Abstract
Heroin addiction is a disease of chronic relapse that harms the individual through devaluation of personal responsibilities in favor of finding and using drugs. Only some recreational heroin users devolve into addiction but the basis of these individual differences is not known. We have shown in rats that avoidance of a heroin-paired taste cue reliably identifies individual animals with greater addiction-like behavior for heroin. Here rats received 5 min access to a 0.15% saccharin solution followed by the opportunity to self-administer either saline or heroin for 6 hours. Large Suppressors of the heroin-paired taste cue displayed increased drug escalation, motivation for drug, and drug loading behavior compared with Small Suppressors. Little is known about the molecular mechanisms of these individual differences in addiction-like behavior. We examined the individual differences in mRNA expression in the nucleus accumbens (NAc) of rats that were behaviorally stratified by addiction-like behavior using next-generation sequencing. We hypothesized that based on the avoidance of the drug-paired cue there will be a unique mRNA profile in the NAc. Analysis of strand-specific whole genome RNA-Seq data revealed a number of genes differentially regulated in NAc based on the suppression of the natural saccharine reward. Large Suppressors exhibited a unique mRNA prolife compared to Saline controls and Small Suppressors. Genes related to immunity, neuronal activity, and behavior were differentially expressed among the 3 groups. In total, individual differences in avoidance of a heroin-paired taste cue are associated with addiction-like behavior along with differential NAc gene expression.
Keywords: Heroin, Reward, Transcription, RNA-Seq
1. Introduction
Heroin addiction is a brain disease of chronic relapse that harms the individual, at least in part, through devaluation of personal duties and natural rewards in favor of finding and using drugs. Despite the intensely rewarding properties of heroin, only a fraction of heroin users transition from recreational use to addiction. In the case of heroin, approximately 50% of individuals who engage in heroin use will devolve into substance dependence [42], a phenomenon also evident in animal model [12, 17]. Given this range of responses, also termed individual differences, understanding why certain individuals develop heroin dependence and others do not would provide specific insight into the addictive process and potential treatments.
One method to identify individual differences in addiction-like behavior is reward comparison between a natural reward and the drug of abuse. Suppression of a natural reward in favor of the drug of abuse is a surrogate of devaluation of normal life activities in favor of drug abuse. Reward comparisons can be performed in humans and animal models and in animal models providing a useful tool for understanding individual differences. For example, rats suppress intake of a palatable saccharin solution when it predicts cocaine access and greater suppression of intake of the saccharin taste cue is associated with greater cocaine seeking and taking [10]. Recently, we have shown that avoidance of a heroin-paired taste cue also reliably identifies individual differences in addiction-like behavior for heroin [12]. Adult male Sprague-Dawley rats received 5 min access to a 0.15% saccharin solution followed by the opportunity to self-administer either saline or heroin for 6 hours. Large Suppressors of the heroin-predictive taste cue displayed increased drug escalation, drug-loading behavior, and relapse-like behaviors compared with Small Suppressors. However, little is known about the molecular mechanisms of individual differences in addiction-like behavior for heroin. Exploring the molecular differences and neuroadaptations that underlie these divergent behavioral phenotypes can help to elucidate the etiology of drug addiction. Understanding the development of heroin addiction is of great interest, particularly given ongoing epidemic of heroin use in the United States partially due to addicts transitioning from prescription opioids given heroin's lower cost and ease of access [6].
One of the primary brain regions of interest in the study of drug addiction has been the nucleus accumbens (NAc) given its role in reward [11] and its sensitivity to drugs of abuse [21] and neuroadaptations with drug abuse [28]. Our laboratory has reported that avoidance of a morphine-paired taste cue is associated with full blunting of NAc dopamine response to an otherwise palatable saccharin reward [9]. Intraoral infusion of a taste cue that predicts later access to cocaine self-administration leads to a shift in the NAc neural code from ‘reward’ to ‘aversion’ [47] and a reduction in accumbal dopamine, as determined by voltammetry [46]. Behaviorally, avoidance of the drug-paired taste cue is linked with shorter latency to take drug, larger drug loading, and greater drug taking. Molecular analyses have only recently begun to examine individual differences in NAc adaptations to drug exposure. For example, microarray studies of the rat NAc reveal that high preference for ethanol is accompanied by a pattern in gene expression that is distinct from those rats that showed low fondness for the drug [3].
To better understand the molecular mechanisms underlying these individual differences in addiction-like behavior following extended access to heroin, we examined the NAc transcriptome of behaviorally stratified rats using next-generation sequencing (RNA-Seq). RNA-Seq has benefits over previous transcriptome analyses, such as microarrays, which are limited to detecting genes and transcripts whose oligonucleotide probes are present on the microarray. Further, given that RNA-Seq utilizes unbiased sequencing to determine differences in gene expression, it is possible to detect gene isoforms that previous technologies would have missed [43]. Importantly, RNA-Seq offers the ability to determine from which parent DNA strand the mRNA is transcribed. This ability to determine the origin of the mRNA provides greater accuracy of expression values in regions where genes may overlap on the forward and reverse genomic strands. Using this advanced tool for detecting differences in gene expression, we hypothesized that individual differences in avoidance of the drug-paired cue would be associated with a specific mRNA profiles in the nucleus accumbens. Here we demonstrate that not only is there a common mRNA response to heroin self-administration regardless of natural reward suppression, a specific signature of Large Suppressors of a heroin-paired taste cue compared to Small Suppressors who accept the heroin-paired saccharin cue as well.
2. Materials and Methods
2.1 Behavioral Paradigm
2.1.2 Subjects and surgeries
The subjects were 20 male, Sprague-Dawley rats obtained from Charles River at approximately 90 days of age. The rats were housed singly in suspended, stainless steel cages in a humidity-controlled environment under a 12/12 h light/dark cycle. Food and water were available ad libitum, except where noted. These subjects are described in greater detail in [12] and are from the second replication of Experiment 2 in that report. The subjects were implanted with jugular catheters and had 2 weeks recovery time before testing began. All animal experiments were executed according to protocols approved by the Penn State University Institutional Animal Care and Use Committee.
2.1.3 Apparatus
Testing was conducted in 12 self-administration chambers as previously described [32]. Each operant chamber was equipped with three retractable sipper tubes that entered the chamber through three holes. A stimulus light was located above each tube. A lickometer circuit was used to monitor licking on the leftmost saccharin spout, the middle inactive spout (the spout upon which responding elicited no consequence), and the rightmost active spout (the spout upon which a set of fixed ratio (FR) responses led to an i.v. infusion of drug). Each chamber also was equipped with a house light (25 W), a tone generator, and a speaker for white noise. Events in the chamber and collection of the data were computer controlled on-line using programs written in the Medstate notation language (MED Associates, Inc., St. Albans, VT).
