Abstract
Parkin is familiar to many because of its link to Parkinson’s disease, and to others because of its well-characterized role as a central factor mediating selective mitophagy of damaged mitochondria for mitochondrial quality control. The genetic connection between Parkin and Parkinson’s disease derives from clinical gene-association studies, whereas our mechanistic understanding of Parkin functioning in mitophagy is based almost entirely on work performed in cultured cells. Surprisingly, experimental evidence linking the disease and the presumed mechanism derives almost entirely from fruit flies; germline Parkin deficient mice do not develop Parkinson’s disease phenotypes. Moreover, genetic manipulation of Parkin signaling in mouse hearts does not support a central role for Parkin in homeostatic mitochondrial quality control in this mitochondria-rich and -dependent organ. Here, I provide an overview of data suggesting that (in mouse hearts at least) Parkin functions more as a stress-induced and developmentally-programmed facilitator of cardiomyocyte mitochondrial turnover.
Parkin and the disease after which it was named
Parkin is a cytosolic E3 ubiquitin ligase of the RING-between-RING family that received its name from the causal association between loss of function genetic mutations (PARK2) and hereditary juvenile onset Parkinson’s Disease. The biochemical function of Parkin is to covalently attach ubiquitin to a large number of protein substrates, thereby targeting either the individual proteins for proteasomal degradation or the entire organelle for mitophagic elimination [1–3]. Parkin’s ubiquitin ligase activity occurs largely at damaged or electrochemically depolarized mitochondria, and is dependent upon activity of the mitochondrial kinase PTEN-induced putative kinase 1 (PINK1), which is encoded by another Parkinson’s disease gene, PARK6 [4]. PINK1 modulates Parkin in several distinct ways: First, its mitochondrial abundance is regulated through proteolysis; healthy mitochondria degrade PINK1, thereby normally maintaining low levels and activity. However, upon mitochondrial depolarization or activation of the unfolded protein response PINK1 passively accumulates, enabling it to phosphorylate substrates [4, 5]. Secondly, stabilized PINK1 phosphorylates the outer mitochondrial membrane fusion protein mitofusin 2 (Mfn2) on critical serine and threonine residues, thereby converting Mfn2 from a mitochondrial tether that promotes organelle fusion into a fusion-incompetent molecule that binds Parkin [6, 7]. Thus, PINK1-mediated conversion of Mfn2 from a mitochondrial fusion protein into a mitochondrial Parkin receptor simultaneously sequesters the damaged organelle (by preventing it from fusing with and contaminating healthy mitochondria) while initiating Parkin-mediated mitophagy that will physically eliminate the damaged organelle. Third, PINK1 activity provides Parkin with a preferred co-factor for ubiquitination of mitochondrial outer membrane proteins, PINK1-phosphorylated ubiquitin [8–10]. Finally, some mitochondrial proteins are themselves transformed by tagging with PINK1-phosphorylated ubiquitin into autophagy receptors that can initiate Parkin-independent mitochondrial autophagy [11]. These actions of PINK1 constitute an “on switch” for mitophagy. Recent work has also identified a mechanism acting in opposition (i.e. as an “off switch”), deubiquitinating enzymes or “DUBs”. Several DUBs, including USP30 [12], USP15 [13], USP8 [14], and USP35 [15], remove ubiquitin chains from Parkin or its substrates and thereby eliminate the mitochondrial marker for autophagic engulfment. This is a rapidly developing area and emerging data have been from Drosophila or cultured cells; the in vivo role of DUBs in mammalian mitophagy requires examination.
Evidence linking genetic mutations of Parkin and PINK1 to hereditary juvenile Parkinson’s disease, together with insights into their functional interactions in mitophagy, suggested a pathological role for impaired Parkin-mediated mitophagy in Parkinson’s disease. Whether or not this inference completely explains the underlying pathophysiology of Parkinson’s disease is currently unclear [16]. Parkinson’s disease was named for Dr. James Parkinson who described six progressive cases in his 1817 report “An Essay on the Shaking Palsy” [17]. Dr. Parkinson’s original clinical description reported the now classic constellation of symptoms relating to motor nerve dysfunction: slow movement with abnormal gait, muscular rigidity, and resting tremor; these symptoms may be accompanied by signs of more generalized neuro-muscular involvement including depression, skeletal myopathy and autonomic dysfunction. Most Parkinson’s disease is sporadic and likely the result of combined environmental and genetic influences (especially genes involved in the electron transport chain, glucose catabolism, or biogenesis factors such as regulated peroxisome proliferator-activated receptor γ coactivator-1a [PCG-1α]) [18]. Fewer than 10% of Parkinson’s disease cases are familial (i.e. primarily genetic in etiology); mutations in Parkin and PINK1 are most commonly associated [19].
