Abstract
There are large differences between the cellular environment and the conditions widely used to study RNA in vitro. SHAPE RNA structure probing in Escherichia coli cells has shown that the cellular environment stabilizes both long-range and local tertiary interactions in the adenine riboswitch aptamer domain. Synthetic crowding agents are widely used to understand the forces that stabilize RNA structure and in efforts to recapitulate the cellular environment under simplified experimental conditions. Here, we studied the structure and ligand binding ability of the adenine riboswitch in the presence of the macromolecular crowding agent, polyethylene glycol (PEG). Ethylene glycol and low molecular mass PEGs destabilized RNA structure and caused the riboswitch to sample secondary structures different from those observed in simple buffered solutions or in cells. In the presence of larger PEGs, longer range loop-loop interactions were more similar to those in cells than in buffer alone, consistent with prior work showing that larger PEGs stabilize compact RNA states. Ligand affinity was weakened by low mass PEGs, but increased with high mass PEGs, indicating that PEG cosolvents exert complex chemical and steric effects on RNA structure. Regardless of polymer size, however, nucleotide-resolution structural characteristics observed in cells were not recapitulated in PEG solutions. Our results reveal that the cellular environment is difficult to recapitulate in vitro; mimicking the cellular state will likely require a combination of crowding agents and other chemical species.
Keywords: RNA, macromolecular crowding, SHAPE, riboswitch, in-cell structure
Graphical Abstract
INTRODUCTION
RNAs function in the complex cellular environment where macromolecules reach concentrations of 300 g/L and occupy at least 30% of the total volume.1 The cellular environment also contains metabolites, small ions, and polyamines, many of which have the potential to affect RNA folding and ligand binding.2–4 Most studies of RNA structure and function have been carried out in simple buffered solutions; however, extensive recent work demonstrates that RNA stability,5–10 structure,6,8,11–18 and dynamics7,10,11,14 in simple buffered solutions can differ substantially from that formed both in the presence of macromolecular crowding agents, such as polyethylene glycol (PEG),7–11,14 and in the complex environment inside cells.5,12,13,15–18
The two broadest effects of macromolecular crowding, whether in cells or in vitro, are steric repulsion, which favors compaction of macromolecules,19,20 and nonspecific chemical interactions, which can be attractive or repulsive.21–23 Repulsive interactions reinforce steric effects, whereas attractive charge-charge interactions favor less compact species. The effects of crowding on proteins have been studied extensively and recently reviewed.24–26
Effects on RNA are now receiving substantial interest, especially studies of RNA folding and structure in the presence of crowding agents in vitro7–11,14 and in cells.12,13,15–18 In general, crowded conditions, either in vitro or in cells, tend to stabilize RNA secondary and tertiary structures but the effects of other features, especially those associated with function, are more variable. High concentrations of synthetic polymers tend to stabilize the formation of compact RNA states, accelerate folding, and promote RNA-mediated activities including catalysis. Steric repulsion is undoubtedly a major component of the influence of the cellular environment on RNA structure but is not the only one. The extent to which macromolecular crowding – as imposed by synthetic cosolvents – recapitulates the cellular environment is not well understood.
We recently used single-nucleotide resolution selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) probing to interrogate the structure of the add A riboswitch aptamer domain RNA in healthy, growing E. coli cells with 2-min time resolution.13 This RNA forms a three-helix junction stabilized by both long-range loop-loop interactions and by a network of interactions in the three-helix junction and ligand-binding pocket (Figure 1). The conformation of the free add A riboswitch aptamer domain in E. coli cells is different from, and more highly structured than, the conformation in buffer at physiological (~1 mM) Mg2+ concentrations.13 Increasing the Mg2+ concentration to high levels (30 mM) did not recapitulate the structure observed in cells. In contrast, the cellular environment had almost no effect on the structure of the ligand-bound aptamer,13 which was organized and compact both in cells and under simplified in vitro conditions, in agreement with high resolution structures.27,28 These in-cell studies provided an initial indication that it may be difficult to mimic the cellular environment using simplified conditions, especially with respect to biologically important, partially folded (in this case, unliganded) RNA structures.
Figure 1.