2.1.4 Drug Acquisition
Rats were habituated in the self-administration chambers 5 min per day for 3 days prior to the start of acquisition. During habituation, the rats were on a water restriction regimen in which they received 5 min access to water through 1 of the 3 spouts in the chambers and 25 ml of water in the home cage overnight. This habituation occurred over 3 days until each animal experienced each of the 3 spouts. Thereafter, water was returned to the rats and acquisition began. Following habituation, rats were given 5 min access to the 0.15% saccharin solution followed immediately thereafter by a 6 h session to self-administer either saline (n=7) or 0.06 mg/0.2 ml infusion of heroin (n=13) on an FR10 schedule of reinforcement. Self-administration trials were once a day, 5 days a week, for a total of 16 trials.
2.1.5 Progressive Ratio
Upon completion of the 16-trial acquisition phase, the willingness to work was assessed in a one-trial progressive ratio (PR) test where a greater number of operant responses were required for each successive infusion of heroin. After 5 min access to the saccharin cue, each subject began on an FR 10 on the active spout for the 1st infusion, with subsequent infusions requiring the completion of the following progression of licks to obtain the next infusion: 10, 12, 16, 22, 30, 40, 52, 66, 82, 100, 120, 142, 166. Breakpoint was defined as the last ratio completed and the trial concluded when 30 min elapsed without having earned an infusion. After the PR test the animals underwent 3 days of maintenance training with the previous FR10 schedule.
2.1.6 Extinction and Reinstatement Test
All rats used for RNA-Seq analysis then were given a one-day extinction and reinstatement test for relapse-like behaviors. Subjects initially received a 5 min access to the saccharin cue. Upon completion of the 5 min availability of the saccharin cue, the extinction phase commenced. During the extinction phase, rats underwent a 6 h extinction session where responding on the active spout was not reinforced. Following the extinction phase, the rats received a single experimenter delivered iv infusion of heroin and reinstatement of heroin seeking was examined across an additional 1 h of extinction testing.
2.1.7 Behavioral Stratification
Using previously established behavioral criteria [10], Small Suppressors were defined as rats that emitted >200 licks/5min during terminal saccharin intake, while Large Suppressors were defined as rats that emitted <200 licks/5min.
2.2 Gene Expression Analysis
2.2.2 Sacrifice and Tissue Dissection
To ensure that no drug was on board at the time of sacrifice, subjects were sacrificed by rapid decapitation 24 h after the extinction/reinstatement test. The nucleus accumbens were dissected as previously described [15]. The nucleus accumbens contained both core and shell, combined.
2.2.3 Subject Selection Criteria
Using terminal saccharin avoidance (Trial 16) as a selection method, the top 5 performers of each group were chosen for RNA-Seq analysis (5 Small Suppressors with the most licks, and the 5 Large Suppressors with the fewest licks). The top 5 from each group to determine the changes that occur in gene expression at the extremes of the behavioral spectrum.
2.2.4 RNA Isolation and cDNA synthesis
RNA was isolated from the nucleus accumbens using AllPrep DNA/RNA Mini Kit (Qiagen) as previously described [23]. RNA quantity and quality (>8 RNA integrity number) were measured using the RNA 6000 Nano Assay with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). Using 1 μg RNA, cDNA was synthesized from purified RNA using ABI High-capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) [24].
2.2.5 Library Construction and Next Generation Sequencing
Library construction was performed in a stranded manner to retain the directionality of the transcripts [37] for Saline, Small Suppressor, and Large Suppressor groups (n=5/group). Each sequencing library was prepared from RNA from a single animal to provide biological variance and for correlation to individual animal behavioral data. Illumina Truseq Stranded HT library generation was performed according to manufacturer's instructions (See Supplemental Figure 1A).Briefly, polyA containing mRNA was purified using oligo-dT attached magnetic beads. mRNA were then chemically fragmented and cDNA synthesized. For strand-specificity, the incorporation of dUTP instead of dTTP in the second strand cDNA synthesis does not allow amplification past this dUTP with the polymerase. Following cDNA synthesis, each product underwent end repair process, the addition of a single ‘A’ base, and finally ligation of adapters. The cDNA products were further purified and was enriched using PCR to make the final library for sequencing. Library sizing was performed by TapeStation (Agilent) and libraries were quantified by qPCR (Kappa Biosystems). The cDNA library was then sequenced using an Illumina Hiseq2500 at Oklahoma Medical Research Foundation Genomics Facility in a 2×100bp fashion.
2.2.6 RNA-Seq Data Analysis
Following sequencing, reads were trimmed, aligned, differential expression statistics and correlation analyses were performed in Strand NGS software package (Agilent). Reads were aligned against the Rnor 5 build of the rat genome (2013.04.03). Alignment and filtering criteria included: fixed 2bp trim from 3’ and 5’ ends, a maximum number of one novel splice allowed per read, a minimum of 90% identity with the reference sequence, a maximum of 5% gap, trimming of 3’ end with Q<10. Alignment was performed directionally with Read 1 aligned in reverse and Read 2 in forward orientation. Reads were filtered based on the mapping status and only those reads that aligned normally (in the appropriate direction) were retained. Normalization was performed with the RPKM algorithm [25]. For the sake of thoroughness alternative normalization methods TMM [34] and DESeq [1] were also utilized and returned highly similar results (data not shown). Transcripts with an average read count value >1 in at least 100% of the samples in at least one group were considered expressed at a level sufficient for quantitation and those transcripts below this level were considered not detected/not expressed and excluded, as these low levels of reads are close to background and are highly variable. One Saline sample failed these filtering and quality control steps and was excluded from subsequent quantitative analyses. For statistical analysis of differential expression, one-way ANOVA followed by Student-Newman Keuls post hoc test was used. For those transcripts meeting this statistical criterion, a fold change >|1.25| cutoff was used to eliminate those genes which were statistically significant but unlikely to be biologically significant and orthogonally confirmable due to their very small magnitude of change. Correlation (Pearson's) of normalized RNA-Seq gene expression to behavioral measures was performed in Stand NGS as were visualizations of hierarchical clustering, principle components analysis, and partition depths. The entirety of the sequencing data is available for download in FASTQ format from NCBI Sequence Read Archive (Accession number SRX1117567).
2.2.7 Tertiary Analysis
For potential disease relationships, functions, signaling pathways and upstream regulators, the Ingenuity Pathway Analysis (Qiagen, Redwood City, CA) database was used. Bioinformatic analysis was conducted individually on all three sets of differentially expressed genes (Large Suppressors vs Small Suppressors, Large Suppressors vs Saline, and Small Suppressors vs Saline). For each gene set an overlap p value and an activation z-score were computed [13]. The p value was calculated using Fisher's Exact Test based on overlap between genes in the differentially expressed list and genes pertaining to the function, pathway or regulator. The activation z-score is used to infer likely activation states of a function or upstream regulator based on the direction of changes in the gene list and literature-derived functional or regulation directions. A positive z-score cutoff signifies activation while a negative z-score indicates inhibition.