Ultrastructurally, Parkinson’s disease is characterized by apoptosis and autophagy of dopaminergic neurons located within the substantia nigra, which is important for motor neuron function [20]. It is worth considering, however, that autophagy could be either a causal pathway contributing to neuronal death or a protective compensatory mechanism [21, 22]. Furthermore, defective mitophagy has not been unequivocally linked to degeneration of motor neurons in this syndrome [16]. Thus, the current understanding of how PINK/Parkin functioning in mitophagy relates mechanistically to the clinical pathophysiology of Parkinson’s disease is unclear and likely to be revised with additional in vivo data.
“Parkin lessons” – Learning how Parkin works
The functional relationship between PINK1, Parkin and mitophagy as introduced above is seemingly straightforward. This is an illusion; as more mechanistic details have been elucidated this mitophagy pathway has revealed itself to have unanticipated complexity. In considering the consequences of such complexity it is important to keep in mind that the PINK1-Parkin pathway is not the only mitophagy mechanism, and that much of the work attributing importance of this pathway in mitochondrial quality control was performed in cultured cells poisoned by application of potent mitochondrial uncoupling agents. There is nothing inherently wrong in this approach, but the mitophagic response to pharmacological mitochondrial depolarization in vitro does not recapitulate homeostatic or physiological mitochondrial quality control mechanisms. Moreover, mitochondrial density, network integrity, turnover, and rate of replication vary in cells from different organs [23], which again is not reflected in cultured cells. Thus, mitophagic mechanisms observed in vitro do not necessarily translate in vivo, and what is observed in vivo almost certainly differs qualitatively and quantitatively between organs, cell types and pathophysiological circumstance. Caution is advised before extrapolating findings from one context to another in the absence of directly relevant experimental data.
An anecdotal example of how new data are disrupting conventional wisdom (and to clarify some misperceptions) is some controversial aspects of our work defining the PINK1-Mfn2-Parkin relationship. The title of our manuscript first describing how Mfn2 phosphorylated through the actions of PINK1 can bind Parkin at mitochondria is “PINK1-phosphorylated Mitofusin 2 is a Parkin receptor for culling damaged mitochondria” [6]. This wording (“a” receptor, not “the” receptor) was carefully considered to account for inferential evidence supporting the existence of other, functionally redundant Parkin receptors. We specifically noted this evidence, consisting of Parkin translocation to depolarized mitochondria of cells lacking Mfn1 and Mfn2 [24], in the discussion, and have since reproduced the finding in Mfn2 null fibroblasts [7]. Nevertheless, this same observation has been cited as evidence that Mfn2 is not a Parkin receptor. Indeed, one manuscript advances only this argument to support their dogmatic claim (as expressed in the manuscript title) that “Phosphorylated ubiquitin chain is the genuine Parkin receptor” (underline added). Whether or not this was just a tactic to enhance the perceived impact of the work, the reasoning is flawed. Just as preservation of mitophagy in Parkin null mice [25, 26] does not rule out a genuine role for Parkin in mitophagy, and preserved mitochondrial fragmentation during apoptosis in Drp1 null cells [27] does not exclude a role for Drp1 in mitochondrial fission, Parkin translocation to mitochondria in cells lacking Mfn2 does not prove that PINK1-phosphorylated Mfn2 is not a Parkin receptor. Rather, each of these findings reveals the presence of one or more redundant compensatory mechanisms. Indeed, independent observations that mitochondrial Parkin localization is defective after in vivo cell type-specific Mfn2 ablation in cardiomyocytes [6] and neurons [28] suggest a broad role for the Mfn2-Parkin interaction. Moreover, recently published results that avoid opportunistic compensation so commonly provoked by gene ablation [29] demonstrate functional preeminence of the PINK1-Mfn2-Parkin pathway in both cultured fibroblasts and in vivo mouse hearts [7]. The accumulated data unequivocally demonstrate that PINK1 phosphorylation of Mfn2, which simultaneously permits mitochondrial Mfn2 to recruit Parkin and functionally sequesters the organelle by abrogating Mfn2-mediated fusion [7], is an important proximal event in mitophagy. It seems likely that PINK1 phosphorylation of mitochondrial ubiquitin on S65 activates Parkin, amplifies its activity and helps to retain it on mitochondria [30]. Binding of Parkin to S65 phosphorylated ubiquitin chains may even be the principal mechanism of Parkin recruitment to peroxisomes or lysosomes, which lack Mfn2 [31]. The in vivo relevance of this alternate Parkin binding mechanism requires formal testing.