Secondary structure models of the ligand-free (left) and ligand-bound (right) add adenine riboswitch aptamer domain. The aptamer RNA (backbone in dark blue) was expressed and analyzed imbedded within a tRNA scaffold.13
Here, we investigate the effects of the widely used macromolecular crowding agent polyethylene glycol (PEG) and its monomer ethylene glycol (EG) on the structure of the adenine riboswitch aptamer domain in the context of a tRNA scaffold (Figure 1). We found that high molecular mass PEGs stabilize interactions involving long-range loop-loop interactions in the RNA but do not significantly affect local interactions in the three-helix junction or the ligand-binding cleft. Low molecular mass PEGs destabilized or partially denatured the RNA, consistent with attractive interactions between the cosolvent and the RNA. PEG did not induce the riboswitch RNA to adopt a structure that fully mimics the structure that predominates in the cellular environment.
EXPERIMENTAL PROCEDURES
RNA construct and in vitro folding
The adenine riboswitch aptamer domain RNA was expressed in E. coli in the context of a tRNA cassette, in which the riboswitch RNA was inserted into the tRNA anticodon stem (Figure 1). The aptamer construct was purified by anion exchange, fast performance liquid chromatography as described.13 The purified aptamer RNA construct was folded by incubation in folding buffer [50 mM HEPES (pH 8.0), 200 mM potassium acetate (pH 8.0), and 1 mM MgCl2] for 30 min at 37 °C. If included, EG (Fisher Scientific, MA) or PEG (Sigma Aldrich, MO) was then added, and incubation was continued for 20 min.
Equilibrium dissociation constants
Ligand dissociation constants were measured by the ability of the riboswitch RNA to quench 2-aminopurine (2AP) fluorescence upon adding RNA to a fixed concentration of 2AP.29 Samples (50 µL) were prepared in folding buffer with aptamer concentrations from 0 to 3 µM, containing 0 or 100 g/L EG or PEG and 40 nM 2AP. Fluorescence intensities were measured in a 40 µL cuvette at 25 °C in a Varian Cary Eclipse fluorimeter. Data were acquired from 330 nm to 450 nm with excitation at 300 nm. The emission was integrated and fit to a single-site binding equation:29 dF/F = ((1 – a) [RNA] / KD,app + [RNA]), where dF is the change in fluorescence intensity, F is the intensity in the absence of RNA, a (< 1.0) is proportional to the ratio of quantum yields of the 2AP-RNA complex and free 2AP, and KD,app is the apparent dissociation constant.
SHAPE probing
The purified aptamer was folded and incubated with EG or PEG as described above. For SHAPE analysis of the ligand-bound aptamer, 2AP (1 mM) was added after folding, and incubation was continued for 10 min. The aptamer construct (8 pmol) was then added to 1/50 volume 300 mM 1-methyl-7-nitroisatoic anhydride (1M7) in DMSO or neat DMSO. The samples were incubated at 37 °C for 3 min. RNA was precipitated with ethanol, washed three times with 70% (vol/vol) aqueous ethanol, and resuspended in 15 µL deionized H2O.30 SHAPE adducts were detected by primer extension, resolved by capillary electrophoresis.13 SHAPE reactivities were quantified using QuShape.31 SHAPE reactivities are reported as the mean of replicate experiments; per-nucleotide variation was typically within ±10%. Full datasets are provided in the Supporting Information.