2.2.8 qPCR Confirmations
For confirmations of genes discovered in the RNA-Seq dataset, quantitative PCR (qPCR) analysis of targets of interest was performed using TaqMan Assay-On-Demand (Life Technologies) gene-specific primers/probe assays (Supplemental Table 1) and a QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems) according to our standard methods [4, 24]. Relative gene expression was calculated with Expressionsuite v 1.0.3 software using the 2−ΔΔCt analysis method with β-actin as an endogenous control. qPCR confirmation analysis was done in the same subjects used in the RNA-Seq analysis (n=5/group). Independent t-test were used to compare results from qPCR analysis based on prior differences found using RNA-Seq.
3. Results
3.1 Drug Taking Behaviors
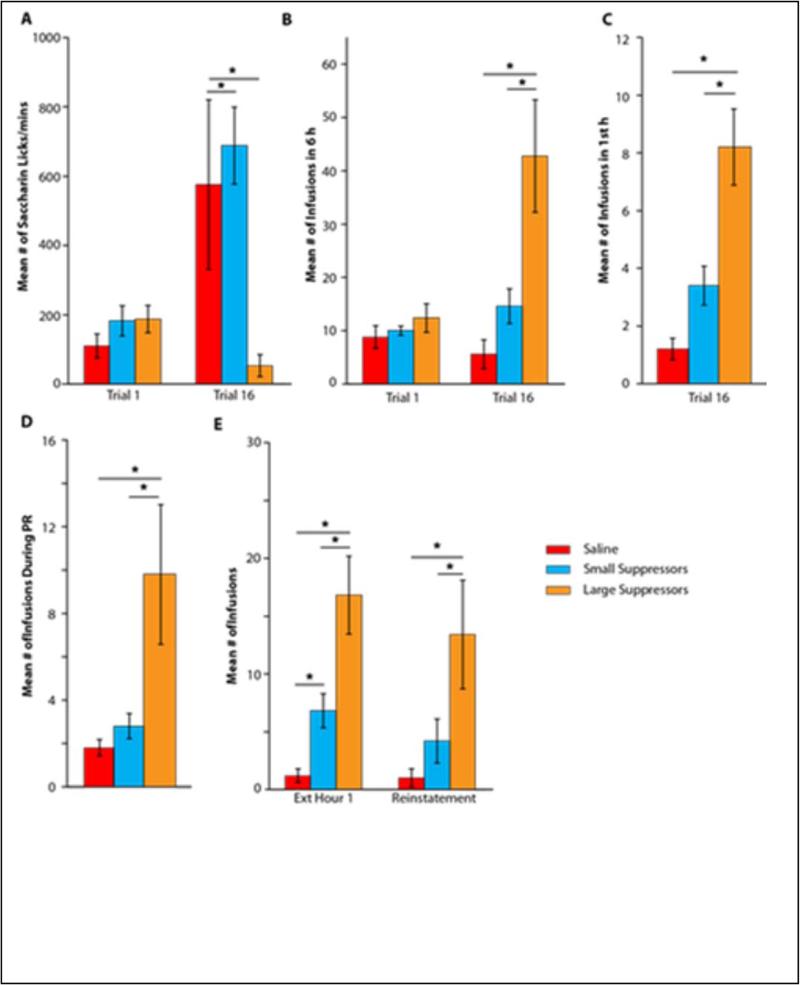
As described in greater detail [12], of 13 rats with heroin access, n=7 met the criteria for Large Suppressors and n=6 for Small Suppressors on their terminal trial 16 saccharin consumption. Given that we are interested in the molecular outcomes in the extreme ends of the behavioral spectrum, we selected the top 5 performers in saccharin consumption in each of the 3 groups for further analysis (n=5/group). It should be noted that this still represents a continuum of behavior. Figure 1A depicts the avoidance of the drug-paired taste cue when it predicts 6 h of heroin self-administration for the extremes within each group (n=5/group). Post hoc test of a significant one-way ANOVA of terminal (Trial 16) saccharin intake (F(2,14)=4.71; p<0.05) revealed that the Large Suppressors showed the greatest avoidance of the drug-paired taste cue compared to Saline and Small Suppressors rats (ps<0.05). In addition, as seen in Figures 1B and 1C, the Large Suppressors also showed the greatest drug taking over 6 h (F(2,14)=8.72; p<0.01) and the greatest escalation of terminal heroin intake (F(2,14)=16.42; p<0.001), as defined as the mean number of infusions taken within the first h of drug access on the Terminal day (Trial 16). A one-way ANOVA revealed significant differences in the motivation to work for drug under progressive ratio testing (F(2,14=5.27; p<0.05). Post hoc analysis demonstrated that Large Suppressors showed the greatest motivation for heroin compared to Small Suppressors (Figure 1D). Following the three-day maintenance phase to reestablish normal FR responding, the subjects underwent a same day extinction and reinstatement test (Figure 1E). Post hoc analysis of a significant mixed factorial 3 × 6 ANOVA varying Group (Saline, Small, and Large Suppressors) × Time (h 1 – 6) interaction (F(12,72)=3.36; p<0.05), revealed that while both heroin groups showed robust drug seeking in the initial hour of extinction testing, as measured by unreinforced infusion attempts, compared to the Saline controls, Large Suppressors showed significantly more drug seeking behaviors than Small Suppressors (p<0.05). Thereafter, responding dropped off precipitously and there were no differences found among any of the 3 groups between hours 2 through 6 of extinction testing (data not shown). After a non-contingent iv infusion of heroin (right panel), Large Suppressors showed a greater amount of drug-seeking behavior than both Small Suppressors and Saline controls (ps<0.05).
Figure 1. Extended Access Addition-like Behavior.