Another source of confusion is the relative role of Mfn2 versus Mfn1 in mitophagy. Although there are differences in GTPase activities, these two proteins are highly similar (63% identity and 80% conserved amino acid sequence), have overlapping functions and mutually interact to promote mitochondrial fusion [32]. However, Mfn2 has at least two functions that do not appear to be shared by Mfn1, Parkin binding (vide supra), and tethering of mitochondria to endo/sarcoplasmic reticulum [33, 34]. The reason why Mfn1 cannot function as an ER-mitochondrial tether is clear; it does not localize to ER. On the other hand, an explanation why Mfn1 does not normally function as a Parkin receptor is unclear. Perhaps it is not phosphorylated by PINK1, or is phosphorylated on the wrong residues. The analogous amino acid to PINK1-phosphorylated Mfn2 T111 is Mfn1 S90, which is a conservative substitution; Mfn2 S442 is retained in Mfn1 as S442. Alternately, structural differences between Mfn2 and Mfn1 may prevent PINK1-phosphorylation of Mfn1 from conferring Parkin binding activity. These possibilities should be testable by determining the effects of mutational mimicry of PINK1 phosphorylation by glutamate (E) substitution for Mfn1 S90 and S442, which for Mfn2 T111 and S442; the analogous substitutions in Mfn2 (T111E and S442E) converted it into a constitutively active Parkin receptor [6, 7].
“No Parkin” – Parkin ablation in flies and mice
Despite the potentially confounding effects of opportunistic compensation from functionally overlapping pathways, germ line or conditional gene ablation have long been considered to be essential approaches for uncovering relevant in vivo functions of specific gene products [35]. The species in which gene manipulation has the longest history is Drosophila melanogaster, the fruit fly. Indeed, studies of Parkin and PINK1 mutant (knockout) flies have been central to our current understanding of mitochondrial PINK1-Parkin signaling. Mutational ablation of Drosophila Parkin provoked striking phenotypes somewhat reminiscent of aspects of Parkinson’s disease: locomoter defects and skeletal myopathy associated with mitochondrial dysmorphology [36]. PINK1 mutational ablation in flies evoked similar changes, and combined mutation of both Parkin and PINK1 genes induced the same overall phenotype [37, 38]. Absence of any additive effects of combinatorial gene deletion suggested that PINK1 and Parkin interacted functionally in a linear pathway, with mitochondrial abnormalities pointing to impaired maintenance of mitochondrial health. Strikingly, transgenic overexpression of PINK1 did not rescue the phenotype provoked by Parkin ablation, whereas transgenic expression of Parkin fully rescued PINK1 null flies [37–39]. These results and others [40] have been widely interpreted as showing that PINK1-Parkin mediated mitophagy is central to degenerative phenotypes in flies, and this notion was extrapolated to mammals and to the clinical human syndrome. However, Parkin does not rescue the mitophagy defect induced by loss of PINK1 in most mammalian systems [4, 7] (likely because PINK1 phosphorylation of ubiquitin is central to Parkin activation [8–10]). For this reason, the underlying cellular defect that is completely rescued by Parkin overexpression in PINK1 deficient flies is uncertain, as is the underlying pathophysiology induced by Parkin and PINK1 mutations in Parkinson’s disease [16].
Further complicating interpretations of Drosophila experimentation [41], the orthologous germ-line gene ablation studies in mice produced minimal phenotypes [42]. Systemic Parkin or PINK1 gene ablation in mice did not recapitulate either the neuromuscular phenotypes seen in flies or the seminal features of clinical Parkinson’s disease. These discrepancies may derive from different functionality of PINK1-Parkin signaling in flies and mammals, or may be the consequence of mammals having more sophisticated alternative mitochondrial quality control mechanisms and/or compensatory strategies [43]. Inferential evidence for opportunistic compensation is provided by the observation that hundreds of cardiac-expressed mRNAs are altered in germ-line Parkin knockout mice [44] (GEO accession # GSE74517), whereas there is virtually no genetic counter-regulation when Parkin is ablated only in cardiomyocytes of adult mouse hearts [29] (GEO accession # GSE74518). Together the data indicate that, as in other signaling pathways exhibiting functional redundancy [45], developmental plasticity in gene expression provoked by absence of Parkin in the early embryo can be avoided by deleting Parkin in the fully developed (and much less transcriptionally malleable) adult. Moreover, extra-cardiac effects can be avoided by cardiomyocyte-specific conditional ablation.
