Abstract
The histone H3 variant, CENP-A, is incorporated into nucleosomes that mark centromere location. We recently reported that CENP-A confers an altered nucleosome shape relative to its counterparts containing conventional H3. Using a single molecule fluorescence resonance energy transfer (FRET) approach with recombinant human histones and centromere DNA, we now find that the nucleosome shape change that CENP-A directs is dominated by lateral passing of the two DNA gyres (gyre sliding). A non-histone centromere protein, CENP-C, binds to and reshapes the nucleosome, sliding the DNA gyres back to positions similar to those in canonical nucleosomes containing conventional histone H3. The model we generate to explain the CENP-A nucleosome transition provides an example of a shape change imposed by external binding proteins, and has important implications for understanding the epigenetic basis for the faithful inheritance of centromere location on the chromosome.
Introduction
In diverse eukaryotes, centromere location is specified by a unique chromatin domain containing CENP-A nucleosomes1–3. CENP-A is one of the best candidates to epigenetically mark the centromere. CENP-A and other constitutive centromere proteins track with newly formed centromeres (i.e. neocentromeres)4–6 lacking the repetitive α-satellite DNA found at typical human centromeres, but they are absent from centromeres that are silenced in pseudo-dicentric chromosomes4–7. The notion that CENP-A, as a histone variant, would carry the epigenetic information to specify centromere location has been bolstered by many lines of evidence (reviewed in refs 1-3), including key experiments to use CENP-A nucleosome assembly (through direct or indirect targeting or reconstitution) to seed new centromeres at ectopic chromosomal loci8–12, on a plasmid13, or in Xenopus extracts14.
Though CENP-A is thought to be the key molecule to specify centromere location, it does not act alone when performing its essential centromere function. For one example, we recently found that CENP-A collaborates with a constitutive centromere protein, CENP-C, to maintain centromere identity15. In the absence of CENP-C, CENP-A directs an altered shape to the octameric histone core, and when CENP-A is directed to chromosome locations lacking a high local concentration of CENP-C it is destabilized15. The nucleosome shape deviation emanates from rotation at the CENP-A–CENP-A interface16, requiring the movement of H2A–H2B dimers away from each other to avoid steric clashing. Indeed, H2A–H2B dimers are 5 Å further away from each other in CENP-A nucleosomes than in canonical H3-containing nucleosomes15, but the nature of the structural rearrangement—central to understanding the altered path of CENP-A nucleosomal DNA—remains unclear (Fig. 1a). The central domain of CENP-C (CENP-CCD) contacts the C-terminal tail of CENP-A, as well as discrete surfaces on histones H2A and H417,18, and reshapes the CENP-A nucleosome15 (Fig. 1a), providing a prime example for the chromatin field that nucleosome shape and function can be modulated in a manner analogous to the allosteric regulation of enzymes.
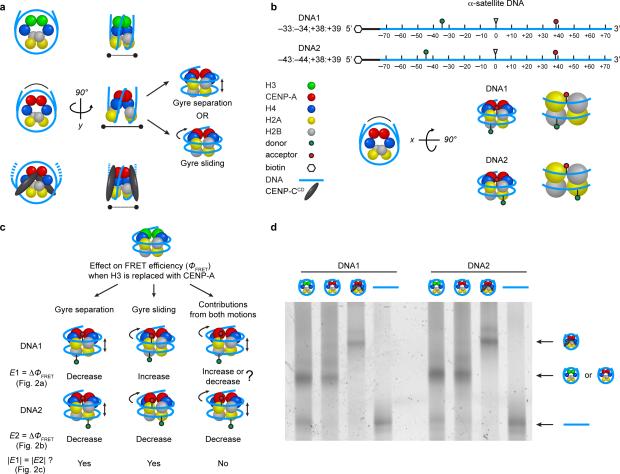
Figure 1. A single molecule FRET approach to measure the DNA wrap of nucleosomes.
