Summary
Cells of the immune system are auxotrophs for most amino acids, including non-essential ones. Arginine and tryptophan are used within the regulatory immune networks to control proliferation and function through pathways that deplete the amino acid, or create regulatory molecules such as nitric oxide or kynurenines. Strategies to harness amino acid auxotrophy to block cancerous lymphocyte growth have been attempted for decades, with limited success. How immune cells integrate information about external essential amino acids supplies and transfer signals to growth and activation pathways remains unclear, but has potential for pathway discovery. Emerging insights may lead to strategies to both degrade amino acids and to block the immunoregulatory pathways controlled by amino acids.
Introduction
Humans and most mammals long ago abandoned producing 9 of the 20 amino acids needed for protein biosynthesis. These are acquired from the diet and microbiota. Most cells of the immune system (with a few notable exceptions discussed herein) have additional requirements for non-essential amino acids such as glutamine and asparagine. The need for external supply of a nutrient is called auxotrophy. Amino acid auxotrophy has evolved to become an immunoregulatory control point to shape immune responses, to control the production of antimicrobial effectors and control their tissue damage, and to temper T cell responses through a variety of mechanisms including amino acid starvation.
One of the earliest investigations about amino acid auxotrophy in immune responses was the discovery that macrophages block tumor growth through the consumption of arginine, or use arginine to make an anti-tumor molecule, later discovered to be nitric oxide (NO)1-6. Therefore, in the late 1970s two features of the biology of the arginine immunoregulatory system had been discovered without knowledge about the proteins and pathways involved. Thirty years later, arginine metabolism through depletion and NO pathways remains in focus. In addition, tryptophan metabolism has emerged an important immunological control mechanism7. I will concentrate on arginine and tryptophan, and note new areas of amino acid auxotrophy under study including exploitation of amino auxotrophy in clinical medicine.
Regulated of amino acid metabolism
Over the course of immune system evolution, two primary amino acid metabolic regulatory nodes were selected: arginine and tryptophan. The regulated enzymes that metabolize arginine are inducible nitric oxide synthase (iNOS), arginase-1 and arginase-2, Arg1 and Arg2, respectively (Fig. 1). Indoleamine 2,3-dioxygenases (IDO1 and IDO2) are enzymes that metabolize tryptophan. At this point it is important to stress these are the regulated enzymes closely linked to immunological control mechanisms. This is not to discount other uses of amino acids in protein biosynthesis, or key metabolic pathways. However, it is curious that there seems no regulated enzyme system to destroy or metabolize leucine or threonine, for example, as these are also essential amino acids. A reasonable model is nature selected the arginine and tryptophan pathways as sufficient, and subsequently refined them into more complex regulatory pathways.
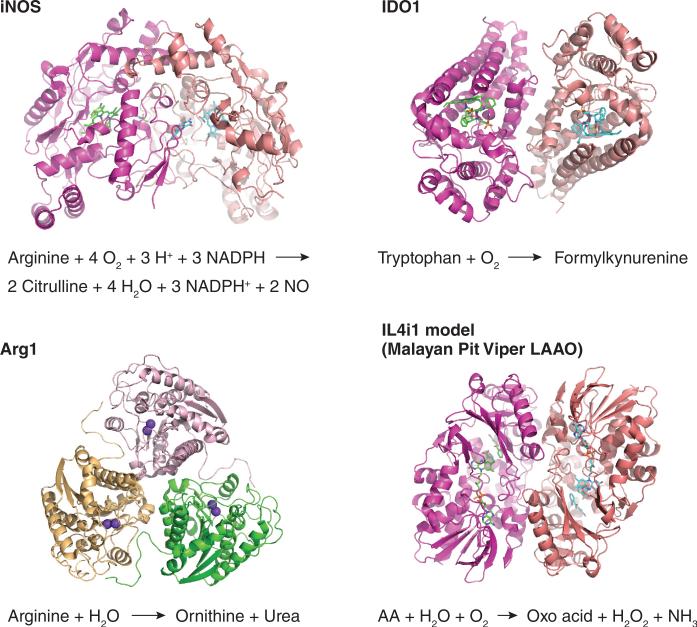
Figure 1. Regulated amino acid metabolizing enzymes in immunity.
Shown are structures and reaction mechanisms of iNOS, IDO1, Arg1 derived from deposited coordinates in the PDB (pdb.org). The structure of IDO2 has not been reported but is anticipated to be similar to IDO1. The structure of Arg2 is similar to Arg1 (Ref.109). The primary sequence structure of mammalian IL4i1 is conserved compared to snake venom L-amino acid oxidases110.
An additional immune-linked amino acid metabolizing protein is the interleukin 4 (IL-4)-inducible IL4i1 encoded by Il4i1 (Fig. 1). IL4i1 is an amino acid oxidase that is possibly secreted or localized to lysosomes8, 9. Little is known about IL4i1, including its precise substrate specificity, although some new work has linked IL4i1 to promotion of regulatory T cells and aberrant T cells responses in cancer8, 10 or suppressing human TH17 cells11. IL4i1 is interesting because it is closely related at the sequence level to L-amino acid oxidases found in high abundance in snake venoms12, making its potential immunological roles intriguing. At this point, Il4i1-deficient mice have yet to be described. As IL4i1 is the only known secreted L-amino acid oxidase encoded in the human or mouse genomes, it seems likely IL4i1 has a key role in one or more immunological processes that await evaluation.
Arginine metabolism in macrophages
Activated macrophages (and lymphocytes) import arginine through cation transporters (especially the inducible SLC7A2 transporter)13. A substantial literature has accumulated about arginine metabolism in macrophages over the last three decades14. However, the immune-specific tasks requiring arginine are often misunderstood. In part, misconceptions about arginine use in activated macrophages have arisen because of efforts to promote dualistic models of arginine metabolism based around NO-producing M1 macrophages versus arginine-hydrolyzing M2 macrophages15 and problems in translating findings in mouse system to human macrophages16.
Macrophage activation (or polarization) is a complex area of research because there are no straightforward means to precisely define how individual macrophages have been stimulated in complex immune microenvironments. In vivo, macrophage activation involves at least three intertwined pathways that establish the final activation state: developmental pathways that lead bone marrow-derived inflammatory monocytes to become macrophages within tissues, tissue microenvironmental cues that vary across organs and inflammatory environments, and the influence of polarizing cytokines or other molecules17. Most researchers are familiar with the latter “polarization” pathways, as bone marrow-derived macrophages (BMDMs) or rested peritoneal inflammatory macrophages can be readily and reproducibly M1 or M2 polarized with interferon-γ (IFN-γ) and Toll-like receptor (TLR) agonists or IL-4 and IL-13, respectively18. The experimental polarization of BMDMs is a limited reflection of what happens in vivo, as the culture conditions are either artificial, or fail to replicate the tissue microenvironment. However, the in vitro evaluation of changes in activation brought about by the different cytokine milieus allowed different groups to establish the concept of polarization linked to function, and that different polarization conditions are associated with differences in protein expression19-24. Dualistic models of macrophage activation were proposed based on inflammation versus wound healing in different mouse backgrounds25. Over time, the means to describe polarization markers, functional consequences and the association of polarization with arginine metabolism soon evolved into a hydra of confusion as far as terminology that is incrementally being addressed by the field18, 26.