It is notable that heart development and functioning appears normal in germ-line Parkin-deficient mice [46] and after cardiomyocyte-specific Parkin deletion in adult hearts [29], whereas conditional cardiomyocyte-specific Parkin deletion in the perinatal period provoked an aggressive and ultimately lethal cardiomyopathy [7]. Finally, germ-line Parkin-deficient mice do not develop Parkinson’s disease-like phenotypes, but these phenotypes are provoked by conditional deletion of Parkin in nigrostriatal dopaminergic neurons of fully grown mice (using stereotaxically injected viral Cre in combination with floxed Parkin alleles) [47, 48]. Thus, multiple examples of tissue-specific gene deletion have revealed Parkin functionality that appears to be suppressed by opportunistic induction of compensatory pathways in germ-line Parkin knockout mice. In the next section I more closely examine the consequences of germ-line versus cardiomyocyte-specific Parkin ablation in mice.
“Where’s the Parkin?” - Parkin in the heart
The initial in vivo full phenotypic characterization of Parkin-deficient hearts was performed by Gustafsson’s group using germ-line Parkin knockout mice. Just as Parkin null mice did not develop expected neurological phenotypes [42], these mice did not exhibit cardiac dysfunction or structural abnormalities at baseline [46]; cardiomyocyte mitochondria of these Parkin null mice were, however, smaller and less organized than normal and developed abnormal electron dense inclusions over time [49]. Studies of cardiomyocyte-specific Parkin deficient (RNAi) Drosophila heart tubes indicated that Parkin is essential to maintaining normal mitochondrial morphology and inner membrane polarization, and to preventing abnormal reactive oxygen species (ROS) production [44]. Thus, either Parkin-independent mitophagy mechanisms induced by germ-line Parkin deficiency in the mouse can compensate for absent Parkin in the heart (and elsewhere), or Parkin is largely dispensable for constitutive housekeeping mitochondrial quality control in the heart, or both.
Given the abundance of mitochondria in mammalian myocardium (30–40% of wet heart weight), and the obvious need to prevent cardiotoxicity from damaged or senescent mitochondria [50], it is remarkable that normal adult mouse hearts contain so little Parkin protein [29]. In our initial mouse heart Parkin immunoblot studies it was necessary to immunoprecipitate Parkin in order to reliably measure it (see Figure 2b of [6]). The observation that Parkin is expressed at very low levels in normal hearts was initially viewed as somewhat controversial, in part because early studies did not emphasize observed stress-mediated Parkin upregulation. Moreover, statements to the effect that Parkin is “abundant” in hearts were generalized rather than applied to the particular contexts of research studies. Our group examined Parkin mRNA and protein levels in normal mouse hearts using RNA sequencing, RT-qPCR, and immunoblotting with multiple available antibodies. We found that Parkin transcripts and protein are an order of magnitude less abundant in normal mouse myocardium than other mitophagy factors, including PINK1 and Mfn2 [29, 51]. Simple mass-action relationships warrant the consideration that scarce Parkin has a correspondingly small physiological role, at least in normal hearts. Absence of functionally meaningful consequences on mouse hearts after genetic deletion of Parkin in either the mouse germ-line [46] or specifically in adult mouse cardiomyocytes [29] is also explained if Parkin is already “mostly not there”.