(a) Cartoon schematic of H3, CENP-A, and CENP-A nucleosomes bound by CENP-CCD. The schematic for CENP-A nucleosomes indicates an altered histone core structure where the H2A–H2B dimers rotate away from each other, which suggests an altered DNA wrap that could lead to either the DNA gyres widening or sliding past each other. (b) Diagram of the two DNAs used in single molecule FRET experiments to investigate differences in the DNA wrap of H3, CENP-A, and CENP-A nucleosomes bound by CENP-CCD. The DNA sequence is derived from human α-satellite DNA and the donor and acceptor fluorophores are represented by green and red lollipops, respectively. Basepair numbering corresponds to the 5’ (−) to 3’ (+) direction relative to the dyad. (c) Predictions for the change in ΦFRET between CENP-A and H3 nucleosomes for the two fluorophore pairs if the DNA gyres widen, slide past each other, or if both movements occur. (d) Native PAGE of indicated samples visualized by Cy3 fluorescence. Uncropped gel image is shown in Supplementary Data Set 1.
Here, we set out to define the CENP-A nucleosome structural transition resulting in an altered path of nucleosomal DNA, because its importance is in understanding both the epigenetic maintenance of centromere identity and the possible ways in which nucleosome structure and function can be modulated.
Results
DNA gyre sliding in the CENP-A nucleosome
The 147 bp of DNA that wrap a canonical nucleosome make ~1.7 turns around the histone core, resulting in two DNA gyres that contact the H2A–H2B dimers in the histone core19. H2A–H2B dimers moving away from each other in CENP-A nucleosomes could cause the two gyres of DNA to move away from each other, resulting in a widening of the DNA gyres (Fig. 1a). Alternatively, the movement of the dimers could result in the DNA gyres moving laterally past each other, resulting in a tightening of the DNA wrap at the point of contact of the dimers (Fig. 1a). In order to measure the relative contribution of these two types of DNA gyre movement, we designed a FRET-based scheme that employs two nucleosomal DNAs (DNA1 and DNA2; Fig. 1b) each derived from a human α-satellite sequence20 where the dyad position precisely matches where CENP-A nucleosomes map at native centromeres21. In the case of DNA1 (Fig. 1c), gyre separation would result in the fluorophores moving away from each other, resulting in a decrease in FRET efficiency (ΦFRET). For lateral DNA gyre passing, the donor fluorophore moves closer to the acceptor fluorophore, resulting in higher ΦFRET. In the case of DNA2 (Fig. 1c), gyre separation also results in the fluorophores moving away from each other, leading to a decrease in ΦFRET. However, if lateral DNA gyre passing occurs, the donor fluorophore moves further away from the acceptor fluorophore, resulting in a decrease in ΦFRET for DNA2. An important aspect of our design is that we expect the absolute change (ΔΦFRET) to be roughly equal for both DNA1 and DNA2 if either gyre separation or lateral passing dominates the structural change, because each pair is located approximately the same distance from one another on the DNA. Therefore, in the case that gyre separation dominates, ΦFRET will decrease for both DNA1 and DNA2, with both decreasing by a similar magnitude (Fig. 1c). If DNA gyre lateral passing dominates, then ΦFRET should increase for DNA1 and decrease for DNA2, but the magnitude of change should be approximately equal (Fig. 1c). If both types of movement substantially contribute to altering the DNA path, then the absolute ΦFRET measured for DNA1 should be different from the measured ΦFRET for DNA2, because DNA gyre separation and lateral DNA gyre passing have opposite effects on ΦFRET for DNA1 but each result in decreasing ΦFRET for DNA2 (Fig. 1c).