RESULTS
RNA Affinity for 2-Aminopurine
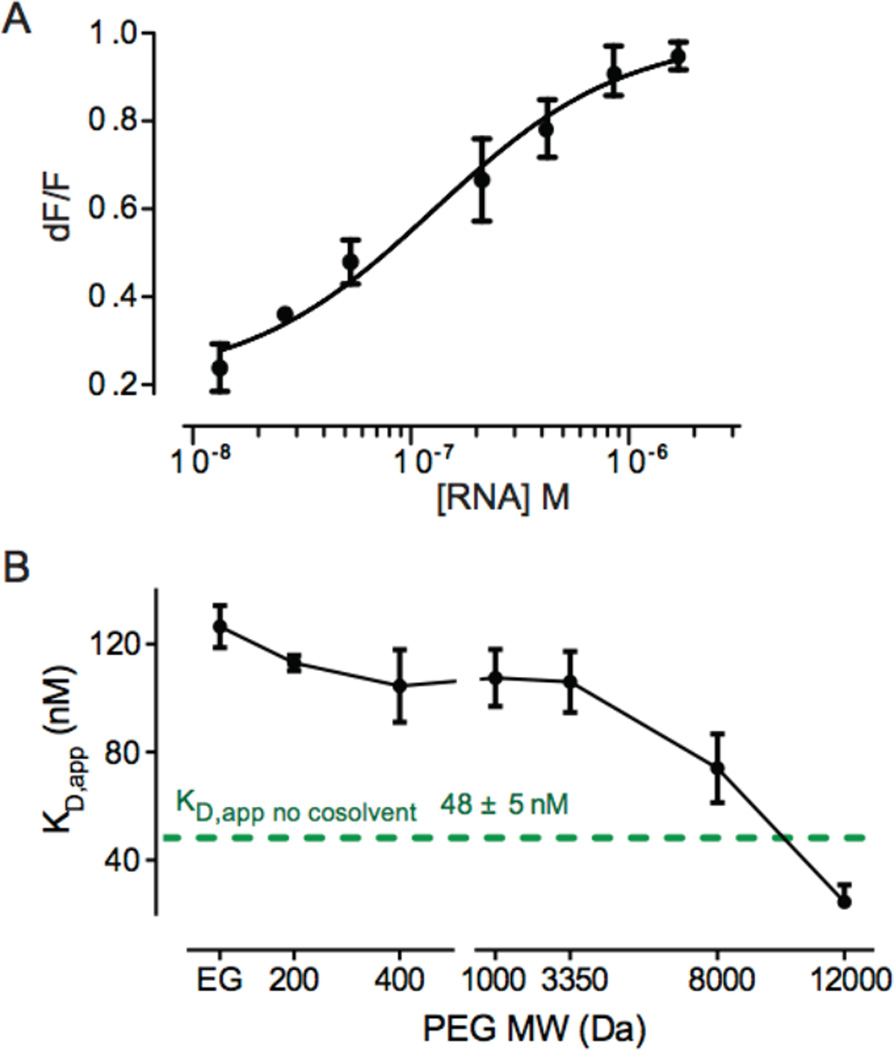
The apparent equilibrium dissociation constant, KD,app, of the complex between the aptamer and 2-aminopurine (2AP) was determined by analysis of fluorescence quenching of 2AP as a function of aptamer concentration. Experiments were performed in 200 mM potassium acetate and 1 mM MgCl2; this is the approximate Mg2+ concentration in E. coli cells (0.8 ± 0.2 mM) as directly measured previously.13 The KD,app for the aptamer-2AP complex in simple buffered solution was 48 ± 5 nM (Figure 2). This value is modestly tighter than that measured for the free riboswitch (120 nM),32 likely reflecting a stabilizing effect of the tRNA cassette.
Figure 2.
RNA-ligand affinities. (A) RNA binding to the fluorescent ligand, 2AP, in a 100 g/L solution of EG. Curve shows fit to single-site binding. (B) Aptamer RNA ligand affinity as a function of PEG molecular mass. KD,app values were determined at cosolvent concentrations of 100 g/L. The curve emphasizes the general trend, but is not of theoretical significance. The affinity at 12000 Da PEG represents an upper limit (because the KD,app is approaching the assay 2AP ligand concentration). Error bars indicate standard deviations from triplicate experiments.
We next performed binding experiments in the presence of ethylene glycol (EG, 62 Da; Figure 2A) and PEGs with mean molecular masses of 200, 400, 1000, 3350, 8000 and 12000 Da. Measurements of binding affinities as a function of crowder were carried out using 100 g/L solutions of EG or a given PEG. The apparent binding affinities showed a polymer size-dependent trend (Figure 2B). In the presence of EG or low molecular mass PEGs, the affinity of the aptamer for 2AP decreased by three-fold compared to the affinity in buffer alone. This effect was diminished with increasing PEG size such that in 12000 Da PEG the affinity was ≥2-fold higher than in buffer (Figure 2B).
These ligand-binding studies suggested that the effects of PEG on the structure of the riboswitch RNA are complex and, importantly, that PEG might destabilize RNA in a way that affects ligand binding. The inhibition of binding by low molecular mass PEGs is consistent with a chemical effect that weakens RNA-ligand interactions. In contrast, higher molecular mass PEGs restore or even modestly enhance ligand affinity relative to experiments performed in the absence of cosolvent, consistent with stabilization of more compact forms of the RNA.