In M2-dominated immunity (worm infections, asthma, tissue repair) macrophages make ornithine but very little or no NO. M2 macrophages have high expression of Arg1, but negligible expression of iNOS, which is regulated by IFN-γ. Deletion of Arg1 is lethal, as Arg1 has an essential role in the urea cycle. Another arginase isoform, Arg2 is mitochondrial and deletion of Arg2 has a limited phenotype27. The influence of Arg2 on immunity and arginine metabolism has yet to be fully explored. However, Arg2 should not be ignored, as the reactions performed by both Arg1 and Arg2 are identical. For example, Arg2 inhibits NO output in Helicobacter pylori infections 28. Therefore, experimental attribution of ornithine production and NO regulation must account for the activity of both enzymes. How Arg2 expression is regulated remains a gap in current knowledge.
Functions of Arg1 in M2 macrophages
Arg1 expression in M2 conditions is controlled by many factors, but the major axis is mediated by IL-4 and IL-13, which activates the transcription factor STAT6 (Fig. 2a)29. STAT6 binds to an enhancer in the Arg1 locus and cooperates with other transcription factors such as C/EBPβ30, 31. In this setting, STAT6 enforces >100-fold increase in Arg1 expression. Other factors increase Arg1 expression including IL-10-mediated increases in expression of the IL-4 receptor32. PPAR transcription factors also regulate Arg1 expression in M2 macrophages33. The PPAR pathway is likely to be a central means to enforce M2-like Arg1+ phenotypes on tissue-resident macrophages, such as adipose macrophages, which use the M2 pathway to balance inflammation in fat tissue and whole organism metabolism34.
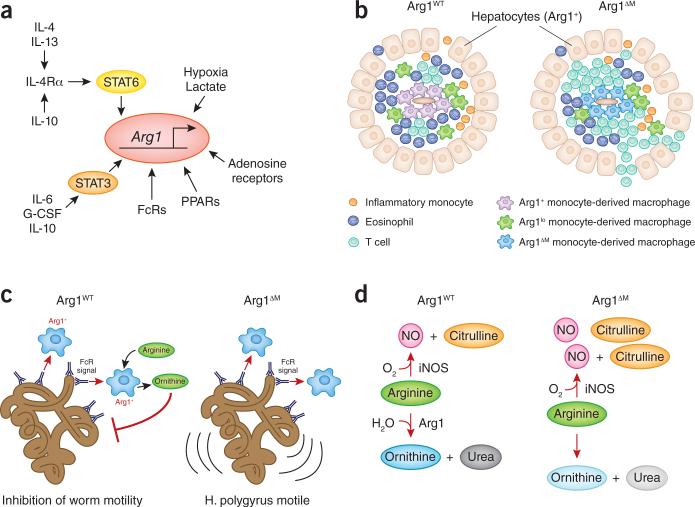
Figure 2. Arg1 in immune responses.
(a) Signaling pathways that increase Arg1 gene expression in the mouse. Two main pathways control Arg1 expression (STAT6 and STAT3). In addition, other pathways modulate the STAT3 and STAT6 pathways such as IL-10-mediated increases in IL-4Rα expression32, or act independently of the main pathways, such as hypoxia, lactate and adenosine pathways111-113. (b) Schematic of the how macrophage Arg1 regulates T cell proliferation in liver granulomas. In normal animals, Arg1+ macrophage encase schistosome eggs. The granulomas are embedded within the liver paranchyma, of which all hepatocytes constitutively express Arg1. When Arg1 is deleted only from macrophages, T cell proliferation is enhanced, leading to a feed-forward lethal non-resolving granulomatous reaction36, 38. (c) Possible mechanism of macrophage Arg1 in controlling worm motility. Macrophages are recruited to deposited antibodies on the surface of worms, which in turn activates Arg1 expression and ornithine production. Ornithine is proposed to contribute to stunting worm movement44. (d) Simplified schematic of the competition mechanism between Arg1 and iNOS when both enzymes are co-expressed35, 52.
A central question in understanding the links between amino acid metabolism and immunity is why do M2-activated macrophages make a protein that destroys arginine? A key advance toward answering this question was the development of macrophage-specific Arg1-deficient mice (Arg1ΔM)35. Using these animals to probe M2 infections, we found Arg1 has organ-specific effects in controlling T cell proliferation36, 37. For example, Arg1ΔM mice infected with Schistosoma mansoni had a lethal T cell-associated immunopathologic non-resolving inflammatory response to worm eggs in the liver36 or in the gut38. In this model, which replicates many features of the human disease bilharzia, macrophages encircle toxic worm eggs, walling them off from the liver parenchyma (Fig. 2b). Activated T cells making IL-4 and IL-13, together with recruited eosinophils, drive a potent TH2 response necessary to kill and dispose of the egg 39. The TH2 response enforces M2 polarization in local inflammatory macrophages, which express high amounts of Arg1. A popular hypothesis was M2 Arg1+ macrophages would produce ornithine, which can contribute to collagen synthesis40. However, schistosome-infected Arg1ΔM mice had increased liver fibrosis and collagen deposition, establishing that macrophage Arg1 was not essential for the production of collagen36. Another finding from the schistosome model was that the effects of macrophage-specific Arg1 on restricting T cell proliferation were confined to the liver, and not found in the lungs when worm eggs were artificially lodged in the lung parenchyma37. One interpretation of this finding concerns the relative perfusion rate of the two organs. Blood flow through the lung provides sufficient arginine that any effects of macrophage Arg1 on hydrolyzing arginine are not seen, while in the liver, the lower rate of blood flow combined with the high expression of Arg1 in hepatocytes (as part of the urea cycle) means the liver has a higher relative rate of arginine hydrolysis compared to the lung. Thus activated T cells drawn to the liver to control worm eggs encounter an arginine-poor microenvironment and cease proliferation, helping to dampen the inflammatory response to the toxic eggs and eventually reduce and resolve the granulomatous inflammatory response. By contrast, the removal of Arg1 in macrophages alone (but not hepatocytes) causes a lethal non-resolving inflammation. Confirmation that macrophages locally deplete arginine from T cells will require means to measure the rate of arginine hydrolysis in the organs of living animals. Beyond the obvious technological challenge of such an experiment, these data raise the key question of exactly how T cells detect how much arginine is necessary for their growth and function. A considerable literature exists on the consequences of arginine deprivation in T cells, and this pathway potentially regulates T cell proliferation and function in vivo41-43. However, isolation and phenotypic characterization of T cells from arginine-deprived immune microenvironments will be essential to trace the decision-making process T cells use to decide if there is sufficient arginine available for proliferation and function.