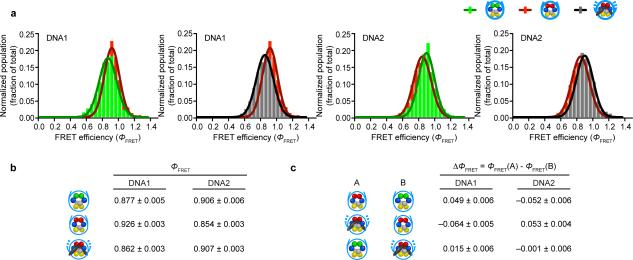
Our single molecule FRET measurement setup is based on one that we have used to address diverse issues in nucleosome structure and dynamics22,23. Nucleosomes (and nucleosomal complexes; see native PAGE in Fig. 1d) exhibited a range of ΦFRET values and we focused our analysis on the high FRET group where nucleosomes are positioned on the DNA as we designed. We performed separate analysis on those in low and medium FRET groups where positioning on the DNA template may vary slightly (see Methods, Supplementary Fig. 1, and Supplementary Table 1). In all bins, CENP-A nucleosomes reconstituted with DNA1 exhibit a significantly increased ΦFRET, but with DNA2 exhibit a significantly decreased ΦFRET, relative to H3 nucleosomes (Fig. 2a-c). Further, the absolute change in ΦFRET is almost identical in these comparisons (Fig. 2a-c). The low and medium FRET sub-groups that we observed may represent sub-populations of the nucleosomes with varying gaps between the two nucleosomal DNA gyres24. Changes in ΦFRET can originate from changes in physical and photophysical properties of the fluorophores, such as rotational freedom and fluorescence quantum yield. As CENP-A is highly unlikely to be in direct contact with the fluorophores (see Fig. 1b for approximate locations of CENP-A and fluorophores), CENP-A in the nucleosomes unlikely affects the rotational freedom or the quantum yield of the fluorophores. Taken together, these data indicate that DNA alteration in CENP-A nucleosomes relative to its conventional counterparts containing canonical H3 is heavily dominated by the gyres laterally passing one another, with a small contribution from the DNA gyres separation.
Figure 2. CENP-A nucleosomes adopt an altered DNA wrap that reverts to canonical wrapping upon CENP-CCD binding.
(a) ΦFRET values for H3, CENP-A, and CENP-A nucleosomes bound by CENP-CCD plotted as histograms and fitted to a Gaussian distribution for DNA1 and DNA2. (b) Summary of ΦFRET values for nucleosomes and DNA indicated. Efficiency values are the center of Gaussian curve fitting in panel a. (c) Summary of change in ΦFRET values between the indicated nucleosomes. The differences in ΦFRET are calculated for three different nucleosome comparisons (indicated in columns A and B) on both DNA1 and DNA2. Error is reported as the uncertainty in the average FRET at a 95% c.i. DNA1: N = 249 (H3), 157 (CENP-A), 355 (CENP-A + CENP-CCD); DNA2: N = 128 (H3), 312 (CENP-A), 145 (CENP-A + CENP-CCD), where N represents the total number of individual nucleosome measurements taken from 3 separate slides.
CENP-C reverts the altered DNA wrap of CENP-A nucleosomes
We predicted that CENP-CCD binding to the CENP-A nucleosome would cause the gyres of the DNA to slide back to a conventional nucleosome orientation. This conclusion is both based on our earlier findings15 and the present findings that the major form of structural alteration in unbound CENP-A nucleosomes occurs via DNA gyre sliding (Figs. 1 and 2). Indeed, CENP-A nucleosomes bound by CENP-CCD have essentially the same ΦFRET as those of isolated canonical nucleosomes containing conventional H3, suggesting that the internal DNA wrap of these two nucleosome complexes are nearly identical (Fig. 2a-c). It is unlikely that the ΦFRET changes upon CENP-C binding are due to altered fluorophore photophysics, given that CENP-C binding exerted opposing effects on ΦFRET in the two different nucleosomes. Therefore, the ΦFRET changes we observed are mainly due to the changes in the distance between the fluorophores. Taken together, our findings provide a view of the starting and ending points of the protein15 and DNA (Figs. 1 and 2) components of the CENP-A nucleosome during the structural transition directed by CENP-CCD that rigidifies and stabilizes it to maintain centromere identity through cell generations.