Effect of PEG on Ligand-Free Riboswitch RNA
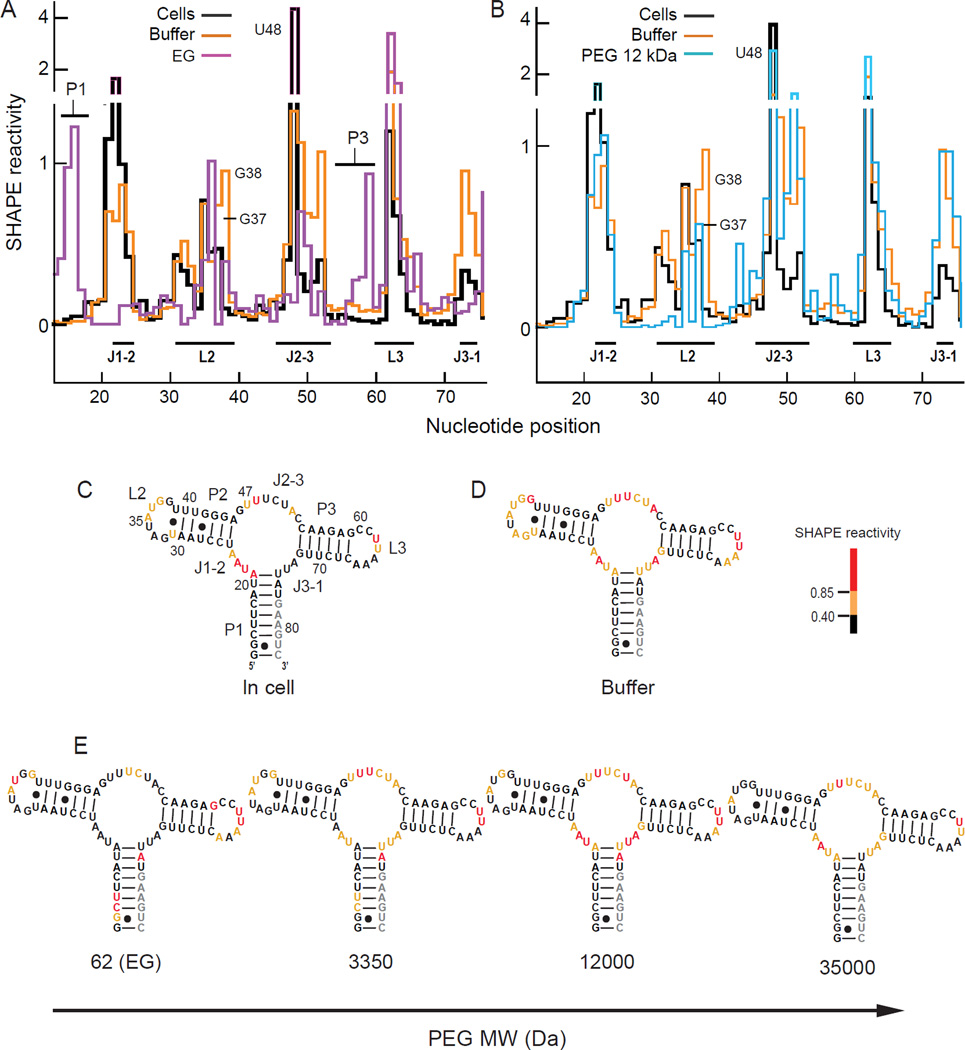
We used nucleotide-resolution SHAPE to probe the nucleotide-resolution aptamer RNA structure in the presence of low (62 Da EG), medium (3350 Da PEG), and high (12000 and 35000 Da PEG) molecular mass crowding agents in buffer containing 1 mM Mg2+. SHAPE has been extensively benchmarked against RNAs of known structure, including as a function of Mg2+ concentration and in the presence of ligands containing hydroxyl groups and amines33 in the presence of high concentrations of proteins and lipids34,35 and inside E. coli cells.13,17,18 These studies strongly support the view that the intrinsic reactivity of the 1M7 SHAPE reagent is insensitive to the solution components present during probing because any 1M7-reactive molecule would have to out-compete reaction with water (at >40 M even under the most crowded conditions) to have a substantial impact. Individual reactivity profiles were normalized using an approach that assumes that a few nucleotides in every RNA are highly dynamic and thus reactive by SHAPE.31,36 In-cell reactivities (Figures 3A, 3B, 3C) were taken from previous work, where a ligand-free form of the riboswitch was shown to predominate in cells in the absence of added 2AP.13 SHAPE reactivity profiles are shown both as histograms (Figures 3A, 3B) and superimposed in color on secondary structure models (Figures 3C–E).