A) Mean (+/− SEM) number of licks/5 min of 0.15% saccharin during Trial 1 and Trial 16 for Saline, Small, and Large Suppressors. Mean (+/− SEM) number of saline or heroin (0.06 mg/0.2ml of heroin) infusions/6 h B) and C) within in the 1st h for Saline, Small, and Large Suppressors during Trial 1 and Trial 16. D) Mean (+/− SEM) number of saline or heroin infusions earned during progressive ratio testing for Saline, Small, and Large Suppressors. E) Mean (+/− SEM) number of drug seeking behaviors/1 h exhibited during Hour 1 of the 6 h extinction period (left panel) and during 1 h following a single experimenter delivered iv dose of saline or heroin for Saline, Small, and Large Suppressors. *p<0.05 (n=5/group)
3.2 RNA-Seq Analysis
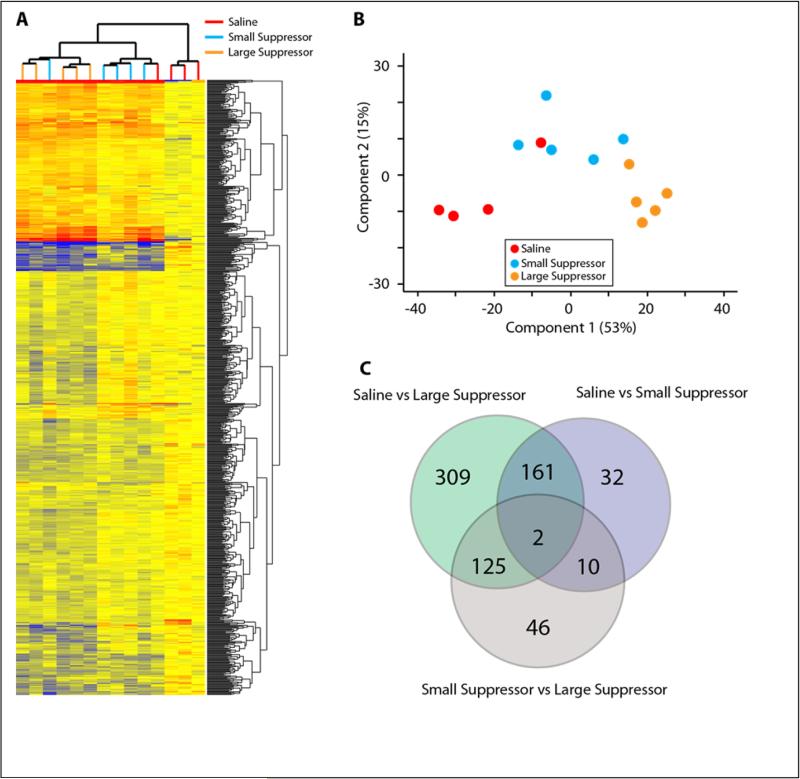
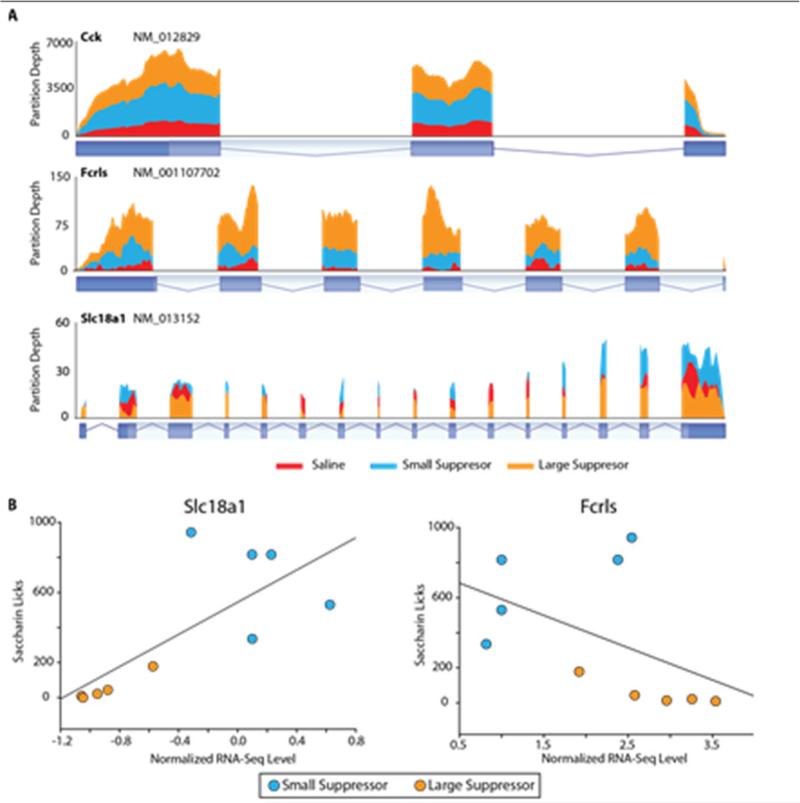
An average of 26.1 million reads per sample passed filtering and alignment criteria. There was no difference in the average number of reads between groups and equivalent numbers of reads mapped to the positive and negative genomic strands (Supplementary Figure 1B). Using a cutoff of >1.0 RPKM for all samples in at least one group, 13,752 of the 31,399 transcripts with rat RefSeq annotations were called as expressed. Differentially expressed transcripts between groups were identified though a one-way ANOVA design with Student Neuman Keuls pair-wise post hoc testing (α<0.05). These transcripts were further filtered to only those that had a >|1.25| fold difference between the groups that had a significant post-hoc test result. This resulted in a total list of 773 transcripts with pairwise numbers of 597 (Saline vs Large Suppressor), 205 (Saline vs Small Suppressor), 183 (Small Suppressor vs Large Suppressor) (Figure 2A). Both hierarchical clustering (Figure 2A) and Principle Components Analysis (PCA) on samples (Figure 2B) demonstrate that the individual samples cluster according to their groups. There was a large degree of overlap in differentially expressed genes between Saline and heroin groups as well as between the Large Suppressor vs Saline and vs Small Suppressor comparisons (Figure 2C). Importantly, for the 161 genes in the overlap between the heroin groups and Saline, every gene showed concordance, i.e., genes were either induced or repressed in both comparisons. Similarly all of the genes in the overlap between the Large Suppressors vs Saline and Large Suppressors vs Small Suppressors agreed in the direction of change. The full list of differentially expressed transcripts is provided in Supplemental Table 2. RNA-Seq assesses differences in abundance across the entire gene and not just at single oligonucleotide binding site. Three examples of differentially expressed genes are presented in Figure 3A. As a goal of this study is to identify molecular changes that may underlie the observed behavioral differences, transcripts identified as differentially expressed were correlated to the following quantitative behavioral measures: saccharin licks, heroin infusions in the 1st hour, heroin infusions in the 6 hour session, infusions during extinction, infusions during reinstatement, and progressive ratio infusions. A number of positive and negative correlations were observed (Pearson's >|0.5|, Supplemental Table 3), two examples are provided in Figure 3B. The approach allows for both statistical comparisons with Large and Small Suppressors are bimodal groups (above) but as a continuum of behavior here. For Slc18a1, greater avoidance of the drug paired taste cue was significantly correlated with decreased mRNA expression (r=0.75; p<0.05). Conversely, larger avoidance of the drug paired taste cue was significantly correlated with increased expression of the Fcrls gene (r=−0.50; p<0.05).
Figure 2. Identification of Differential Gene Expression by RNA-Seq.
A) Hierarchical clustering of genes and samples for the 685 differentially expressed transcripts (ANOVA, SNK pairwise posthoc, fold change >|1.25|. B) Relationship between samples through principal compenent analysis based on the differentially expressed genes. Samples segregated along the first compenet by group. C) Venn diagram representation of the differentially expressed transcripts. A number of common changes were observed between heroin groups and Saline and in the comparisons between Large Supressors and Saline and Small Suppressors. n=4-5/group
Figure 3. Differential Expression Across Exons and Correlation of Gene Expression to Behavioral Measures.