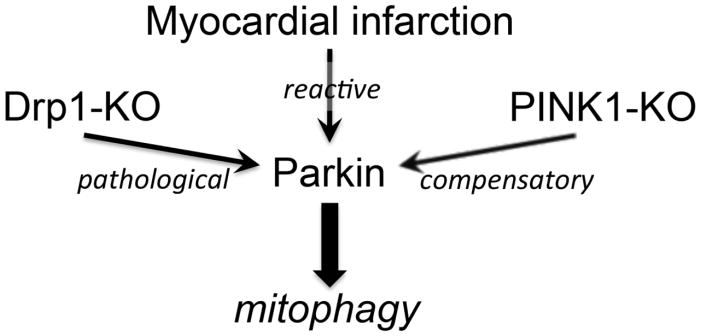
Parkin is scarce in normal adult hearts, but its abundance is increased by different forms of cardiac stress (Figure 1). Parkin protein is upregulated within 8 hours, and is sustained for at least 48 hours, in the border zone of infarcted mouse hearts [46]. Parkin mRNA and protein are likewise concordantly and chronically increased after interrupting cardiomyocyte mitochondrial fission through conditional cardiac-specific genetic inactivation of the fission factor, dynamin-related protein 1 (Drp1) [29]. Furthermore, myocardial Parkin protein is markedly increased in PINK1-deficient hearts, compared to normal controls [52]. Together, these observations suggest that Parkin is upregulated in response to mitochondrial stress, with or without apparent cardiac dysfunction. The remarkable degree to which Parkin protein is increased in Drp1-deficient and PINK1-deficient hearts compared to normal controls, and the reversal of Parkin upregulation by concomitant Parkin gene ablation, was difficult to appreciate in some figures from the original publications, but is self-evident on immunoblots containing equal amounts of cardiac protein from all groups, when viewed in their entirety [51]. Stress-related Parkin upregulation has been functionally linked to pathological mitophagy through reversal (by cardiomyocyte-specific Parkin ablation) of hyper-mitophagy and the cardiomyopathy evoked by cardiomyocyte-specific Drp1 ablation [29]. Likewise, germ-line Parkin ablation interrupted the compensatory mitophagy that normally clears damaged mitochondria from peri-infarct border zones after cardiac ischemia [46].
Figure 1. Conditions under which Parkin is upregulated in mouse hearts and its consequences on mitophagy.
Finally, in contrast to normal homeostasis, any alternate mitochondrial quality control mechanisms that are invoked by Parkin ablation are clearly inadequate to compensate for interrupted mitophagy under conditions of increased mitochondrial stress. Three cardiac conditions have been described in which myocardial Parkin is reactively increased, and either germ-line or cardiac-specific Parkin deletion reverses stress-mediated increases in mitophagy: after myocardial infarction [46], when mitochondrial fission is rendered defective [29, 53], and with PINK1 deficiency [52]. Together, these data support a central, but conditional, role for Parkin in the inducible removal of cardiomyocyte mitochondria under conditions of mitochondrial stress. By inference, Parkin-independent mechanisms may play the major role in maintaining homeostatic mitochondrial quality control in normal adult hearts.
“Parallel Parkin” – Parkin-independent mitophagy
The PINK1-Parkin mechanism is the best characterized pathway for mitophagic mitochondrial quality control, but direct and indirect evidence support the existence of alternate mechanisms. As described above, the absence of strong cardiac phenotypes or detectable mitochondrial dysfunction in un-stressed hearts of germ-line or cardiac-specific Parkin-deficient adult hearts is a compelling argument favoring parallel pathways sufficient to preserve the general fitness of cardiomyocyte mitochondria under normal conditions. Additional evidence revealing Parkin-independent mitophagy in cardiac myocytes accrued from studies of mouse hearts deficient in PINK1 and Mfn2, the two upstream factors in PINK1-Mfn2-Parkin mediated mitophagy.
As introduced above, PINK1 is essential to Parkin-dependent mitophagy because it phosphorylates Mfn2 that recruits Parkin to damaged mitochondria (Parkin translocation) [6], and it phosphorylates ubiquitin that becomes the preferred co-factor for Parkin-mediated mitochondrial outer membrane protein ubiquitination (Parkin activation) [8–10]. Thus, deletion of PINK1 interrupts Parkin-mediated mitophagy at two distinct steps. Accordingly, Parkin translocation to pharmacologically-depolarized mitochondria is virtually absent in cultured MEFs derived from PINK1 null mice, and in cultured M17 neuroblastoma cells in which PINK1 was suppressed using a specific shRNA [4]. Surprisingly, Parkin was still capable of localizing to cardiomyocyte mitochondria of PINK1 null mice, although mitophagy appeared blunted [52]. These findings are consistent with the existence of one or more PINK1-independent pathways for mitochondrial Parkin recruitment and activation.