Figure 3.
SHAPE reactivity profiles of the ligand-free RNA aptamer (A) in cells,13 in buffer, and in EG and (B) 12000 Da PEG. SHAPE reactivities overlaid on the secondary structure of the free aptamer (C) in cells, (D) in buffer, and (E) in buffer containing the indicated PEGs at 100 g/L. Nucleotides are colored by SHAPE reactivity (see scale); gray indicates no data. The complete data are available in supporting Tables S1 and S2.
In buffer, the RNA formed a state in which the three major helices were stably folded but in which nearly all nucleotides in the loop and joining regions showed medium and high SHAPE reactivities and were thus unstructured (Figures 3A, 3B, 3D). Addition of EG yielded a SHAPE reactivity pattern that differed strongly from that observed either in buffer alone or in cells13.(Figure 3E). In EG, nucleotides in the P1 and P3 helices were reactive, whereas some nucleotides in each of the L2, L3, J1-2 and J2-3 loops were unreactive. The reactivity of the P1 helix in the presence of EG is especially striking given that the RNA is linked to the (presumably stabilizing) tRNA scaffold. These observations suggest that EG causes the RNA to sample conformations that are quite different from the native structure.
When the aptamer structure was probed in the presence of 12000 and 35000 Da PEGs, nucleotides in each of the three primary helices were unreactive, with the exception of a few nucleotides at helix termini, indicating stable formation of these helices (Figures 3B and 3E). In addition, multiple nucleotides in the L2 and L3 loops showed low SHAPE reactivities (implying increased structure), roughly similar to the in-cell state (Figure 3). In particular, nucleotides G37 and G38, which form base pairs with C60 and C61, and the 3' AA sequence in L3, which forms an extended network of tertiary interactions with L2 in the high resolution structure,28 were either less reactive or unreactive in the presence of the high molecular mass PEGs (Figure 3E). In clear contrast however, nucleotides in the J2-3 and J3-1 joining segments remained reactive in the presence of the high molecular mass cosolvents. These reactivity patterns support the interpretation that the L2 and L3 loops do interact stably but that the J2-3 and J3-1 regions do not adopt in-cell-like structures in the presence of high mass PEGs. With 3350 Da PEG, the RNA showed features intermediate between those observed in EG (62 Da) and high mass PEGs. Nucleotides in P1 were reactive, suggesting partial destabilization, whereas the pattern of reactivity in the L2 and L3 loops resembled that of the in-cell state (Figure 3E).
To further quantify the effects of PEG, we calculated correlation coefficients (R) comparing in-cell SHAPE reactivities with those measured in vitro in the presence of PEG cosolvents (Figure 4). Correlation coefficients were calculated over the loop, joining, and terminal base-paired nucleotide positions because SHAPE reactivities at these nucleotides are specifically diagnostic of higher-order RNA folding in the adenine riboswitch RNA (Figures 1 & 3). For the ligand-free RNA, the correlation between the in-cell reactivities and reactivities in aqueous buffer was 0.6 (Figure 4A, open symbol). Upon adding EG, the correlation with in-cell reactivities was much lower at 0.1. Structural correlation between in-cell reactivities and those determined in vitro increased with PEG molecular mass. The correlation between in-cell reactivities and reactivities in 35,000 Da PEG reached 0.8 (Figure 4A, solid symbols). The lower correlation largely reflects the substantial structural differences measured for the J2-3 and J3-1 regions. Taken together with the nucleotide-resolution SHAPE reactivities (Figure 3), this analysis supports a model in which 12,000 and 35,000 Da PEGs promote net formation of native-like long-range tertiary interactions between the L2 and L3 loops. However, the high mass PEGs do not stabilize native structure in the region of the three-helix junction to the same extent. EG largely destabilizes the RNA structure, and the 3350 Da PEG induces a conformation roughly intermediate between these two extremes.
Figure 4.