A) With RNA-Seq differentially expressed genes are evident through different read depths across the exons of the transcript. B) Gene expression of differentially expressed genes was correlated (Pearson's) to behavioral measures. Two examples are given demonstrating positive and negative correlations to saccharin licks. Only heroin groups were included in the correlation analysis. n=4-5/group
3.3 Tertiary Analysis
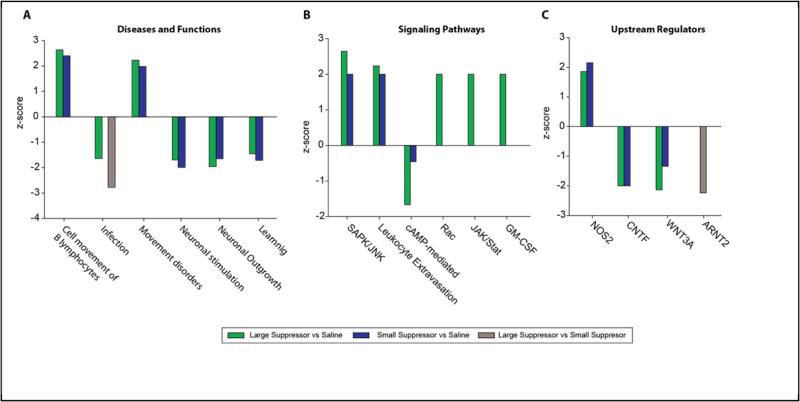
Analysis of diseases and functions, signaling pathways, and upstream regulators (Figure 4A-C) of the three differentially expressed gene sets identified common responses in the heroin self-administration groups as compared to the Saline controls. Of note were both the commonalities in categories and the fact that the z-scores were in the same direction with a slightly higher |value| for the high suppressor comparisons, suggestive of a greater effect (Supplemental Tables 5-7). Fewer significant gene sets were identified in the Large Suppressor vs Small Suppressor comparison. However, there was a common and unique response of impaired inflammatory/infection response in the Large Suppressors as compared to both of the other groups.
Figure 4. Tertiary Analysis of Differentially Expressed Transcripts.
Differentially expressed transcripts in each of the three pair-wise comparisons were assessed for over-representation of disease relationships and functional categories (A), signaling pathways (B), and upstream regulators (C) using the Ingenuity Pathway Analysis database. Selected categories of interest that passed statistical thresholds (Fisher's Exact Test, p<0.05) are presented.
3.4 qPCR Confirmations
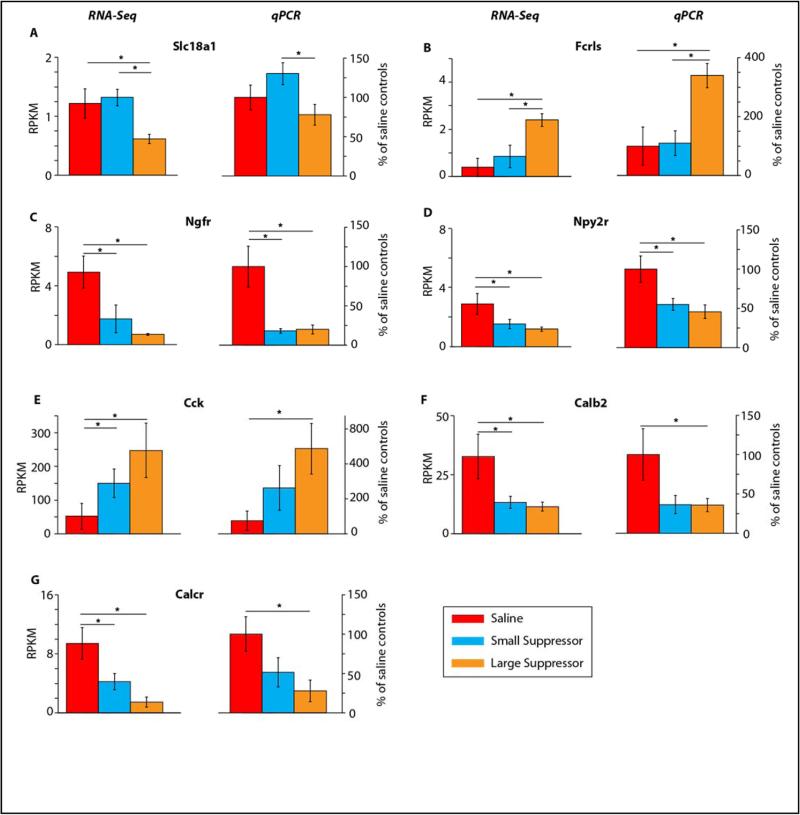
To confirm changes in gene expression from the RNA-Seq analysis, confirmatory qPCR was conducted on a number of genes. A sampling of genes was selected based on Ingenuity pathway analysis, fold change, and potential role in drug abuse. Supplemental Table 1 lists the 9 genes selected for qPCR analysis. Since we sought to replicate the findings from the RNA-Seq analysis using qPCR, independent t-tests were conducted on selected pairwise comparisons from the RNA-Seq dataset. Seven of these genes met statistical criteria for confirmation and are presented in Figure 5 with paired RNA-Seq results. These results confirm the variety of pairwise expression differences observed in the discovery RNA-Seq experiments including: reductions (Slc18a1 and inductions (Fcrls in Large Suppressors as compared to Small Suppressors; induction (Cck or reduction (Calcr, Calb2, Npyr2, Ngfr in one or both of the heroin groups as compared to Saline controls. For the genes that did not meet our confirmation criteria: Slc17a7 showed a 5 fold increase in the heroin groups, similarly to the RNA-seq data, but a high degree of variability which prevented reaching of statistical significance, and Pip5k1c did not correspond with our RNA-Seq data (Supplemental Table 4). In total, these orthogonal confirmations demonstrate the analytical reproducibility of the discovery experiment and suggest that other genes identified in the RNA-Seq analysis are also differentially expressed.
Figure 5. Confirmatory qPCR of genes selected from RNA-Seq analysis of the nucleus accumbens.
Left panels depict mean (+/− SEM) changes in RPKM (Reads per Kilobase per Million mapped reads). Right panel depicts mean (+/− SEM) changes found to be significant through qPCR. n=4-5/group *p<0.05
4. Discussion
The data presented here examine the gene expression profile in the nucleus accumbens (NAc) in animals stratified by their addiction-like behaviors, namely in their avoidance of a natural reward that predicts heroin self-administration. Individual differences in the avoidance of a drug-paired taste cue was associated with greater drug taking, drug escalation, and relapse-like behaviors [12]. The human literature has shown that only a fraction of individuals who engage in drug use transition to drug addiction, with heroin showing some of the greatest addiction liability [42]. Given these large individual differences in the response to drugs of abuse, we sought to discover the mRNA changes that occur in the NAc of rats that were behaviorally stratified by their addiction-like behavior.