Findings in Mfn2-deficient mouse hearts also suggested the presence of Parkin-independent mechanisms for mitophagy. Because PINK1-phosphorylated Mfn2 (but not Mfn1) is important for mitochondrial localization of Parkin, and Mfn1 redundantly promotes mitochondrial fusion with Mfn2, the major functional impact of cardiomyocyte deficiency of Mfn2 was impaired mitophagy, rather than inhibition of mitochondrial fusion [6]. Mfn2-deficient mice exhibit defective mitochondrial Parkin localization and develop a slowly progressing cardiomyopathy associated with mitochondrial enlargement, an increased burden of double stranded mitochondrial DNA breaks, mitochondrial hypopolarization, and increased mitochondrial-derived ROS [6, 34, 54, 55]. In examining the potential for cytotoxic effects of mitochondrial-derived ROS to cause the cardiomyopathy that develops in these mitophagically impaired hearts, we used a transgenically-expressed mitochondrial-targeted catalase to suppress cardiomyocyte ROS levels; this approach had previously improved other cardiomyopathies linked to mitochondrial dysfunction [56]. As expected, normalization of mitochondrial ROS with mitochondrial-directed catalase expressed at modest levels improved mitochondrial polarization and largely corrected mitochondrial respiratory dysfunction and dysmorphology in these mitophagically-impaired hearts [55]. However, ROS suppression to well below normal basal levels (achieved by expressing higher levels of mitochondrial-directed catalase) did not improve either mitochondrial or cardiac defects provoked by Mfn2 ablation [55]. We interpreted partial phenotypic rescue by ROS normalization, but not ROS supersuppression, as failure of secondary homeostatic mitochondrial autophagy mechanisms that are ROS-stimulated and Mfn2-Parkin independent.
Whereas descriptive studies such as those described above can infer the presence of PINK1-Parkin independent pathways leading to mitophagy, they do not reveal molecular details. Given the importance of mitochondrial quality control to the heart it is likely that multiple parallel and redundant pathways leading to mitophagy exist, some of which involve PINK1 and Parkin but not Mfn2, some of which involve Parkin but not PINK1 [52], and some of which involve PINK1, but not Mfn2 or Parkin. Indeed, mechanistic details of an example of the latter category were recently described: PINK1-mediated phosphorylation of ubiquitin on two mitochondrial outer membrane proteins converts them into LC3-binding receptors that can directly recruit autophagosomes independent of Parkin [11]. Moreover, compelling evidence supports an important role for pro-apoptotic Nix and Bnip3, which can also act as mitochondrial adaptor proteins for p62 and autophagosomal LC3 [57], in homeostatic cardiomyocyte mitochondrial quality control [52, 58].
“Parkin saves fuel” – Metabolic consequences of Parkin deficiency in perinatal hearts
As introduced above, conditional cardiomyocyte-specific ablation of Parkin in adult mouse hearts had no measurable consequences on cardiac function [29], whereas ablation of Parkin in the perinatal period using the same cardiomyocyte-specific system was almost uniformly lethal [7]. Intriguingly, elimination of Parkin from neonatal hearts was associated with abonormal retention of mitochondria having the distinct morphological characteristics of fetal cardiomyocyte mitochondria beyond the time (3 weeks of age; weaning) when they would normally have been replaced with adult mitochondria [7]. Given that a major known function of Parkin in diseased or metabolically stressed hearts is to eliminate damaged mitochondria (vide supra) we hypothesized that Parkin also played a role in the normal perinatal metabolic transformation of the heart.