Relationships between in-cell and in-vitro SHAPE reactivities for the aptamer RNA as a function of PEG molecular mass. Pearson correlation coefficients (R) for the (A) ligand-free and (B) 2AP-bound aptamer are shown. The reference state in each case is the structure probed in E. coli cells.13 Open symbols indicate correlations in the absence of added PEG cosolvent. Correlation coefficients were calculated based on reactivities of nucleotides A21-U25, U31-U39, A45-C54, G59-C67, and G72-U75.13
Effect of PEG on the Ligand-Bound Aptamer State
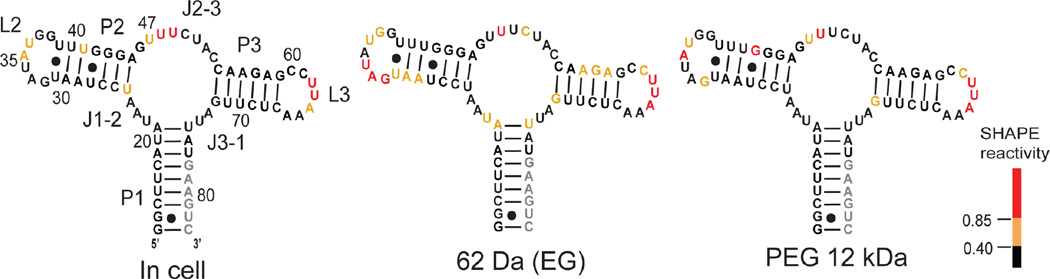
Finally, we used SHAPE probing to examine the effect of EG or PEG cosolvent in the presence of saturating concentrations of the 2AP ligand. SHAPE reactivities in the presence of 3350, 12000, and 35000 Da PEGs were essentially identical and were highly similar to reactivities in the ligand-bound states in buffer and in cells (Figures 5 and S1). The correlation coefficients relating the in-cell state with 12000 and 35000 Da PEG were 0.9 (Figure 4B). In contrast, the SHAPE profile in EG was very different from the other ligand-bound states. The correlation coefficient relating in-cell and EG SHAPE reactivities was 0.4 (Figure 4B) and nucleotide reactivities in the P1 and P2 helices and in the L2 loop were higher in EG than those observed in cells (Figure 5). These data emphasize that binding by the 2AP ligand strongly promotes formation of the fully folded state such that higher molecular mass PEGs no longer significantly affect the final (largely folded) RNA structure. However, interactions between the riboswitch RNA and EG remain strong enough to disrupt native RNA folding.
Figure 5.
SHAPE reactivities for the ligand-bound aptamer in cells,13 in buffer with 100 g/L EG, and in buffer with 100 g/L 12000 Da PEG plotted on the standard secondary structure.
DISCUSSION
The structure of the adenine riboswitch aptamer domain RNA is significantly stabilized in the crowded intracellular environment relative to its structure in simple buffered solutions,13 emphasizing that it is important to consider the contributions of the cellular environment when exploring RNA structure and function. We initiated this work with the goal of identifying in vitro solution conditions that might do a good job of mimicking the in-cell structure of this RNA. The net result of these efforts, however, has been to accomplish the opposite: to show that the cellular environment is difficult to mimic with PEG, the most commonly employed macromolecular crowding agent.7–11,14,37–39
The effects on ligand binding affinity and riboswitch RNA structure depended on the molecular mass of PEG, and were much more important for the ligand-unbound state. EG decreased ligand-binding affinity by 3-fold relative to the affinity in the absence of cosolvent (Figure 2) and disrupted both the free and ligand-bound aptamer structures (Figures 3 and 5), consistent with work showing that EG destabilizes RNA structure.6 These observations suggest that relatively strong attractive interactions between cosolvent and RNA destabilize ligand binding and folding. Similar interactions have been observed between PEG and DNA.37,38 In the presence of PEGs of increasing size, binding affinity increased to surpass the affinity in buffer alone by ≥2-fold (Figure 2). The RNA also became more highly structured and interactions between the L2 and L3 loops were at least partially formed in the presence of high molecular mass PEG (Figures 3 and 4).13 Taken together, these data suggest that steric repulsion is enhanced by increasing crowder size, as has been observed with both RNA and DNA,8,9,38 and that PEG may partially pre-organize RNA structure leading to increased ligand affinity.
However, despite these notable stabilizing effects on a subset of interactions, especially in the L2 and L3 loops, and increased overall correlations in SHAPE reactivities between the PEG and in-cell data, high concentrations of EG or any PEG did not result in formation of an RNA structure equivalent to that visualized in cells. Specifically, the J2-3, and J3-1 strands that comprise the ligand-binding pocket and form other tertiary interactions remained reactive by SHAPE, and thus relatively unstructured, even in the presence of high mass PEGs (Figure 3). These differences presumably reflect the large differences between PEG and the intracellular environment. For example, PEG forms a random coil or a random-flight chain in solution,40,41 but the cytoplasm primarily contains globular macromolecules and relatively stiff nucleic acids.24,42 In addition, although PEG interacts with nucleic acids,37,38 only a small subset of potential in-cell chemical interactions are available to the methylene, ether, and hydroxyl groups in PEG.