Recently there has been a surge in discovery of neuroadaptations associated with individual differences in response to drugs of abuse [28]. For example, individual differences in responding to heroin-primed reinstatement were linked with differences in gene expression of vasopressin, D2 receptor, and orexin [49]. Based on the RNA-Seq analysis here, a large number of mRNA expression changes occurred among the three conditions in the NAc with the majority of the changes in gene expression observed when comparing the heroin groups to the Saline controls. Pathway analysis revealed that many of these genes were related to movement disorders, neuronal stimulation, neuronal outgrowth, and learning. Moreover, Large Suppressors showed significant differences in gene expression compared to Saline controls that were not evident in Small Suppressors versus Saline subjects potentially indicative of different amounts of heroin self-administration. Importantly however, there were a number of differences in gene expression between Large and Small Suppressors that were only evident in this comparison between two groups with heroin self-administration. These data demonstrate that changes in mRNA expression occur not only with heroin self-administration but also between rats with a history of heroin self-administration that have been stratified based on their avoidance of a drug-paired natural reward. Importantly, 189 genes demonstrated significant correlations with saccharin drinking but only 105 significant correlations to 6 h heroin intake would help boost their argument. This supports the concept that these individual differences reflect in part devaluation of natural reward and not simply difference in heroin intake. Additionally, 19 different genes had correlations to four or more of the, albeit related, behavioral measures (with r-values of greater than 0.6). This suggests that there could be gene dosage effects across the continuum of behavior.
Two genes of focus, Slc18a1 and Fcrls, were uniquely associated with the degree of avoidance of the drug-paired natural reward. It may be worth noting that, aside from a smaller correlation of Slc18a1 with hour one infusions, neither one of these genes demonstrates an association with measures of self-administration behavior. Slc18a1, the vesicular monoamine transporter (VMAT1), was originally thought to be only expressed in the periphery, however recent evidence has shown that VMAT1 may serve a role in the brain [26]. Deletion of Slc18a1 in mice reduces hippocampal neurogenesis and produces neurocognitive deficits [26]. Furthermore mutations in the Slc18a1 gene also have been linked to an increased likelihood of developing schizophrenia [33] and type 1 bipolar disease [20]. The decreased expression of this gene in the Large Suppressor group may reflect blunted synaptic plasticity in the nucleus accumbens; a difference which could contribute to the addiction phenotype. Of note, it has long been postulated that the etiology of schizophrenia and bipolar disorder stems from dysregulation of dopamine transmission [22, 38]. These data demonstrate, for the first time, that Slc18a1 may also be involved in the addictive processes, a disease in which dopamine plays a central role. In contrast to the expression pattern of Slc18a1, the gene expression of Fcrls was increased in the Large Suppressors compared to Saline controls and Small Suppressors. Fcrls is a scavenging receptor that is has been shown to be preferentially expressed on microglia [2]. The increase in Fcrls expression in the Large Suppressors may reflect the ability of opioids to influence immune function [35] although a growing body of literature indicates that immune molecules also function as neurotransmitters [31]. To our knowledge, this is the first time that Fcrls has been found to be associated with addiction-like behavior. These findings further support that individual differences in drug induced avoidance of a natural reward is linked not only to changes in addiction-like behavior but also to changes in gene expression. A number of additional differences specific to the Large Suppressor vs Small Suppressor category were identified and these may serve as targets for further studies in the future.
Selected additional genes with differences in expression between heroin and Saline groups were also confirmed including Ngfr and Npy2r, which both showed significant decreases in gene expression in both small and Large Suppressors compared to Saline controls. Ngfr, also known as p75 neurotrophin receptor, has been implicated in the development of analgesic tolerance, as null mice for Ngfr show a lack of analgesic tolerance to repeated injections of morphine [40]. The neuropeptide y (NPY) system has been implicated in the drug abuse field. Previously we have reported that heroin exposure and enforced drug abstinence decreased NPY expression in the medial prefrontal cortex [16]. Single nucleotide polymorphisms of the NPY2R have been implicated in the development of alcohol addiction [45]. Given that both genes showed decreased mRNA expression in out studies, it appears that Ngfr and Npy2r may be more sensitive to heroin self-administration regardless of the amount taken.
Cholecystokinin (Cck) showed significantly greater gene expression in the Large Suppressors compared to the Saline controls. Cck is a neuropeptide that has been implicated in the role of satiety [48]. Moreover Cck has been shown to oppose many opiate induced effects such as analgesia and locomotor activity [8, 36]. Of note, the Large Suppressors showed a robust increase in Cck expression compared to Saline subjects. Given that Cck has been shown to attenuate the rewarding effects of opiates, the increase in Cck expression in the Large Suppressors could counteract the effects of heroin self-administration and return the subject back to homeostasis [44]. This pattern of activity may serve to ameliorate the deleterious effects that opiates have on neuronal growth and survival [27]. Additionally, given its role in satiety, changes in Cck may contribute to suppression of intake of the heroin-paired saccharin cue in these subjects.
Confirmatory qPCR found 2 genes (Calb2 and Calcr) to be significantly lower in expression in the Large Suppressors than in the Saline controls. Calb2, also known as calretinin, is a neuroprotective calcium binding protein [39]. Heroin exposure in rats has been shown to decrease the amount of immunoreactive neurons for Calb2 in the dentate gyrus [5]. A similar effect was observed here, showing that Large and Small Suppressors displayed decreased Calb2 gene expression compared to Saline subjects. The Calcr gene encodes the calcitonin receptor, which is a G-coupled protein receptor that normally plays a role in calcium homeostasis [30]. Studies have shown that the calcitonin receptor gene may also serve a role in controlling ingestive behaviors [18]. Furthermore heroin exposure in rats has been previously shown to disrupt intracellular calcium homeostasis, which correlated with an increase in neuronal cell damage when challenged with drug [19]. Although the role of Calcr gene in addiction has not been elucidated it is worth noting the lower expression of the gene in Large Suppressors versus Saline controls may reflect a heroin-induced defect in calcium homeostasis.
One concern regarding the findings presented here is that the changes observed might be an effect of the amount of drug taken. Indeed, this is a possible explanation for the differences in gene expression since Large Suppressors do show the greatest drug taking behaviors compared to Small Suppressors. However, these subjects were freely able to decide how much heroin to infuse and how much to avoid of a natural reward. Therefore this choice in self-administration mimics the human condition in which the individual decides how much drug to consume. Yoked delivery may be used to address this concern [14], but does have some drawbacks. Twining et al. [41] demonstrated that yoked delivery of cocaine was aversive to the subject and caused behaviors that protected the rat from working for drug. Therefore, we simply asked the question what transcriptomic changes do occur in behaviorally stratified subjects based on their avoidance of a taste cue and their heroin self-administration history. Taken together, it appears that the Large Suppressors of a drug-paired taste cue show a unique mRNA profile that is distinct from Saline controls and distinct from that found in Small Suppressors of the natural reward vs Saline controls.