A model of CENP-A structural transitions
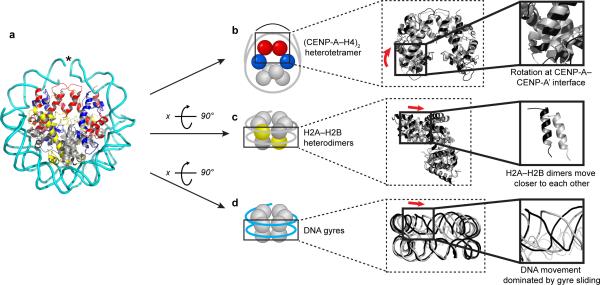
To visualize the CENP-A nucleosome structural transition, we constructed a model of its favored state in solution prior to binding to CENP-C (Fig. 3a). Our model of the CENP-A nucleosome in the absence of CENP-C (Fig. 3a) integrates compaction of the (CENP-A–H4)2 heterotetramer at the CENP-A–CENP-A interface16 (Fig. 3b), movement of the H2A–H2B dimers away from each other15 (Fig. 3c), and lateral passing of the DNA gyres (Fig. 3d) relative to the crystallized form of the CENP-A nucleosome25.
Figure 3. CENP-A nucleosomes adopt both an altered histone core and DNA wrap in solution prior to CENP-C binding.
(a) Model of the CENP-A nucleosome in solution prior to CENP-C binding. (b-d) The CENP-A nucleosome model (black) in solution prior to CENP-C binding superimposed on the CENP-A nucleosome crystal structure with DNA mutated to match the sequence used in this study (grey, PDB ID 3AN2; representative of the CENP-A nucleosome structure upon CENP-C binding). Solid black boxes indicate enlarged regions where notable histone–DNA movements occur upon CENP-C binding, including rotation of the CENP-A–CENP-A’ interface outward (b), H2A–H2B dimers moving closer together (c), and DNA gyre sliding (d). Red arrows indicate the direction of histone–DNA movement in the CENP-A nucleosome upon CENP-C binding. Only the α1 helices of one H2B subunit are shown in the enlargement for (c) for ease of viewing.
The rotation and compaction that initiates at the CENP-A–CENP-A four-helix bundle is simply propagated through the H2A–H2B dimer and leads to DNA gyre sliding. Interestingly, we note that even in the crystallized form of the CENP-A nucleosome25 the CENP-A–CENP-A four-helix bundle is rotated but not compacted relative to the H3–H3 four-helix bundle in canonical nucleosomes (Supplementary Fig. 2). For the nucleosomal DNA in our model, the major alteration is at or near the dyad (i.e. near the CENP-A–CENP-A interface), where there is a decreased radius of curvature around the histone core in CENP-A nucleosomes prior to CENP-C binding. We adjusted the path of DNA using the FRET measurements (Fig. 2) on a high-resolution CENP-A nucleosome structural model25 (PDB ID 3AN2). One chain of each histone (CENP-A, H4, H2A, and H2B) on one half of the dyad axis of symmetry was fixed, and the other chains (CENP-A’, H4’, H2A’, and H2B’) were rotated. After simple model minimization, we note that the DNA contacts to each half of the nucleosome were maintained, which causes unrealistic bond distances at the dyad site of the nucleosomal DNA (Fig. 3a, marked by asterisk; note the break in the continuity of the DNA ribbon diagram). DNA compression at the dyad is expected in order to accommodate the reduced radius of gyration. In addition to compression, kinking and stretching at histone contact points throughout the nucleosome would also distribute the changes required by the altered CENP-A nucleosome structural state, as has been well noted when analyzing canonical nucleosomes crystallized on various DNA sequences26–28. Indeed, independent evidence of altered DNA conformation in CENP-A nucleosomes is seen by increased intercalation and reactivity of N-(2,3-epoxypropyl)-1,8-naphthalimide (ENA) at a GG dinucleotide located 1.5 turns of DNA from the dyad (Supplementary Fig. 3). Thus, the DNA near the dyad is substantially altered while the gyres simply pass by one another on the opposite side of the nucleosome without any other extraordinary bends of the DNA. CENP-CCD binds near the dyad15,18 and stabilizes the form of the CENP-A nucleosome where the CENP-A–CENP-A four-helix bundle is rotated to a conventional shape, and the gyres of the DNA slide back to the path found in canonical nucleosomes (Fig. 3 and Supplementary Video 1).
