi: 10.3892/mmr.2012.1247 (2013). [DOI] [PubMed] [Google Scholar]
- Lin G. T. et al. Combinational polymorphisms of seven CXCL12-related genes are protective against breast cancer in Taiwan. OMICS 13, 165–172, doi: 10.1089/omi.2008.0050 (2009). [DOI] [PubMed] [Google Scholar]
- Mazur G. et al. The CXCL12-3′A allele plays a favourable role in patients with multiple myeloma. Cytokine 64, 422–426, doi: 10.1016/j.cyto.2013.05.004 (2013). [DOI] [PubMed] [Google Scholar]
- Pemberton N. C. et al. The SDF-1 G > A polymorphism at position 801 plays no role in multiple myeloma but may contribute to an inferior cause-specific survival in chronic lymphocytic leukemia. Leuk. Lymphoma 47, 1239–1244, doi: 10.1080/10428190600562112 (2006). [DOI] [PubMed] [Google Scholar]
- Razmkhah M. & Ghaderi A. SDF-1alpha G801A polymorphism in Southern Iranian patients with colorectal and gastric cancers. Indian J. Gastroenterol. 32, 28–31, doi: 10.1007/s12664-012-0283-0 (2013). [DOI] [PubMed] [Google Scholar]
- Shi M. D. et al. CXCL12-G801A polymorphism modulates risk of colorectal cancer in Taiwan. Arch. Med. Sci. 9, 999–1005, doi: 10.5114/aoms.2013.39211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Jaiswal P. K., Kapoor R., Kapoor R. & Mittal R. D. Impact of chemokines CCR532, CXCL12G801A, and CXCR2C1208T on bladder cancer susceptibility in north Indian population. Tumour Biol. 35, 4765–4772, doi: 10.1007/s13277-014-1624-7 (2014). [DOI] [PubMed] [Google Scholar]
- Teng Y. H. et al. Contribution of genetic polymorphisms of stromal cell-derived factor-1 and its receptor, CXCR4, to the susceptibility and clinicopathologic development of oral cancer. Head Neck 31, 1282–1288, doi: 10.1002/hed.21094 (2009). [DOI] [PubMed] [Google Scholar]
- Theodoropoulos G. E. et al. Analysis of the stromal cell-derived factor 1-3′A gene polymorphism in pancreatic cancer. Mol. Med. Rep. 3, 693–698, doi: 10.3892/mmr_00000319 (2010). [DOI] [PubMed] [Google Scholar]
- Vairaktaris E. et al. A DNA polymorphism of stromal-derived factor-1 is associated with advanced stages of oral cancer. Anticancer Res. 28, 271–275 (2008). [PubMed] [Google Scholar]
- Vazquez-Lavista L. G., Lima G., Gabilondo F. & Llorente L. Genetic association of monocyte chemoattractant protein 1 (MCP-1)-2518 polymorphism in Mexican patients with transitional cell carcinoma of the bladder. Urology 74, 414–418, doi: 10.1016/j.urology.2009.04.016 (2009). [DOI] [PubMed] [Google Scholar]
- Wong H. L. et al. Cytokine signaling pathway polymorphisms and AIDS-related non-Hodgkin lymphoma risk in the multicenter AIDS cohort study. AIDS 24, 1025–1033, doi: 10.1097/QAD.0b013e328332d5b1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracci P. M. et al. Chemokine polymorphisms and lymphoma: a pooled analysis. Leukemia & lymphoma 51, 497–506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavi S. R. et al. The SDF-1 3′A Genetic Variation Is Correlated with Elevated Intra-tumor Tissue and Circulating Concentration of CXCL12 in Glial Tumors. J. Mol. Neurosci. 50, 298–304 (2013). [DOI] [PubMed] [Google Scholar]
- Boffetta P. Human cancer from environmental pollutants: the epidemiological evidence. Mutat. Res. 608, 157–162, doi: 10.1016/j.mrgentox.2006.02.015 (2006). [DOI] [PubMed] [Google Scholar]
- Brownson R. C., Alavanja M. C., Caporaso N., Simoes E. J. & Chang J. C. Epidemiology and prevention of lung cancer in nonsmokers. Epidemiol. Rev. 20, 218–236 (1998). [DOI] [PubMed] [Google Scholar]
- Liyanage U. K. et al. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J. Immunol. 169, 2756–2761 (2002). [DOI] [PubMed] [Google Scholar]
- Brooks J. D. et al. Variation in genes related to obesity, weight, and weight change and risk of contralateral breast cancer in the WECARE Study population. Cancer Epidemiol. Biomarkers Prev 21, 2261–2267, doi: 10.1158/1055-9965.epi-12-1036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti J. M. et al. Alterations in the PITSLRE protein kinase gene complex on chromosome 1p36 in childhood neuroblastoma. Nat. Genet. 7, 370–375, doi: 10.1038/ng0794-370 (1994). [DOI] [PubMed] [Google Scholar]
- Michailidou K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361, 361e351-352, doi: 10.1038/ng.2563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K. et al. The CXCL12 G801A polymorphism is associated with cancer risk: a meta-analysis. PLoS One 9, e108953, doi: 10.1371/journal.pone.0108953 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D. et al. CXCL12 G801A polymorphism and cancer risk: An updated meta-analysis. J Huazhong Univ. Sci. Technolog. Med. Sci. 35, 319–326, doi: 10.1007/s11596-015-1431-4 (2015). [DOI] [PubMed] [Google Scholar]
- Ioannidis J. P., Ntzani E. E., Trikalinos T. A. & Contopoulos-Ioannidis D. G. Replication validity of genetic association studies. Nat. Genet. 29, 306–309, doi: 10.1038/ng749 (2001). [DOI] [PubMed] [Google Scholar]
- Sterne J. A. & Davey Smith G. Sifting the evidence-what’s wrong with significance tests? BMJ 322, 226–231 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F. R. The chemokine system and cancer. J. Pathol. 226, 148–157, doi: 10.1002/path.3029 (2012). [DOI] [PubMed] [Google Scholar]
- Burger J. A. & Kipps T. J. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood 107, 1761–1767, doi: 10.1182/blood-2005-08-3182 (2006). [DOI] [PubMed] [Google Scholar]
- Winkler C. et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 279, 389–393 (1998). [DOI] [PubMed] [Google Scholar]
- Jiang Y. X., Li G. M., Yi D. & Yu P. W. A meta-analysis: The association between interleukin-17 pathway gene polymorphism and gastrointestinal diseases. Gene 572, 243–251, doi: 10.1016/j.gene.2015.07.018 (2015). [DOI] [PubMed] [Google Scholar]
- Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L. & Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer. Inst. 96, 434–442 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A. J. et al. Methods for meta-analysis in medical research Vol. 348 (Wiley Chichester, 2000). [Google Scholar]