Although the focus of this study was to determine individual differences in gene expression among behaviorally stratified subjects on their addiction-like behavior, future studies should examine additional aspects of the transcriptome. Transcription from the DNA plus strand produces antisense RNAs that are complementary to their related gene transcripts and have been shown to alter gene expression due to changes in the environment [29]. Furthermore antisense RNAs have been implicated in various human disease such as Alzheimer's disease and cancer [7]. Moreover, additional experiments should be devoted to seeing if mRNA changes are evident in other brain regions associated with drug addiction such as the medial prefrontal cortex and ventral tegmental area. Given that drug addiction affects multiple regions of the brain as shown in Kuntz et al. [15] and Zhou et al. [49], it is worth investigating if region specific differences in mRNA expression exist following heroin self-administration and avoidance of the drug-paired taste cue. Future experiments also should examine if these changes in mRNA are persistent, or even enhanced during abstinence. Previously our laboratories have shown persistent mRNA changes following 14 days of drug-enforced abstinence after heroin self-administration [15]. Given that addiction is a chronic relapsing disease that can manifest itself after prolonged periods of abstinence, exploring whether individual differences in gene expression can contribute to this vulnerability towards relapse or if certain patterns of mRNA expression may be protective could inform treatment decisions and potentially lead to better therapeutics. The data reported here demonstrate changes in gene expression that are specific to a behavioral measure of reward devaluation that goes beyond just pharmacological measures of gene response to drugs of abuse. In addition, these data serve as a reference for other investigations examining molecular alterations in other opioid administration paradigms, both investigator initiated and self-administered.
5. Conclusions
In sum, in addition to demonstrating that heroin self-administration has profound effects on the rodent brain, our data show that individual differences in mRNA expression do occur in subjects stratified by their addiction-like behavior, specifically, avoidance of a drug-paired taste cue. To our knowledge this is the first report of its kind that examines individual differences in gene expression based on avoidance of a natural reward that predicts heroin self-administration. Large Suppressors of the natural reward showed significant differences in mRNA expression compared to Saline controls and these differences were not found between Small and Saline controls. Furthermore, differences in gene expression also were found between Large Suppressors and Small Suppressors. Many of these genes underlie the biological processes of behavior, neuronal function, and immunity. Understanding the individual differences in gene expression that are associated with greater addiction-like behavior will assist in tailoring therapies that target these differences. Here, we demonstrate that greater avoidance of a drug-paired natural reward is associated with a unique mRNA profile in the NAc that is distinct from that of the Small Suppressors (i.e., from those rats who readily ingest the saccharin cue even when it predicts the opportunity to self-administer drug) and distinct from that of the Saline controls.
Supplementary Material
Highlights.
Suppression of a natural reward is associated with greater addiction-like behavior.
Large suppressors of a heroin-paired taste cue showed a unique mRNA prolife compared to small avoiders.
Avoidance of a natural reward is linked with differences in gene expression.
Acknowledgements
The authors thank Robert Brucklacher and Georgina Bixler in the Genome Sciences Facility at the Penn State Hershey College of Medicine for quantitative PCR assistance, and the Oklahoma Medical Research Foundation Genomics Facility Core for RNA-Seq assistance, E.M Bishop for assistance with figure preparation. The authors declare no financial conflicts of interest. This project was also funded, in part, by a grant from the Pennsylvania Department of Health using Tobacco CURE Funds to P.S.G, K.E.V and W.M.F., the Donald W. Reynolds Foundation and the National Institute on Aging (P30AG050911) to W.M.F, the National Institute on Drug Abuse F31DA036322 to C.G.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard A, Tremblay P, Chernomoretz A, Vallieres L. Identification of genes preferentially expressed by microglia and upregulated during cuprizone-induced inflammation. Glia. 2007;55:777–789. doi: 10.1002/glia.20477. [DOI] [PubMed] [Google Scholar]
- 3.Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bixler GV, Vanguilder HD, Brucklacher RM, Kimball SR, Bronson SK, Freeman WM. Chronic insulin treatment of diabetes does not fully normalize alterations in the retinal transcriptome. BMC Med Genomics. 2011;4:40. doi: 10.1186/1755-8794-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casini A, Vivacqua G, Pontieri FE, Kimura H, Bellier JP, D'Este L, Renda TG. Choline acetyltransferase of the common type immunoreactivity in the rat brain following different heroin treatments: a pilot study. J Chem Neuroanat. 2011;41:111–121. doi: 10.1016/j.jchemneu.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Cicero TJ, Ellis MS, Surratt HL, Kurtz SP. The changing face of heroin use in the United States: a retrospective analysis of the past 50 years. JAMA psychiatry. 2014;71:821–826. doi: 10.1001/jamapsychiatry.2014.366. [DOI] [PubMed] [Google Scholar]
- 7.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faris PL. Opiate antagonistic function of cholecystokinin in analgesia and energy balance systems. Ann N Y Acad Sci. 1985;448:437–447. doi: 10.1111/j.1749-6632.1985.tb29939.x. [DOI] [PubMed] [Google Scholar]
- 9.Grigson PS, Hajnal A. Once is too much: conditioned changes in accumbens dopamine following a single saccharin-morphine pairing. Behav Neurosci. 2007;121:1234–1242. doi: 10.1037/0735-7044.121.6.1234. [DOI] [PubMed] [Google Scholar]
- 10.Grigson PS, Twining RC. Cocaine-induced suppression of saccharin intake: a model of drug-induced devaluation of natural rewards. Behav Neurosci. 2002;116:321–333. [PubMed] [Google Scholar]
- 11.Ikemoto S, Panksepp J. The role of nucleus accumbens dopamine in motivated behavior: a unifying interpretation with special reference to reward-seeking. Brain Res Brain Res Rev. 1999;31:6–41. doi: 10.1016/s0165-0173(99)00023-5. [DOI] [PubMed] [Google Scholar]