The use of the Arg1ΔM mice has also revealed other important aspects of macrophage-specific Arg1 expression. For example, Arg1 is required for direct anti-worm activity of macrophages (Fig. 2c). In one experimental setting using H. polygyrus, macrophages encased worms that had been previously coated with worm-specific antibodies and blocked their motility44. However, macrophages from Arg1ΔM mice could not inhibit worm movement. While the mechanism involved remains unclear, ornithine production from macrophages was required. The involvement of M2 macrophages in direct anti-worm functions is consistent with evolution of the M2 pathway to fight helminth infections and at the same time suppress excessive T cell responses.
Ornithine production and Arg1 have long been associated with wounding45. In one study where Arg1ΔM mice were used in a wound repair model, macrophage Arg1 was important for the rate of healing, without an excessive T cell response46. These data argue M2-linked Arg1 has multiple roles in immunity but the most consistent theme concerns the ability of macrophage M2 macrophages to deprive other cells of arginine.
Arginine metabolism in M1-type activated macrophages
A key function of M1-type macrophages is NO production from arginine and oxygen catalyzed by iNOS47. NO is directly toxic to many microbes and especially intracellular pathogens in addition to its myriad signaling and regulatory roles47, 48. Upon activation by IFN-γ and microbial products in a closed experimental system in vitro, M1-type macrophages import, via the regulated SLC7A2 transporter13, 49, all available arginine and create NO50. These data suggest M1 macrophages do not conserve arginine once provoked to make NO. As the final fate of the vast majority of inflammatory monocyte-derived macrophages is death, NO-producing M1 macrophages seem to be on a suicide mission to counter microbial infection. Consistent with this model, NO poisons the respiratory chain of mitochondria, forcing activated macrophages to import glucose and create energy from a Warburg-type metabolism51. A fundamental question in myeloid cell biology concerns the final fates of inflammatory monocytes that are activated to make NO and consume glucose: do they all die? Is the process reversible in a fraction of cells, and, if so, how do surviving macrophages rebuild their mitochondria?
A problem for the host is cytotoxicity of NO to host tissues, prompting a need to temper NO output. Arginases are the chief means macrophages use to suppress NO production once the decision to activate iNOS is made. Arg1 and iNOS, when expressed in the same cell, compete with each other for arginine52. Mycobacterial-infected macrophages, which would be classified as on the M1 part of the polarization spectrum by virtue of the many TLR- and NLR-activating ligands from the bacteria, induce expression of Arg1 (and probably Arg2) but via a pathway completely independent of the STAT6-mediated M2 pathway (Fig. 2a). Instead, TLR signaling induces the cytokines IL-6, G-CSF and IL-10, which signal through an autocrine-paracrine way to their cognate receptors, which activates STAT3 to upregulate Arg1 (ref.53). M1 macrophages therefore co-express iNOS and Arg1, a concept diametrically opposed to the dualistic model where macrophages express iNOS or Arg1 (ref.54). Deletion of macrophage Arg1 causes increased NO output and provided an advantage to the host in terms of controlling experimental Mycobacterium tuberculosis (Mtb)35. However, Arg1 (and probably Arg2) also regulate NO output by controlling how much iNOS protein is made by macrophages. Arg1 can block translation of the iNOS mRNA55, 56. Translating the relative Arg1-dependent effect of arginine competition versus other NO regulation pathways to in vivo settings will likely require single-cell techniques where each individual parameter can be quantified.
Expression of Arg1 in M1 polarized infections also has a key immunoregulatory role in blocking T cell proliferation in Mtb infection. To separate this pathway from Arg1's regulatory effect on NO output, the use of a unique Mtb model was required57. Although seemingly counterintuitive, Mtb infection of iNOS-deficient mice via the ear dermis induces a limited granulomatous reaction in the lung following transit of Mtb bacilli, or infected cells through the lymph57. The dermal infection model allows dissection of iNOS-independent modes of immunity to Mtb. However, in the absence of iNOS, Arg1 expression was increased in granuloma macrophages, raising the question of its function in this setting (where NO does not require tempering)58. To determine the reason for the increased Arg1 expression, mice lacking both iNOS and macrophage Arg1 were infected via the ear dermis. The outcome was a lethal T cell-associated immunopathological reaction occurred in the lung58. Thus, macrophage Arg1 was required, like in the M2-dominated schistosome egg model, to suppress T cell proliferation. Macrophage Arg1 plays two essential and linked roles in immune responses: tempering NO output and/or T cell proliferation.
Regeneration of arginine in immune cells
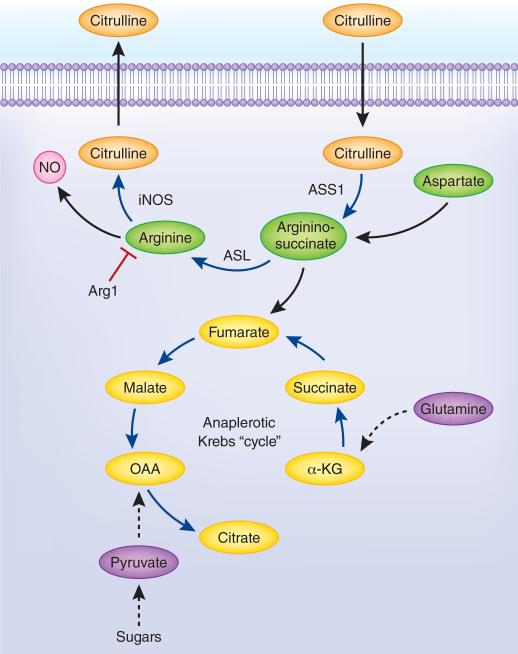
Activated T cells and myeloid cells import substantial amounts of arginine to sustain growth or for NO and ornithine production. Are immune cells then truly arginine auxotrophs? Answering this question revealed unexpected complexity in how arginine is metabolized in immune responses, in contrast to liver arginine metabolism as part of the urea cycle (Fig. 3). The urea cycle eliminates excess nitrogen, especially excess ammonia, which has differential toxicity to different cells. The urea cycle begins with the condensation of ammonia with CO2 to carbamoyl phosphate within mitochondria, which donates its carbamoyl group to ornithine to form citrulline. Through the progressive activity of argininosuccinate synthase (ASS1), argininosuccinate lyase (ASL) and Arg1, arginine is created and then destroyed to make urea and ornithine. Urea is excreted and thus ridding the body of the excess nitrogen. Carbamoyl phosphate synthase is not expressed in immune cells. However, ASS1 and ASL are expressed in immune cells, and ASS1 is regulated by TLR signaling50. The function of ASS1 and ASL was suspected to use the product of NO biosynthesis, citrulline, to recreate arginine via sequential energy-dependent condensation and lyase reactions40, 59. The enzymes required for this reaction (in macrophages) are iNOS, ASS1 and ASL, which presents a conundrum: why do immune cells import every available molecule of arginine if they can make their own? When we used heavy atom tracing of arginine metabolism in activated NO-producing macrophages we found citrulline was exported into the extracellular space50; presenting a second conundrum: how can the ASS1-ASL reaction occur if there is no substrate? To solve this problem we used macrophages lacking full ASS1 activity and manipulated the conditions in which NO was made. When arginine was depleted from the media and converted to NO, citrulline was reimported and converted via the action of ASS1 and ASL into arginine, providing fuel for more NO production 50. Thus, activated macrophages import any available arginine for NO biosynthesis. When arginine becomes limited, citrulline is imported to sustain NO output (Fig. 3). Consistent with this idea, mice lacking ASS1 in bone marrow-derived macrophages were lethally infected with Mtb compared to cognate control mice, arguing for an essential role of the ASS1-ASL-mediated arginine regeneration system in infection immunity50. Recent experiments manipulating the amounts of arginine and citrulline in the media revealed when M1 macrophages produce NO from citrulline using the arginine regeneration pathway, Arg1 is no longer capable of blocking NO production60. These and other data argue activated macrophages compartmentalize arginine metabolism in some way, possibly by sub-cellular partitioning of the key enzymes. If so, new cell biological principles could emerge from analysis of the flux of arginine and its products inside and outside cells.