Discussion
The structural variations of the CENP-A nucleosome and their subsequent changes upon CENP-C binding provide insight into the basis of epigenetic inheritance of the centromere. Here, we have identified the major change to the path of nucleosomal DNA and generated a molecular model to integrate past and current data sets from solution and crystal studies.
CENP-A is found at a high local concentration at the centromere that, in turn, recruits CENP-C to the centromere, binding directly to the CENP-A nucleosome and changing its shape. We envision that when CENP-A is distributed in the genome—as may be the case when found overexpressed in some cancers29—CENP-C molecules could still bind, but limiting amounts of CENP-C or other essential non-histone centromere proteins30,31 do not support the formation of a stable centromere chromatin domain29,32–34. New centromere formation can be stimulated by artificially directing a high local concentration of nascent CENP-A nucleosome assembly8–13, and we envision that rare neocentromeres occurring in the human population4–6,35 arise at chromosome arm sites where a cluster of CENP-A nucleosomes recruits CENP-C molecules and together initiate formation of a heritable centromere. By altering the nucleosome in this way, CENP-C binding stabilizes CENP-A nucleosomes at the centromere and helps solidify the foundation of centromeric chromatin. When CENP-C is removed from centromeres, the stability is compromised15. Thus, the CENP-A nucleosome structural transition is intimately linked to its function in epigenetically marking centromere location over the long timescales that are biologically relevant. Further, our study provides an important example of how nucleosome shape alteration could be coupled to function for other types of nucleosomes—either containing canonical or variant histones—in diverse chromatin contexts.
Online Methods
DNA preparation
Nucleosomal DNA was prepared by ligating oligonucleotides as described elsewhere22. For both DNA1 and DNA2, a 20 base single stranded DNA linker with biotin at one end was added to a 147 bp human α-satellite sequence20. Each DNA construct was prepared by ligating 6 oligonucleotides (Integrated DNA Technologies, Coralville, IA), and both fluorophores are on the forward strand and attached via a six-carbon linker (Integrated DNA Technologies, Coralville, IA). The FRET donor (Cy3) is attached at the −33 base in DNA1and the −43 base in DNA2, and the FRET acceptor (Cy5) is attached at the +38 base in both DNA constructs.
Protein preparation
Human histones and CENP-A were prepared as described elsewhere16. Recombinant human CENP-CCD consisting of the central domain only (a.a. 426-537) was GST-tagged and purified over a GST column followed by PreScission protease cleavage (GE Healthcare) and ion-exchange chromatography and prepared in a buffer containing 20 mM Tris pH 7.5, 200 mM NaCl, 0.5 mM EDTA, 1 mM DTT.
Nucleosome reconstitutions
Nucleosomes were reconstituted as described previously36. Briefly, labeled DNA was mixed with H2A–H2B dimers and (H3–H4)2 or (CENP-A–H4)2 tetramers in a TE buffer (10mM Tris-HCl pH 8.0, and 1mM EDTA) with 2 M NaCl. The mixture was dialyzed stepwise against TE buffers with 850, 650, 500, and 2.5 mM NaCl for 1 h each step at 4°C. Nucleosome assembly was assessed with native PAGE and fluorescence imaging (Typhoon 9410, GE Healthcare). CENP-CCD was incubated with CENP-A nucleosomes for 1 h at room temperature and complex formation was confirmed with native PAGE.