The primary biological imperative of fetal hearts is to grow; cardiac contraction is less vigorous and bioenergetically demanding in the fetal heart. Fatty acids and amino acids that are the preferred mitochondrial substrates for ATP production in adult hearts [59] can therefore be reserved in the fetus for building cell phospholipid membranes and proteins. Consequently, in fetal hearts ATP is largely generated by metabolizing carbohydrates. Between birth and weaning, when cardiomyocyte proliferation ceases and the primary nutrient source is fat-rich milk, cardiac metabolism transitions to a preference for fatty acids [60]. A characteristic change in cardiac gene expression during this transitional period (orchestrated in part by the transcriptional co-regulators peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) α and β [61]) contributed to the notion that carbohydrate-utilizing fetal mitochondria are “re-programmed” into fatty acid-metabolizing adult organelles. However, interruption of normal mitochondrial maturation when Parkin was ablated suggested a central role for Parkin in mitochondrial removal and replacement. To address this apparent quandry we specifically interrupted Parkin translocation to cardiomyocyte mitochondria by conditionally expressing in cardiomyocytes an engineered Mfn2 mutant lacking the two PINK1 phorphorylation sites essential for its binding of Parkin, threonine 111 and serine 442 (which we mutated to non-phosphorylatable alanines). This Parkin binding defective mutant was designated Mfn2 AA. Mfn2 AA suppressed Parkin-mediated mitophagy in vitro and in vivo without affecting the abundance of PINK1 or Parkin [7]. Mimicking the effects previously observed with adult cardiac-specific Parkin gene deletion, expression of Mfn2 AA in adult hearts had no adverse effects. By contrast, its expression in the perinatal period provoked a lethal cardiomyopathy characterized by cardiomyocyte retention of fetal mitochondria. Strikingly, perinatal interruption of Parkin-mediated mitophagy in cardiomyocytes (with Mfn2AA) also suppressed expression of PGC-1α-associated fatty acid metabolism genes, interrupted myocardial fatty acid metabolism, and produced adult cardiomyocytes with a greatly impaired ability to utilize the fatty acid intermediate palmitoyl carnitine as a substrate for ATP synthesis [7]. These findings revealed an unexpected crucial role for Parkin in the normal developmental replacement of fetal by adult cardiomyocyte mitochondria. This cell-wide developmental functioning of Parkin is physiologically distinct from, but employs the same biochemical mechanisms as, canonical PINK1-Mfn2-Parkin mediated targeting of individual damaged mitochondria in diseased hearts. Because normal mitochondrial gene expression (i.e. mitochondrial biogenesis) and normal maturation of cardiac metabolism required Parkin-mediated mitophagy we conclude that accelerated removal of fetal mitochondria is essential to their ultimate replacement by biogenically-derived mature organelles. In short, mitochondrial maturation is not simply a matter of “re-programming” [62]. Taken together with previously reviewed evidence that Parkin is upregulated in diseased or metabolically stressed hearts, these data suggest the possibility that Parkin-mediated generalized mitochondrial turnover can contribute to pathological reverse metabolic remodeling [59]; this intriguing possibility requires formal testing.
Summary
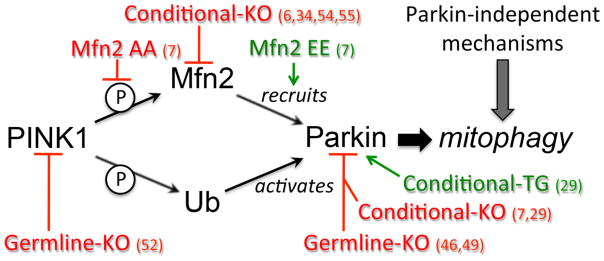
It may be time to take a fresh look at Parkin functionality in the heart and elsewhere, carefully considering that fibroblasts do not faithfully recapitulate many aspects of more developmentally specified cell types, and that cultured cells are imperfect models of in vivo organ functionality. As represented in Figure 2, in vivo modeling using different approaches to modulating PINK1-Mfn2-Parkin signaling is uncovering new and unanticipated roles for Parkin in normal maturational transformation of perinatal cardiac metabolism.
Figure 2. Schematic representation of the diversity of approaches utilized to perturb in vivo PINK1-Mfn2-Parkin signaling in mouse hearts.
References for the different models are provided. Red is loss-of-function and green is gain-of-function. Ub is ubiquitin; encircled P is phosphorylation.
Highlights.