Figure 3. Arginine regeneration and the anaplerotic Krebs cycle in M1 macrophages.
Key regulatory events in the links between the pathways include the supply of citrulline as a substrate, the role of NO in blocking mitochondrial respiration, the expression of the key enzymes and the flux through both pathways at a given time50, 51, 61. Many features of this reaction cycle remain to be uncovered.
A new twist in the story of arginine regeneration involves the way M1 macrophages alter their Krebs cycle. Glutamine and glucose tracing showed M1 macrophages use an interrupted Krebs cycle at the point where citrate is converted to α-ketoglutarate61. Once activated, M1 macrophages use anaplerosis reactions (‘filling up’) to feed metabolites into the segmented Krebs cycle; one of which is fumarate generated from the lyase reaction of ASL breaking argininosuccinate into fumarate and arginine (Fig. 3). NO, arginine and citrulline metabolism are connected to the incomplete Krebs cycle in cells where mitochrondria are unable to make ATP (from NO poisoning)51, 61. A limitation of this study was the use of a single time point, and the failure to recognize citrulline is exported and then reimported, increasing the complexity and dynamism of the system.
Tryptophan metabolism and IDO proteins
Like arginine metabolism, tryptophan degradation contributes to immunity through substrate depletion, product supply, or both pathways working together. Three enzymes degrade tryptophan: IDO1, IDO2 and tryptophan dioxygenase (TDO, a liver-specific enzyme) and each has been implicated in numerous immune responses7, 62, 63. The products of the tryptophan degradation pathway: kynureines and its metabolites, have wide physiological effects. Kynureine itself appears to regulate T cells via binding to the aryl hydrocarbon receptor. The physiology of kynureine production, metabolism and effects is a complex field and the reader is referred to comprehensive reviews64, 65.
The IDO1 pathway of tryptophan degradation has received the most attention because IDO1 expression, like iNOS, is controlled by IFN-γ. Several reports have suggested that IDO1 is immunosuppressive, because myeloid cells expressing IDO1 import and degrade tryptophan, depleting the local amounts of the amino acid, and producing kynurenine. The physiology of tryptophan depletion versus product generation remains debatable66. The notion that IDO1 is immunosuppressive has propelled the idea that IDO inhibitors such as 1-methyl tryptophan (1-MT) could be useful in overcoming local immunosuppression, as found in many cancers. Several problems underpin current research into the tryptophan pathway. First, IDO1 and IDO2 are encoded by linked genes (Ido1 and Ido2) arranged in a head-to-tail orientation. Their apposition prevents generation of an efficient meiotic cross to create mice lacking IDO1 and IDO2. Therefore, results from the IDO1 or IDO2-deficient mice need to account for the activity of the other protein. Second, commercially available antibody reagents against IDO1 lack specificity67 (and our unpublished data), and as yet there are no reporter mice that can faithfully track expression of IDO1 or IDO2 in vivo. Finally, the commonly used IDO inhibitor, 1-MT, was shown to be contaminated with tryptophan68, or to induce IDO169. 1-MT is often used experimentally to ‘rescue’ immunosuppressive effects of IDO+ cells that are considered to deplete local environments of tryptophan: if 1-MT has sufficient tryptophan as a contaminant, then rescue of growth or function attributed to IDO inhibition may also have a contribution from supply of tryptophan. A related issue concerns compounds similar to 1-MT that are IDO inhibitors and also inhibit the necroptotic kinase RIPK1 through an entirely different mechanism70, 71. The necroptosis field has begun to systematically appraise more specific RIPK1 inhibitors that appear to have lower activity against IDO proteins. Nevertheless, the specificity of anti-IDO drugs would be best systematically tested using cells from Ripk1−/− mice versus mice lacking both IDO proteins to truly establish their on-target range.
One of the key reports in understanding how IDO1 suppresses T cell growth showed activated T cells sense tryptophan depletion by IDO1+ myeloid cells via the GCN2 pathway, which leads to growth arrest72, 73. In the absence of GCN2 in CD8+ T cells, the tryptophan sensing mechanism was absent, and T cells entered into the cell cycle, even when tryptophan was limiting. However, GCN2 is not required for CD4+ T cell-mediated amino acid sensing 74. Furthermore, neither the GCN2- or IDO1-deficient mice have been reported to manifest any type of autoimmunity, and loss of IDO1 has a modest effect on the outcomes of genetically driven lung cancer, but a substantial effect on the outcomes of graft-versus-host response and checkpoint inhibitor efficacy75-77. It remains to be determined if the phenotypes in the IDO1-deficient mice are mediated by tryptophan catabolism, or other pathways. Taken together, the GCN2-IDO1 connection is replete with unresolved mechanistic links to amino acid auxotrophy. The aforementioned issues with 1-MT, antibody specificity issues and interpreting the phenotypes of single IDO-deficient cells raise the specter that much research on the IDO pathway needs to be reevaluated with defined and validated reagents.