Single molecule FRET measurements
Single molecule measurements were carried out as described elsewhere22,23. A quartz microscope slide was coated with a 99:1 mixture of polyethyleneglycol (PEG) and biotin-PEG-silane (Laysan Bio, Arab AL), followed by incubation with a 100 pM streptavidin solution. Nucleosomes were immobilized on the slide surface through streptavidin-biotin conjugation and the measurements were completed within 30 min upon immobilization to avoid nucleosome disassembly. Fluorescence signals were collected with an electron multiplying CCD camera (EMCCD; iXon EM+ DU-897, Andor Technology, Belfast, UK) from a home-built prism coupled total internal reflection fluorescence microscope based on a commercial microscope (TE2000; Nikon, Tokyo, Japan). The FRET donor was excited with a 532 nm laser (CrystaLaser, GCL-150-L, Reno NV). The fluorescence signal was separated into two spectral regions, 550-650 and 650-750 nm, for Cy3 and Cy5, respectively, using a dichroic mirror (650DCXR, Chroma Technology Corp., Bellows Falls, VT) and a filter (HQ650/200m, Chroma Technology Corp.). The signals from the two fluorophores were collected simultaneously at a rate of 250 ms/frame. FRET efficiency (ΦFRET) at each time point was calculated with the formula ΦFRET = Icy5/(Icy3 + Icy5), where I is the signal intensity of the corresponding fluorophore. Measurements were taken from a minimum of 3 separate slides.
Single molecule FRET analysis
For each nucleosome class (H3, CENP-A, CENP-A bound by CENP-CCD on either DNA1 or DNA2), the average ΦFRET per trace was calculated for individual nucleosomes and binned into three groups (low, medium, or high FRET). The FRET efficiency distribution in each group was plotted as a histogram, which was normalized by the sample size. The distribution of the FRET efficiencies in a histogram is due to the Poissonian fluorescence photon emission statistics (fluorescence intensity fluctuation over time) that is inherent to any fluorescent signal. The reported uncertainties in the average FRET values are at a 95% c.i. Nucleosomes that contained a malfunctioning Cy5 fluorophore or were aggregates (6–21% of the total fluorescent particles in each experiment) based on multiple photobleaching events were not included in the analysis.
Nucleosome modeling
The molecular model of the CENP-A nucleosome in solution (without CENP-C bound) was generated using a high-resolution CENP-A nucleosome structure25 (PDB ID 3AN2) as a starting model. The DNA sequence was modified to match the DNA sequence that was used in the smFRET experiments, and chains A, B, C, D, I (residues −60 to −1) and J (residues 0 to 60) are moved as a rigid body to satisfy the rotation between chains A and E observed in the (CENP-A–H4)2 crystal structure16 (PDB ID 3NQJ) and the DNA gyre sliding movements observed in smFRET between DNA J −33 and +38 (DNA1) and J −43 and +38 (DNA2). Chains E, F, G, H, I (residues 0 to 60) and J (residues −60 to −1) were kept fixed. The energy of the final model was minimized using an annealing procedure in the CNS program37.
DNA intercalation and footprinting with ENA
Reactions were carried out as described38. Briefly, ENA was added from a 4 mM stock (dissolved in DMSO) at 2:1 or 20:1 molar excess over free DNA or nucleosome complexes, respectively. All DNA and nucleosome samples were reconstituted with a 147 bp non-palindromic form of α-satellite DNA sequence (the same sequence used in our smFRET experiments) that is hexachlorofluorescein (HEX) labeled at the 5’ end of the top strand only. Samples were incubated overnight at room temperature shielded from light. After incubation, addition of 4 M NaCl, followed by phenol-chloroform and subsequent chloroform-only extractions were carried out to remove unreacted ENA, proteins, and excess phenol. DNA was then ethanol precipitated and resuspended in 20 μL of TE buffer (10 mM Tris-Cl, 0.1 mM EDTA). Samples were heated at 95°C for 30 min, followed by addition of 20 μL 1 M piperidine and an additional 30 min incubation at 95°C to induce chemical cleavage of alkylated guanines. 1.2 mL of butanol were added to each sample, followed by brief vortexing and centrifugation at 12,000 × g for 2 min at 4°C. The supernatant was removed, pellets were resuspended in 150 μL 1% SDS and 1 mL butanol, vortexed briefly, and centrifuged at 12,000 × g for 2 min at room temperature. The supernatant was discarded and samples were lyophilized 20 min to remove excess liquid. Samples were resuspended in 10 μL loading buffer (1X TBE [88 mM Tris-borate, 2 mM EDTA], 90% formamide, 0.1% bromophenol blue) boiled at 95°C, and separated by denaturing PAGE (10% polyacrylamide, 7 M urea, 88 mM Tris–borate, 2 mM EDTA, pH 8.3). Maxam-Gilbert purine sequencing standards were prepared as previously described39. Gels were imaged on a Typhoon 9200 imager (GE Healthcare).