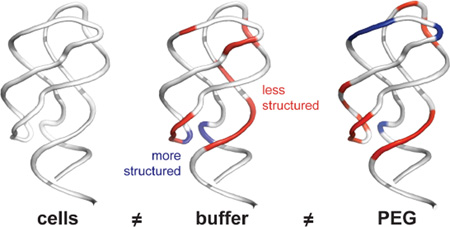
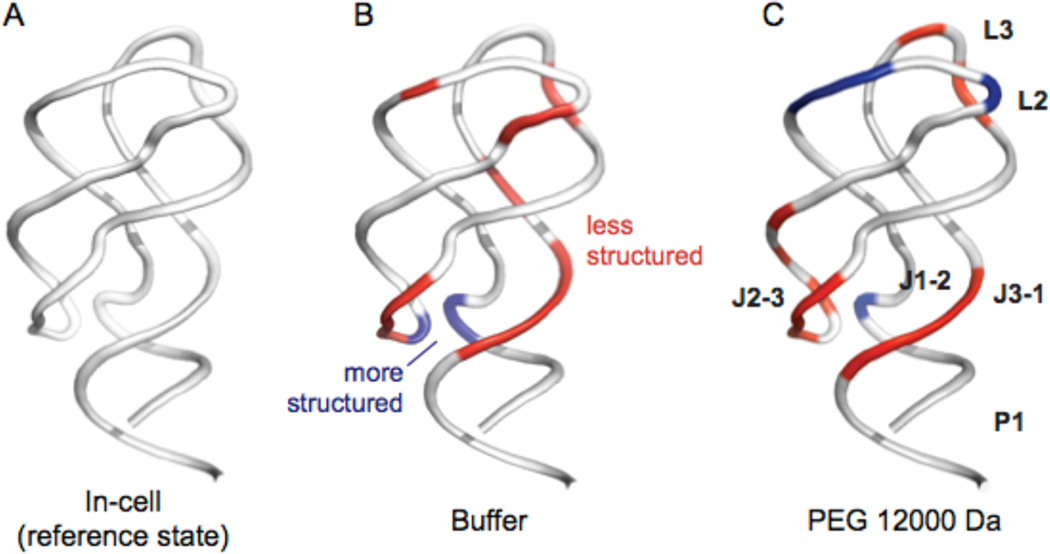
In general, the cellular environment has a strongly structuring effect on the ligand-free aptamer RNA relative to that of an aqueous buffer with roughly physiological ion concentrations (Figures 6A and 6B). In cells, the free RNA samples a state that shares some structural similarity with the ligand-bound state. High molecular mass PEGs appear to specifically stabilize the L2 and L3 loop regions. This effect is sufficiently strong that PEG resulted in a conformation in the L2 and L3 loops that was moderately less reactive by SHAPE (and thus more structured) than that in cells (Figure 6C, blue backbone). In contrast, in PEG, binding pocket regions of the ligand-free aptamer (J2-3 and J3-1) remained reactive by SHAPE (Figure 6C, red backbone). The physical basis for these differences likely reflects, first, that chemical interactions with PEG cosolvents destabilize some structures, interactions that are most clearly seen with EG and 3350 Da PEG (Figures 2–4) Second, the steric repulsive effect contributed by a random-flight PEG chain may be better at stabilizing longer-range RNA-RNA interactions, as occur between the L2 and L3 loops, than more localized interactions such as those in the RNA three-helix junction of the aptamer (Figure 6).
Figure 6.
Nucleotide-resolution effects of the intracellular environment and high molecular mass PEG cosolvent on adenine riboswitch RNA structure. SHAPE reactivity data from experiments performed in the absence of ligand are superimposed on a ribbon diagram of the ligand-bound RNA (PDB ID 1y26).28 In cells, the structure of the free RNA is similar to that of the ligand-bound RNA, especially in base paired and the L2, L3, J2-3 and J3-1 regions. Backbone regions colored red and blue indicate differences in SHAPE reactivities (≥ |0.24 SHAPE units|) that suggest less or more structure, respectively, than the in-cell state. (A) The in-cell structure reference state structure. Comparisons of the reference state with the aptamer domain RNA (B) in dilute buffer, (C) in buffer plus 12000 PEG. RNAs were studied imbedded in a tRNA scaffold.