Key roles of glutamine in immunity
Glutamine is the most abundant circulating amino acid in the body, and an integral requirement for normal metabolism78. Yet antigen-stimulated T cells have an obligatory requirement for not only for glutamine per se, but large quantities of the amino acid, which need to exceed a concentration threshold for effective proliferation79. TCR stimulation increases the activity of amino acid transporters, which import glutamine (ASCT2, encoded by Slc1a5), leucine (SLC7A5) as well as the aforementioned cation transporters80, 81. While it is straightforward to accept proliferating T cells need a lot of amino acids, the selectivity for glutamine is perplexing, especially as proliferating T cells convert to glycolytic metabolism to generate ATP82. Perhaps T cells use glutamine in anaplerotic sustenance of Krebs cycle components? Glutamine is required for mTOR activation and stimulation of the anabolic growth machinery needed to division and conversion into TH1 and TH17 cells, but not Treg or TH2 cells but starvation of glutamine propels CD4+ cells to a Treg cell-like phenotype83. This model correlates with data showing the T cell-specific deletion of mTOR results in a default Treg cell-like phenotype84. At this point, however, the challenge will be linking the observations about glutamine metabolism in T cells with the specific biochemistry of T cell subsets.
In macrophages, another glutamine paradox exists. Glutamine is essential for M2 macrophage development, as measured by signature gene expression. Glutamine feeds the supply of UDP-GlcNac, and subsequent N-linked glycolysis of the M2 secretory protein pool61. Deprivation of glutamine from IL-4-polarized M2 macrophages blunted the expression signature M2 genes. How this occurs is unknown, but suggests a connection between the transcriptional machinery needed for M2 polarization and glutamine. By contrast, glutamine deprivation did not affect M1 signature genes, NO production or viability61. How the selective differences between M1 and M2 macrophages arise in terms of glutamine metabolism is sure to be important in establishing the signals controlling the polarization of macrophages in vivo.
How do immune cells sense essential amino acids?
The most basic requirement for amino acids is to build proteins. How do cells determine how much amino acids are available for growth and function? At this point it is worth considering modes of cell division and their potential amino acid requirements. Some cells divide rarely if at all (neurons in adults), while other cells have a regimented division cycle to replace lost cells (epithelia). Unabated cell division with a shortened G1 phase of the cell cycle occurs in embryonic stem cells, which rapidly divide to create body mass. In the immune system, another mode of cell division is used by activated lymphocytes emerging from quiescence following antigen stimulation, which create hundreds of copies of themselves to fight infection; after which a fraction of cells enter memory programs while the unneeded cells die. The TORC1 pathway is closely tied to amino acid sensing from work predominantly performed on transformed cells like HeLa85. In these systems, TORC1 signaling integrates signals from inside and outside the cell about how much amino acids are available. How this works is unclear, as TORC1-assocated complexes like the ‘Ragulator’ have been implicated in amino acid sensing, but are yet to be definitively shown to be a ‘sensor’86. Recently, a lysosomal transporter called SLC38A9 was implicated in amino acid activation of TORC1, but again through an unknown mechanism87, 88. Although TORC1 signaling is linked to amino acid availability in transformed cells, these links are likely far more complex in the immune system where amino acid auxotrophy is so crucial for cell cycle decisions.
Perhaps a clearer way to think about mTOR and TORC1 signaling is consider their roles in cell growth. TORC1 is required for the emergence of T cells from quiescence, and required for the proliferation of thymic Treg cells: this is because TORC1 signals the accumulation of biomass required to generate daughter cells89, 90. By contrast, mTOR is dispensable for T cell development91. Instead, T cells (created by Cd4-Cre mediated deletion of Mtor) populated the body, but develop into cells like Treg cells. Why does this happen? The rapid proliferation of T cells requires TORC1, but when cells do not need to undergo explosive growth, TORC1 signaling is not required and instead mTOR-TORC1 helps T cells determine their phenotypic fate. The role of mTOR in fate decisions in immune responses is remarkable, as inhibition of T or B cell TORC1 via rapamycin enhances CD8+ memory, or B cell production of broadly neutralizing IgM. Rapamycin also ‘resets’ Treg cell function if given for a defined time window92, and controls macrophage polarization93. In the case of the latter, TORC1 is needed to macrophages to M1 polarize, but why? Likely the answer lies because the amount of biomass (rather than materials for cell division) needed to combat intracellular pathogens, perform antigen presentation and create a robust secretory function are all anabolic activities dependent on TORC1. However, it is unclear if any of the above examples are tied to direct amino acid sensing. Indeed, in the case of M1 macrophages consuming arginine to make NO, there is no apparent sensing system for tempering arginine consumption as M1 macrophages import all available arginine and oxidize or hydrolyze it until they exhaust the external supply50. In summary, the mechanistic basic of amino acid sensing by immune cells remains an open area of research.
Clinical exploitation of amino acid auxotrophy
Early work on the metabolic requirements of transformed cells demonstrated cell-type dependence for asparagine94. The mechanism of asparagine auxotrophy in cancer cells has generally been an assumption that the gene encoding the enzyme that converts glutamine to asparagine (Asparagine synthase, ASNS, encoded by ASNS) is mutated, silenced or deleted95, 96. However, the precise mechanisms involved in asparagine metabolism in cancer remain unresolved. Nevertheless, asparagine auxotrophy was the basis for administration of bacterial asparaginases in the ‘induction’ phase of acute lymphoblastic leukemia (ALL) chemotherapy97. ASNases are systemically administered with the rationale of depriving leukemic cells of asparagine and weakening their ability to survive chemotherapy (Fig. 4). ASNase treatment is now standard-of-care for ALL97. However, ASNase therapy comes with two penalties. First, ASNases are bacterial and elicit (in some patients) neutralizing antibodies voiding the efficiancy subsequent ASNase therapy. Second, some ALL patients develop life-threatening allergic responses to ASNases98. Therefore, a principle limitation of using microbial enzymes to degrade amino acids will be host responses to the foreign protein (Fig. 4). Cancerous cells are also arginine auxotrophs. To exploit this property, another microbial enzyme, arginine deimindase (ADI) from Mycoplasma, has been pegylated to improve in vivo pharmacodynamics. ADI is currently being tested in hepatocellular carcinoma and a wide range of other tumors. It will be of considerable interest to determine the extent of anti-ADI host immunity and dose-limiting toxicities.
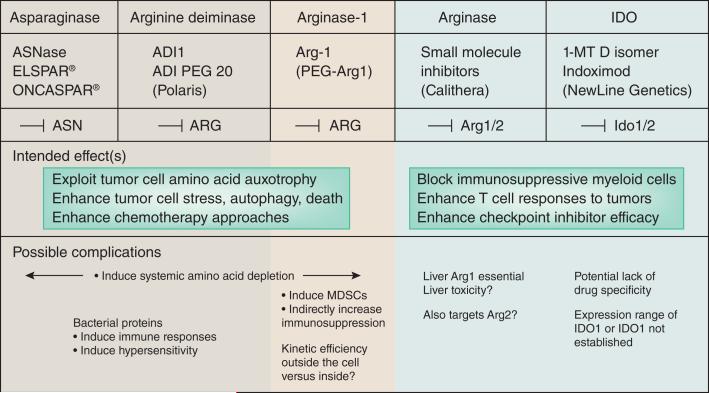
Figure 4. Clinical exploitation of amino acid auxotrophy.