Parkin appears not to be the primary mechanism for homeostatic mitophagy in hearts
Parkin-mediated mitophagy is a stress-response in hearts
The PINK1-Mfn2-Parkin axis is central to perinatal cardiac metabolic remodeling
Parkin may have broad roles in generalized mitochondrial turnover
Acknowledgments
GWD is supported by NIH/NHLBI R01 HL108943, HL87871, HL059888, and HL128071.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 2.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–6. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–7. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–5. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong G, Song M, Kelly DP, Matkovich SJ, Dorn GW., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 9.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–4. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–5. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 13.Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–42. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014;33:2473–91. doi: 10.15252/embj.201489729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Serricchio M, Jauregui M, Shanbhag R, Stoltz T, Di Paolo CT, et al. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–24. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lees AJ. Unresolved issues relating to the shaking palsy on the celebration of James Parkinson’s 250th birthday. Mov Disord. 2007;22(Suppl 17):S327–34. doi: 10.1002/mds.21684. [DOI] [PubMed] [Google Scholar]
- 18.Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 21.Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108–17. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329–44. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, et al. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics. 2012;11:1586–94. doi: 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–37. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams JA, Ni HM, Haynes A, Manley S, Li Y, Jaeschke H, et al. Chronic Deletion and Acute Knockdown of Parkin Have Differential Responses to Acetaminophen-induced Mitophagy and Liver Injury in Mice. J Biol Chem. 2015;290:10934–46. doi: 10.1074/jbc.M114.602284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–66. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Sterky FH, Mourier A, Terzioglu M, Cullheim S, Olson L, et al. Mitofusin 2 is necessary for striatal axonal projections of midbrain dopamine neurons. Hum Mol Genet. 2012;21:4827–35. doi: 10.1093/hmg/dds352. [DOI] [PubMed] [Google Scholar]
- 29.Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ Res. 2015;117:346–51. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A. 2015;112:6637–42. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–33. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 33.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–10. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circ Res. 2012;111:863–75. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker KD, Gottshall KR, Chien KR. Strategies for studying cardiovascular phenotypes in genetically manipulated mice. Hypertension. 1996;27:495–501. doi: 10.1161/01.hyp.27.3.495. [DOI] [PubMed] [Google Scholar]
- 36.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 38.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–8. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–5. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo M. Drosophila as a model to study mitochondrial dysfunction in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2(11):009944. doi: 10.1101/cshperspect.a009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee Y, Dawson VL, Dawson TM. Animal models of Parkinson’s disease: vertebrate genetics. Cold Spring Harb Perspect Med. 2012;2(10):009324. doi: 10.1101/cshperspect.a009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochim Biophys Acta. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW., 2nd Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circ Res. 2014;114:257–65. doi: 10.1161/CIRCRESAHA.114.302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song M, Matkovich SJ, Zhang Y, Hammer DJ, Dorn GW., 2nd Combined cardiomyocyte PKCdelta and PKCepsilon gene deletion uncovers their central role in restraining developmental and reactive heart growth. Sci Signal. 2015;8:ra39. doi: 10.1126/scisignal.aaa1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–26. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens DA, Lee Y, Kang HC, Lee BD, Lee YI, Bower A, et al. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc Natl Acad Sci U S A. 2015;112:11696–701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubli DA, Quinsay MN, Gustafsson AB. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. Commun Integr Biol. 2013;6:e24511. doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorn GW., 2nd Mitochondrial dynamics in heart disease. Biochim Biophys Acta. 2013;1833:233–41. doi: 10.1016/j.bbamcr.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorn GW., 2nd Parkin-dependent mitophagy in the heart. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.023. S0022-2828(15)30127-9 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson AB. PINK1 Is Dispensable for Mitochondrial Recruitment of Parkin and Activation of Mitophagy in Cardiac Myocytes. PLoS One. 2015;10:e0130707. doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song M, Mihara K, Chen Y, Scorrano L, Dorn GW., 2nd Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015;21:273–85. doi: 10.1016/j.cmet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Sparks M, Bhandari P, Matkovich SJ, Dorn GW., 2nd Mitochondrial genome linearization is a causative factor for cardiomyopathy in mice and Drosophila. Antioxid Redox Signal. 2014;21:1949–59. doi: 10.1089/ars.2013.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song M, Chen Y, Gong G, Murphy E, Rabinovitch PS, Dorn GW., 2nd Super-suppression of mitochondrial reactive oxygen species signaling impairs compensatory autophagy in primary mitophagic cardiomyopathy. Circ Res. 2014;115:348–53. doi: 10.1161/CIRCRESAHA.115.304384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai DF, Johnson SC, Villarin JJ, Chin MT, Nieves-Cintron M, Chen T, et al. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ Res. 2011;108:837–46. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dorn GW., 2nd Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–83. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 60.Bartelds B, Knoester H, Smid GB, Takens J, Visser GH, Penninga L, et al. Perinatal changes in myocardial metabolism in lambs. Circulation. 2000;102:926–31. doi: 10.1161/01.cir.102.8.926. [DOI] [PubMed] [Google Scholar]
- 61.Dorn GW, 2nd, Vega RB, Kelly DP. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 2015;29:1981–91. doi: 10.1101/gad.269894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191–8. doi: 10.1111/j.1749-6632.2009.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]