In sum, both chemical interactions and steric repulsion appear to contribute to the effects of PEG on the aptamer RNA structure studied here. The chemical effects that destabilize RNA structure are partially overcome in the presence of high molecular mass PEGs. The high mass PEG crowders induced the aptamer RNA to adopt a more compact state that correlated better with the conformation in cells13 than did the conformation in buffer alone. Nevertheless, SHAPE reactivities for the ligand-free riboswitch RNA obtained under crowding conditions in vitro were distinct from the in-cell RNA structure (Figure 6). These results emphasize that the cellular environment – which contains diverse macromolecules, ions, and ligands – has an effect on RNA structure that is not readily recapitulated by PEG cosolvents and remains difficult to simulate in vitro.
Supplementary Material
Acknowledgments
Funding This work was supported by grants from the National Science Foundation, MCB-1051819 and MCB-1410854 (to G.J.P.) and MCB-1121024 (to K.M.W.) and a National Science Foundation Graduate Research Fellowship (DGE-0646083 to J.T.).
Footnotes
Supporting Information
A figure showing SHAPE reactivity profiles of the 2AP-bound aptamer RNA in cells, dilute buffer, and high molecular mass PEG; plus two tables providing complete per-nucleotide SHAPE data for the free and ligand-bound aptamer RNA as a function of PEG cosolvent. This material is available free of charge via the internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 2.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol. Mol. Biol. Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyamoto S, Kashiwagi K, Ito K, Watanabe S, Igarashi K. Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch. Biochem. Biophys. 1993;300:63–68. doi: 10.1006/abbi.1993.1009. [DOI] [PubMed] [Google Scholar]
- 4.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 6.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J. Mol. Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J. Amer. Chem. Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 8.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 2011;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA. 2014;20:331–347. doi: 10.1261/rna.042747.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuis NF, Holmstrom ED, Nesbitt DJ. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. SciU.SA. 2014;111:8464–8469. doi: 10.1073/pnas.1316039111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strulson CA, Yennawar NH, Rambo RP, Bevilacqua PC. Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions. Biochemistry. 2013;52:8187–8197. doi: 10.1021/bi400816s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paudel BP, Rueda D. Molecular crowding accelerates ribozyme docking and catalysis. J. Am. Chem. Soc. 2014;136:16700–16703. doi: 10.1021/ja5073146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mrna structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 17.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53:3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGinnis JL, Liu Q, Lavender CA, Devaraj A, McClory SP, Fredrick K, Weeks KM. In-cell SHAPE reveals that free 30s ribosome subunits are in the inactive state. Proc. Natl. Acad. SciU.SA. 2015;112:2425–2430. doi: 10.1073/pnas.1411514112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minton AP. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers. 1981;20:2093–2120. [Google Scholar]
- 20.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E. Quinary structure modulates protein stability in cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112 doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ. Macromolecular crowding and protein stability. J. Am. Chem. Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- 24.Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr. Opin. Struct. Biol. 2010;20:196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar MLC, Pielak GJ. Soft interactions and crowding. Biophys. Rev. 2013;5:187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AE, Zhang Z, Pielak GJ, Li C. NMR studies of protein folding and binding in cells and cell-like environments. Curr. Opin. Struct. Biol. 2015;30:7–16. doi: 10.1016/j.sbi.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Lee MK, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuSHAPE: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in trnaasp revealed by single-nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gherghe C, Lombo T, Leonard CW, Datta SAK, Bess JW, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buscaglia R, Miller MC, Dean WL, Gray RD, Lane AN, Trent JO, Chaires JB. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Research. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT., Jr Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuffee SR, Elcock AH. Diffusion, crowding, & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winzor DJ, Wills PR. Molecular crowding effects of linear polymers in protein solutions. Biophys. Chem. 2006;119:186–195. doi: 10.1016/j.bpc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Bhat R, Timasheff SN. Steric exclusion is the principal source of the preferential hydration of proteins in the presence of polyethylene glycols. Protein Sci. 1992;1:1133–1143. doi: 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savelyev A, Materese CK, Papoian GA. Is DNA’s rigidity dominated by electrostatic or nonelectrostatic interactions? J. Amer. Chem. Soc. 2011;133:19290–19293. doi: 10.1021/ja207984z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.