Shown are three examples of proteins that degrade amino acids (ASNase, ADI and Arg1) that can be used to starve target cells of amino acids. Two examples of inhibition strategies for amino avid degrading enzymes are shown: arginases and IDO proteins. In the case of inhibition, the goal is to block the immunoregulatory effects of arginases or IDOs and enhance T cell responses. Possible complications are listed below.
A tactic to bypass the host response to microbial amino acid degrading enzymes is to use human ones. In this regard efforts have been to create Arg1 for injection99 (Fig. 4). Although human enzyme formulations should, at least partially avoid host responses, a different complication is whether the enzymes function efficiently in the extracellular milieu as opposed to an intracellular environment where substrate concentrations are high, and enzyme kinetics optimized. The performance of human enzymes in terms of degradative activity versus pharmacologic properties in whole animals must be carefully determined. Strong rational exists to pursue this approach for Arg1, because it can be disgorged from myeloid cells to locally deplete arginine100, 101. The fundamental question is whether human cytoplasmic enzymes are sufficiently powerful to limit essential amino acids in cancer therapy.
Activated myeloid cells are immunoregulatory in that they use amino acid degradation to deprive neighboring cells of essential amino acids. While essential amino acid degradation by myeloid cells is a key evolutionary adaptation to prevent host tissue damage, it is also thought to be a barrier to natural and enforced T cell activity against malignant cells because of the potent immunoregulatory activity of enzymes such as Arg1 (Refs.102). Therefore, therapeutic usefulness approaches such as checkpoint blockade must overcome the endogenous immunosuppressive microenvironments rich in myeloid cells expressing enzymes such as Arg1, Arg2, iNOS and IDO. Two branches of clinical development have emerged to block arginases or IDO proteins (Fig. 4). As noted previously, many questions surround the biology of IDO proteins in cancer, and the range and purity of 1-MT.
Perspective
Although substantial advances have been made in understanding regulated amino acid metabolism in immunity, key questions remain. For example, the concept of amino acid starvation leading to a ‘spreading’ regulatory environment coined infectious tolerance has yet to be confirmed using genetic approaches to manipulate a key essential amino acid74, 103. While we have a platform of knowledge to link macrophage-mediated arginine starvation to inhibition of T cell proliferation, whether how pathway enforces Treg development and function, and its significance to immune responses, is a major question. A second aspect of amino acid auxotrophy in immunity that deserves greater evaluation concerns intracellular pathogens that are themselves amino acid auxotrophs (for example, Chlamydia requires host tryptophan104) or express their own amino acid degrading enzymes (for example, Leishmania has its own arginase105) infecting macrophages that also require exogenous amino acids. Perhaps amino acid competition forms an integral part of the ‘macrophage paradox’ where the cells needed to kill pathogens are also their safe harbor106. An opportunity for the field is the ability to mix and match mutant macrophages with mutant pathogens: for example, what would be the outcome if arg-deficient Leishmania infect Arg1-deficient macrophages? A third question concerns the influence of non-immune cells in microenvironments enforcing amino acid depletion. For example, expression of Arg2 in neuroblastomas is thought to contribute to evasion of immune recognition107. However, at the same time, many tumor types are amino acid auxotrophs themselves108. Therefore, the complexity of inflammatory microenvironments such as in cancer, should yield to new techniques. However, an impediment is how to precisely measure amino acid concentrations in microenvironment, inside and outside cells, and in living animals.
Acknowledgements
Research in the author's laboratory is supported by grants from the NIH, Calithera Biosciences, NCI Cancer Core Grant P30 CA21765, and the American Lebanese Associated Charities.
Footnotes
Disclosure
The author's laboratory has a collaborative agreement with Calithera Biosciences to investigate aspects of arginine metabolism in immune responses.
References
- 1.Currie GA. Activated macrophages kill tumour cells by releasing arginase. Nature. 1978;273:758–759. doi: 10.1038/273758a0. [DOI] [PubMed] [Google Scholar]
- 2.Currie GA, Gyure L, Cifuentes L. Microenvironmental arginine depletion by macrophages in vivo. Br. J. Cancer. 1979;39:613–620. doi: 10.1038/bjc.1979.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meltzer MS, Ruco LP, Leonard EJ. Macrophage activation for tumor cytotoxicity: mechanisms of macrophage activation by lymphokines. Adv. Exp. Med. Biol. 1979;121B:381–398. doi: 10.1007/978-1-4684-8914-9_36. [DOI] [PubMed] [Google Scholar]
- 4.Ruco LP, Meltzer MS. Macrophage activation for tumor cytotoxicity: development of macrophage cytotoxic activity requires completion of a sequence of short-lived intermediary reactions. J. Immunol. 1978;121:2035–2042. [PubMed] [Google Scholar]
- 5.Keller R, Geiges M, Keist R. L-arginine-dependent reactive nitrogen intermediates as mediators of tumor cell killing by activated macrophages. Cancer Res. 1990;50:1421–1425. [PubMed] [Google Scholar]
- 6.Mills CD. Molecular basis of “suppressor” macrophages. Arginine metabolism via the nitric oxide synthetase pathway. J. Immunol. 1991;146:2719–2723. [PubMed] [Google Scholar]
- 7.McGaha TL, Huang L, Lemos H, Metz R, Mautino M, et al. Amino acid catabolism: a pivotal regulator of innate and adaptive immunity. Immunol. Rev. 2012;249:135–157. doi: 10.1111/j.1600-065X.2012.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulland ML, Marquet J, Molinier-Frenkel V, Moller P, Guiter C, et al. Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood. 2007;110:220–227. doi: 10.1182/blood-2006-07-036210. [DOI] [PubMed] [Google Scholar]
- 9.Mason JM, Naidu MD, Barcia M, Porti D, Chavan SS, et al. IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J. Immunol. 2004;173:4561–4567. doi: 10.4049/jimmunol.173.7.4561. [DOI] [PubMed] [Google Scholar]
- 10.Cousin C, Aubatin A, Le Gouvello S, Apetoh L, Castellano F, et al. The immunosuppressive enzyme IL4I1 promotes FoxP3 regulatory T lymphocyte differentiation. Eur. J. Immunol. 2015;45:1772–1782. doi: 10.1002/eji.201445000. [DOI] [PubMed] [Google Scholar]
- 11.Santarlasci V, Maggi L, Capone M, Querci V, Beltrame L, et al. Rarity of human T helper 17 cells is due to retinoic acid orphan receptor-dependent mechanisms that limit their expansion. Immunity. 2012;36:201–214. doi: 10.1016/j.immuni.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson RW, Pesce JT, Ramalingam T, Wilson MS, White S, et al. Cationic amino acid transporter-2 regulates immunity by modulating arginase activity. PLoS Pathog. 2008;4:e1000023. doi: 10.1371/journal.ppat.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grohmann U, Bronte V. Control of immune response by amino acid metabolism. Immunol. Rev. 2010;236:243–264. doi: 10.1111/j.1600-065X.2010.00915.x. [DOI] [PubMed] [Google Scholar]