- 12.Imperio CG, Grigson PS. Greater avoidance of a heroin-paired taste cue is associated with greater escalation of heroin self-administration in rats. Behavioral neuroscience. 2015;129:380–388. doi: 10.1037/bne0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuntz KL, Patel KM, Grigson PS, Freeman WM, Vrana KE. Heroin self-administration: II. CNS gene expression following withdrawal and cue-induced drug-seeking behavior. Pharmacol Biochem Behav. 2008;90:349–356. doi: 10.1016/j.pbb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuntz KL, Twining RC, Baldwin AE, Vrana KE, Grigson PS. Heroin self-administration: I. Incubation of goal-directed behavior in rats. Pharmacol Biochem Behav. 2008;90:344–348. doi: 10.1016/j.pbb.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuntz-Melcavage KL, Brucklacher RM, Grigson PS, Freeman WM, Vrana KE. Gene expression changes following extinction testing in a heroin behavioral incubation model. BMC Neurosci. 2009;10:95. doi: 10.1186/1471-2202-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenoir M, Cantin L, Vanhille N, Serre F, Ahmed SH. Extended heroin access increases heroin choices over a potent nondrug alternative. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:1209–1220. doi: 10.1038/npp.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine AS, Morley JE. Reduction of feeding in rats by calcitonin. Brain Res. 1981;222:187–191. doi: 10.1016/0006-8993(81)90957-4. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Wang G, Pu H, Jing H. Abnormal intracellular calcium homeostasis associated with vulnerability in the nerve cells from heroin-dependent rat. Brain Res. 2014;1572:40–49. doi: 10.1016/j.brainres.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Lohoff FW, Dahl JP, Ferraro TN, Arnold SE, Gallinat J, Sander T, Berrettini WH. Variations in the vesicular monoamine transporter 1 gene (VMAT1/SLC18A1) are associated with bipolar i disorder. Neuropsychopharmacology. 2006;31:2739–2747. doi: 10.1038/sj.npp.1301196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luscher C, Malenka RC. Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron. 2011;69:650–663. doi: 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manji HK, Lenox RH. Signaling: cellular insights into the pathophysiology of bipolar disorder. Biol Psychiatry. 2000;48:518–530. doi: 10.1016/s0006-3223(00)00929-x. [DOI] [PubMed] [Google Scholar]
- 23.Masser DR, Berg AS, Freeman WM. Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing. Epigenetics Chromatin. 2013;6:33. doi: 10.1186/1756-8935-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masser DR, Bixler GV, Brucklacher RM, Yan H, Giles CB, Wren JD, Sonntag WE, Freeman WM. Hippocampal subregions exhibit both distinct and shared transcriptomic responses to aging and nonneurodegenerative cognitive decline. J Gerontol A Biol Sci Med Sci. 2014;69:1311–1324. doi: 10.1093/gerona/glu091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Multani PK, Hodge R, Estevez MA, Abel T, Kung H, Alter M, Brookshire B, Lucki I, Nall AH, Talbot K, Doyle GA, Lohoff FW. VMAT1 deletion causes neuronal loss in the hippocampus and neurocognitive deficits in spatial discrimination. Neuroscience. 2013;232:32–44. doi: 10.1016/j.neuroscience.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa S, Kim JE, Lee R, Malberg JE, Chen J, Steffen C, Zhang YJ, Nestler EJ, Duman RS. Regulation of neurogenesis in adult mouse hippocampus by cAMP and the cAMP response elementbinding protein. J Neurosci. 2002;22:3673–3682. doi: 10.1523/JNEUROSCI.22-09-03673.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nestler EJ. Cellular basis of memory for addiction. Dialogues Clin Neurosci. 2013;15:431–443. doi: 10.31887/DCNS.2013.15.4/enestler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passalacqua KD, Varadarajan A, Weist C, Ondov BD, Byrd B, Read TD, Bergman NH. Strandspecific RNA-seq reveals ordered patterns of sense and antisense transcription in Bacillus anthracis. PLoS One. 2012;7:e43350. doi: 10.1371/journal.pone.0043350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pondel M. Calcitonin and calcitonin receptors: bone and beyond. Int J Exp Pathol. 2000;81:405–422. doi: 10.1046/j.1365-2613.2000.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prossin AR, Zalcman SS, Heitzeg MM, Koch AE, Campbell PL, Phan KL, Stohler CS, Zubieta JK. Dynamic interactions between plasma IL-1 family cytokines and central endogenous opioid neurotransmitter function in humans. Neuropsychopharmacology. 2015;40:554–565. doi: 10.1038/npp.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puhl MD, Boisvert M, Guan Z, Fang J, Grigson PS. A novel model of chronic sleep restriction reveals an increase in the perceived incentive reward value of cocaine in high drug-taking rats. Pharmacol Biochem Behav. 2013;109:8–15. doi: 10.1016/j.pbb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards M, Iijima Y, Kondo H, Shizuno T, Hori H, Arima K, Saitoh O, Kunugi H. Association study of the vesicular monoamine transporter 1 (VMAT1) gene with schizophrenia in a Japanese population. Behav Brain Funct. 2006;2:39. doi: 10.1186/1744-9081-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saurer TB, Ijames SG, Carrigan KA, Lysle DT. Neuroimmune mechanisms of opioid-mediated conditioned immunomodulation. Brain Behav Immun. 2008;22:89–97. doi: 10.1016/j.bbi.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnur P, Cesar SS, Foderaro MA, Kulkosky PJ. Effects of cholecystokinin on morphine-elicited hyperactivity in hamsters. Pharmacol Biochem Behav. 1991;39:581–586. doi: 10.1016/0091-3057(91)90131-k. [DOI] [PubMed] [Google Scholar]
- 37.Sultan M, Dokel S, Amstislavskiy V, Wuttig D, Sultmann H, Lehrach H, Yaspo ML. A simple strandspecific RNA-Seq library preparation protocol combining the Illumina TruSeq RNA and the dUTP methods. Biochem Biophys Res Commun. 2012;422:643–646. doi: 10.1016/j.bbrc.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 38.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Mol Psychiatry. 2005;10:27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 39.Tirumalai PS, Howells RD. Regulation of calbindin-D28K gene expression in response to acute and chronic morphine administration. Brain Res Mol Brain Res. 1994;23:144–150. doi: 10.1016/0169-328x(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 40.Trang T, Koblic P, Kawaja M, Jhamandas K. Attenuation of opioid analgesic tolerance in p75 neurotrophin receptor null mutant mice. Neurosci Lett. 2009;451:69–73. doi: 10.1016/j.neulet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 41.Twining RC, Bolan M, Grigson PS. Yoked delivery of cocaine is aversive and protects against the motivation for drug in rats. Behav Neurosci. 2009;123:913–925. doi: 10.1037/a0016498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S.A.a.M.H.S.A. U.S. Dept of Health and Human Services, Center for Behavioral Health Statistics and Quality Results from the 2012 National Survey on Drug Use and Health: Summary of National Findings. 2012 [Google Scholar]
- 43.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen D, Cong B, Ma C, Yang S, Yu H, Ni Z, Li S. The effects of exogenous CCK-8 on the acquisition and expression of morphine-induced CPP. Neurosci Lett. 2012;510:24–28. doi: 10.1016/j.neulet.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 45.Wetherill L, Schuckit MA, Hesselbrock V, Xuei X, Liang T, Dick DM, Kramer J, Nurnberger JI, Jr., Tischfield JA, Porjesz B, Edenberg HJ, Foroud T. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol Clin Exp Res. 2008;32:2031–2040. doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler RA, Aragona BJ, Fuhrmann KA, Jones JL, Day JJ, Cacciapaglia F, Wightman RM, Carelli RM. Cocaine cues drive opposing context-dependent shifts in reward processing and emotional state. Biol Psychiatry. 2011;69:1067–1074. doi: 10.1016/j.biopsych.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler RA, Twining RC, Jones JL, Slater JM, Grigson PS, Carelli RM. Behavioral and electrophysiological indices of negative affect predict cocaine self-administration. Neuron. 2008;57:774–785. doi: 10.1016/j.neuron.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 48.Williams KW, Elmquist JK. From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat Neurosci. 2012;15:1350–1355. doi: 10.1038/nn.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, Leri F, Cummins E, Kreek MJ. Individual differences in gene expression of vasopressin, D2 receptor, POMC and orexin: vulnerability to relapse to heroin-seeking in rats. Physiol Behav. 2015;139:127–135. doi: 10.1016/j.physbeh.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.