Movie construction
Movie segments were made in PyMOL (version 1.7.4, http://www.pymol.org) and assembled in QuickTime Pro (version 7.6.6)
Supplementary Material
Acknowledgements
We thank K. Gupta (University of Pennsylvania) for advice on modeling, C. Davey (Nanyang Technical University) and U. Surana (Agency for Science, Technology and Research, Singapore) for advice on ENA experiments, and D. Cleveland (University of California, San Diego), K. Luger (University of Colorado), and A. Straight (Stanford University) for plasmids. This work was supported by the US National Institutes of Health grants GM082989 (B.E.B.) and GM097286 (T.-H.L.) and a postdoctoral fellowship from the American Cancer Society (N.S.). We acknowledge support from the University of Pennsylvania Genetics Training Grant (US National Institutes of Health grant GM008216, S.J.F.).
Footnotes
Author Contributions
S.J.F., J.L., N.S., T.-H.L. and B.E.B. designed experiments. M.A.S. performed preliminary experiments that informed the design of the study. S.J.F. and J.L. performed experiments. S.J.F., J.L., N.S., T.-H.L. and B.E.B. analyzed data. N.S. performed modeling. S.J.F., J.L., N.S., T.-H.L. and B.E.B. wrote and edited the paper. T.-H.L. and B.E.B. directed the study.
References
- 1.Sekulic N, Black BE. Molecular underpinnings of centromere identity and maintenance. Trends Biochem. Sci. 2012;37:220–229. doi: 10.1016/j.tibs.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westhorpe FG, Straight AF. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 2015;7:a015818. doi: 10.1101/cshperspect.a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 5.Amor DJ, et al. Human centromere repositioning ‘in progress’. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassett EA, et al. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 8.Barnhart MC, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hori T, Shang W-H, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-C, et al. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logsdon GA, et al. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 2015;208:521–531. doi: 10.1083/jcb.201412011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tachiwana H, et al. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 14.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falk SJ, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H, et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Harp JM, et al. X-ray diffraction analysis of crystals containing twofold symmetric nucleosome core particles. Acta Crystallogr. D Biol. Crystallogr. 1996;52:283–288. doi: 10.1107/S0907444995009139. [DOI] [PubMed] [Google Scholar]
- 21.Hasson D, et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JY, Lee T-H. Effects of DNA methylation on the structure of nucleosomes. J. Am. Chem. Soc. 2012;134:173–175. doi: 10.1021/ja210273w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JY, Wei S, Lee T-H. Effects of histone acetylation by Piccolo NuA4 on the structure of a nucleosome and the interactions between two nucleosomes. J. Biol. Chem. 2011;286:11099–11109. doi: 10.1074/jbc.M110.192047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngo TTM, Ha T. Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 2015;43:3964–3971. doi: 10.1093/nar/gkv276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 26.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 27.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 28.Tan S, Davey CA. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacoste N, et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 31.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 32.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 34.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
Online Methods References
- 36.Wei S, Falk SJ, Black BE, Lee T-H. A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic Acids Res. 43 doi: 10.1093/nar/gkv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 20152007:e111 2, 2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 38.Davey GE, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosome facilitates alkylation by an intercalating antitumour agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.