- 15.Mills CD. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012;32:463–488. doi: 10.1615/critrevimmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- 16.Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J. Leukocyte Biol. 2011;89:557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J. Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- 22.Munder M, Eichmann K, Moran JM, Centeno F, Soler G, et al. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J. Immunol. 1999;163:3771–3777. [PubMed] [Google Scholar]
- 23.Adams DO, Hamilton TA. The cell biology of macrophage activation. Annu. Rev. Immunol. 1984;2:283–318. doi: 10.1146/annurev.iy.02.040184.001435. [DOI] [PubMed] [Google Scholar]
- 24.Mackaness G. B. The Immunological Basis of Acquired Cellular Resistance. J. Exp. Med. 1964;120:105–120. [PubMed] [Google Scholar]
- 25.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi O, Morris SM, Jr., Zoghbi H, Porter CW, O'Brien WE. Generation of a mouse model for arginase II deficiency by targeted disruption of the arginase II gene. Mol. Cell. Biol. 2001;21:811–813. doi: 10.1128/MCB.21.3.811-813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, et al. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, et al. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J. Immunol. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- 30.Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM., Jr. Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, et al. Enhancer-mediated control of macrophage-specific arginase I expression. J. Immunol. 2004;172:7565–7573. doi: 10.4049/jimmunol.172.12.7565. [DOI] [PubMed] [Google Scholar]
- 32.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 33.Chawla A. Control of macrophage activation and function by PPARs. Circ. Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, et al. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barron L, Smith AM, El Kasmi KC, Qualls JE, Huang X, et al. Role of arginase 1 from myeloid cells in th2-dominated lung inflammation. PLoS One. 2013;8:e61961. doi: 10.1371/journal.pone.0061961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbert DR, Orekov T, Roloson A, Ilies M, Perkins C, et al. Arginase I suppresses IL-12/IL-23p40-driven intestinal inflammation during acute schistosomiasis. J. Immunol. 2010;184:6438–6446. doi: 10.4049/jimmunol.0902009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat. Rev. Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris SM., Jr. Arginine metabolism: boundaries of our knowledge. J. Nutr. 2007;137:1602S–1609S. doi: 10.1093/jn/137.6.1602S. [DOI] [PubMed] [Google Scholar]
- 41.Fletcher M, Ramirez ME, Sierra RA, Raber P, Thevenot P, et al. l-Arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer Res. 2015;75:275–283. doi: 10.1158/0008-5472.CAN-14-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–1573. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez PC, Zea AH, DeSalvo J, Culotta KS, Zabaleta J, et al. L-arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J. Immunol. 2003;171:1232–1239. doi: 10.4049/jimmunol.171.3.1232. [DOI] [PubMed] [Google Scholar]
- 44.Esser-von Bieren J, Mosconi I, Guiet R, Piersgilli A, Volpe B, et al. Antibodies trap tissue migrating helminth larvae and prevent tissue damage by driving IL-4Ralpha-independent alternative differentiation of macrophages. PLoS Pathog. 2013;9:e1003771. doi: 10.1371/journal.ppat.1003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albina JE, Mills CD, Henry WL, Jr., Caldwell MD. Temporal expression of different pathways of 1-arginine metabolism in healing wounds. J. Immunol. 1990;144:3877–3880. [PubMed] [Google Scholar]
- 46.Campbell L, Saville CR, Murray PJ, Cruickshank SM, Hardman MJ. Local arginase 1 activity is required for cutaneous wound healing. J. Invest. Dermatol. 2013;133:2461–2470. doi: 10.1038/jid.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 48.Schindler H, Bogdan C. NO as a signaling molecule: effects on kinases. Int. Immunopharmacol. 2001;1:1443–1455. doi: 10.1016/s1567-5769(01)00089-3. [DOI] [PubMed] [Google Scholar]
- 49.Nicholson B, Manner CK, Kleeman J, MacLeod CL. Sustained nitric oxide production in macrophages requires the arginine transporter CAT2. J. Biol. Chem. 2001;276:15881–15885. doi: 10.1074/jbc.M010030200. [DOI] [PubMed] [Google Scholar]
- 50.Qualls JE, Subramanian C, Rafi W, Smith AM, Balouzian L, et al. Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host & Microbe. 2012;12:313–323. doi: 10.1016/j.chom.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modolell M, Corraliza IM, Link F, Soler G, Eichmann K. Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow-derived macrophages by TH1 and TH2 cytokines. Eur. J. Immunol. 1995;25:1101–1104. doi: 10.1002/eji.1830250436. [DOI] [PubMed] [Google Scholar]
- 53.Qualls JE, Neale G, Smith AM, Koo MS, DeFreitas AA, et al. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Science Signaling. 2010;3:ra62. doi: 10.1126/scisignal.2000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills CD. Macrophage arginine metabolism to ornithine/urea or nitric oxide/citrulline: a life or death issue. Crit. Rev. Immunol. 2001;21:399–425. [PubMed] [Google Scholar]
- 55.El-Gayar S, Thuring-Nahler H, Pfeilschifter J, Rollinghoff M, Bogdan C. Translational control of inducible nitric oxide synthase by IL-13 and arginine availability in inflammatory macrophages. J. Immunol. 2003;171:4561–4568. doi: 10.4049/jimmunol.171.9.4561. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Ryu H, Ferrante RJ, Morris SM, Jr., Ratan RR. Translational control of inducible nitric oxide synthase expression by arginine can explain the arginine paradox. Proc. Natl. Acad. Sci. USA. 2003;100:4843–4848. doi: 10.1073/pnas.0735876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reece ST, Loddenkemper C, Askew DJ, Zedler U, Schommer-Leitner S, et al. Serine protease activity contributes to control of Mycobacterium tuberculosis in hypoxic lung granulomas in mice. J. Clin. Invest. 2010;120:3365–3376. doi: 10.1172/JCI42796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duque-Correa MA, Kuhl AA, Rodriguez PC, Zedler U, Schommer-Leitner S, et al. Macrophage arginase-1 controls bacterial growth and pathology in hypoxic tuberculosis granulomas. Proc. Natl. Acad. Sci. USA. 2014 doi: 10.1073/pnas.1408839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mori M. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J. Nutr. 2007;137:1616S–1620S. doi: 10.1093/jn/137.6.1616S. [DOI] [PubMed] [Google Scholar]
- 60.Rapovy S, Zhao J, Bricker RL, Schmidt SM, Setchell KDR, et al. Differential requirements for L-citrulline and L-arginine during anti-mycobacterial macrophage activity. J. Immunol. 2015;195:3293–3300. doi: 10.4049/jimmunol.1500800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 65.Stone TW, Stoy N, Darlington LG. An expanding range of targets for kynurenine metabolites of tryptophan. Trends Pharmacol. Sci. 2013;34:136–143. doi: 10.1016/j.tips.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol. Cell Biol. 2003;81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- 67.Thomas S, DuHadaway J, Prendergast GC, Laury-Kleintop L. Specific in situ detection of murine indoleamine 2, 3-dioxygenase. J. Cell. Biochem. 2014;115:391–396. doi: 10.1002/jcb.24674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt SK, Siepmann S, Kuhlmann K, Meyer HE, Metzger S, et al. Influence of tryptophan contained in 1-Methyl-Tryptophan on antimicrobial and immunoregulatory functions of indoleamine 2,3-dioxygenase. PLoS One. 2012;7:e44797. doi: 10.1371/journal.pone.0044797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Opitz CA, Litzenburger UM, Opitz U, Sahm F, Ochs K, et al. The indoleamine-2,3-dioxygenase (IDO) inhibitor 1-methyl-D-tryptophan upregulates IDO1 in human cancer cells. PLoS One. 2011;6:e19823. doi: 10.1371/journal.pone.0019823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Degterev A, Maki JL, Yuan J. Activity and specificity of necrostatin-1, small-molecule inhibitor of RIP1 kinase. Cell Death Differ. 2013;20:366. doi: 10.1038/cdd.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohrig UF, Majjigapu SR, Vogel P, Zoete V, Michielin O. Challenges in the Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors. J. Med. Chem. 2015 doi: 10.1021/acs.jmedchem.5b00326. In press. [DOI] [PubMed] [Google Scholar]
- 72.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, et al. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 74.Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2012;2:722–735. doi: 10.1158/2159-8290.CD-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jasperson LK, Bucher C, Panoskaltsis-Mortari A, Taylor PA, Mellor AL, et al. Indoleamine 2,3-dioxygenase is a critical regulator of acute graft-versus-host disease lethality. Blood. 2008;111:3257–3265. doi: 10.1182/blood-2007-06-096081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J. Exp. Med. 2013;210:1389–1402. doi: 10.1084/jem.20130066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat. Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 79.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klysz D, Tai X, Robert PA, Craveiro M, Cretenet G, et al. Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Science Signaling. 2015;8:ra97. doi: 10.1126/scisignal.aab2610. [DOI] [PubMed] [Google Scholar]
- 84.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeng H, Yang K, Cloer C, Neale G, Vogel P, et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 2015;36:13–20. doi: 10.1016/j.it.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Procaccini C, De Rosa V, Galgani M, Abanni L, Cali G, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weichhart T, Hengstschlager M, Linke M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broome JD. Studies on the mechanism of tumor inhibition by L-asparaginase. Effects of the enzyme on asparagine levels in the blood, normal tissues, and 6C3HED lymphomas of mice: differences in asparagine formation and utilization in asparaginase-sensitive and -resistant lymphoma cells. J. Exp. Med. 1968;127:1055–1072. doi: 10.1084/jem.127.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan WK, Lorenzi PL, Anishkin A, Purwaha P, Rogers DM, et al. The glutaminase activity of L-asparaginase is not required for anticancer activity against ASNS-negative cells. Blood. 2014;123:3596–3606. doi: 10.1182/blood-2013-10-535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang J, Fan J, Venneti S, Cross JR, Takagi T, et al. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol. Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 98.Chen SH, Yang W, Fan Y, Stocco G, Crews KR, et al. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2011;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yau T, Cheng PN, Chan P, Chan W, Chen L, et al. A phase 1 dose-escalating study of pegylated recombinant human arginase 1 (Peg-rhArg1) in patients with advanced hepatocellular carcinoma. Invest. New Drugs. 2013;31:99–107. doi: 10.1007/s10637-012-9807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Munder M. Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol. 2009;158:638–651. doi: 10.1111/j.1476-5381.2009.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sahin E, Haubenwallner S, Kuttke M, Kollmann I, Halfmann A, et al. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J. Immunol. 2014;193:1717–1727. doi: 10.4049/jimmunol.1302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qin S, Cobbold SP, Pope H, Elliott J, Kioussis D, et al. “Infectious” transplantation tolerance. Science. 1993;259:974–977. doi: 10.1126/science.8094901. [DOI] [PubMed] [Google Scholar]
- 104.Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 1994;62:3705–3711. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gaur U, Roberts SC, Dalvi RP, Corraliza I, Ullman B, et al. An effect of parasite-encoded arginase on the outcome of murine cutaneous leishmaniasis. J. Immunol. 2007;179:8446–8453. doi: 10.4049/jimmunol.179.12.8446. [DOI] [PubMed] [Google Scholar]
- 106.Price JV, Vance RE. The macrophage paradox. Immunity. 2014;41:685–693. doi: 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 107.Mussai F, Egan S, Hunter S, Webber H, Fisher J, et al. Neuroblastoma arginase activity creates an immunosuppressive microenvironment that impairs autologous and engineered immunity. Cancer Res. 2015;75:3043–3053. doi: 10.1158/0008-5472.CAN-14-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scott L, Lamb J, Smith S, Wheatley DN. Single amino acid (arginine) deprivation: rapid and selective death of cultured transformed and malignant cells. Br. J. Cancer. 2000;83:800–810. doi: 10.1054/bjoc.2000.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cama E, Colleluori DM, Emig FA, Shin H, Kim SW, et al. Human arginase II: crystal structure and physiological role in male and female sexual arousal. Biochemistry. 2003;42:8445–8451. doi: 10.1021/bi034340j. [DOI] [PubMed] [Google Scholar]
- 110.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, et al. EMBO J. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Csoka B, Selmeczy Z, Koscso B, Nemeth ZH, Pacher P, et al. Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 2012;26:376–386. doi: 10.1096/fj.11-190934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.El Kasmi KC, Pugliese SC, Riddle SR, Poth JM, Anderson AL, et al. Adventitial fibroblasts induce a distinct proinflammatory/profibrotic macrophage phenotype in pulmonary hypertension. J. Immunol. 2014;193:597–609. doi: 10.4049/jimmunol.1303048. [DOI] [PMC free article] [PubMed] [Google Scholar]